DOI:
10.1039/C8RA04065F
(Paper)
RSC Adv., 2018,
8, 25519-25525
Characterization of interaction between scoparone and bovine serum albumin: spectroscopic and molecular docking methods
Received
13th May 2018
, Accepted 2nd July 2018
First published on 17th July 2018
Abstract
Scoparone is a major biological active substance derived from the traditional Chinese herbal medicine called Artemisia capillaris. It has been confirmed that scoparone has anti-inflammatory, anti-tumor, hepatoprotective and antioxidant effects. However, the binding interaction of scoparone with bovine serum albumin (BSA) still remains unknown. Therefore, the present study was conducted to clarify the binding interaction of scoparone with BSA under simulated physiological conditions (pH = 7.4) by utilizing spectroscopic and molecular docking methods. The formation of the scoparone–BSA complex was identified by UV-vis absorption spectroscopy experiment results. The fluorescence experiment results revealed that the quenching mechanism was static quenching and the binding procedure was spontaneous mainly driven by hydrophobic interaction. At 310 K, the number of binding sites was approximately equal to 1 and the binding constant was 6.79 × 105 mol L−1. The binding distance (4.81 nm) between scoparone and BSA was determined by Förster's non-radiative energy transfer theory. Molecular docking and site marker competitive experiment results verified that scoparone was more likely to be located in site I of BSA. In addition, the results of synchronous fluorescence spectroscopy and circular dichroism spectroscopy experiments proved that scoparone slightly changed the conformation of BSA by binding interaction with BSA. These findings would be useful for understanding the pharmacokinetics of scoparone in vivo, including scoparone transport, distribution, metabolism and excretion.
Introduction
After entering the blood circulation, drugs need to be stored and transported by plasma to reach the target tissue and exert pharmacological functions.1 Serum albumin is the most abundant carrier protein in plasma. It can combine with many drugs to store and transport these drugs in plasma.2 Therefore, the study of the mechanism on the interaction between drugs and serum albumin is helpful to understand the transportation and distribution of drugs in the body on the molecular level, and provide theoretical guidance and valuable information for designing more effective drugs, which has become a very focused research field in chemicobiology, life sciences and medicine.3
BSA is a heart-shaped protein consisting of 583 amino acid residues. It has three domains: domains I, II and III. Each domain has two subdomains, A and B. Two main drug binding sites (site I and site II) are respectively located in sub-domain IIA and sub-domain IIIA.4,5 Since the amino acid sequences of BSA and human serum albumin (HSA) are very similar and both have a high homology (approximately 76%), experimental studies often use BSA instead of HSA to study the binding interaction of serum albumin with drugs.6
Scoparone is a major biological active substance derived from the traditional Chinese herbal medicine called Artemisia capillaris. It has been reported that scoparone possesses anti-inflammatory, anti-tumor, hepatoprotective and antioxidant effects, and is usually used to treat neonatal jaundice.7,8 Currently, many studies concerning the binding interaction of drugs with serum albumin have been carried out. However, the study about the interaction of serum albumin with scoparone has not been reported, which is very meaningful for understanding the mechanism of scoparone action and pharmacokinetics in the human body.
In this paper, fluorescence spectroscopy, UV-vis absorption spectroscopy and circular dichroism spectroscopy were used to study the spectral characteristics about the binding interaction between scoparone and BSA in detail under simulated physiological conditions. Some important information about pharmacological mechanisms and pharmacokinetics of scoparone in vivo will be provided.
Experimental section
Chemicals and reagents
Scoparone (purity ≥ 98%) was obtained from Chengdu Must Bio-Technology Co., Ltd (Chengdu, China). It was dissolved in dimethyl sulfoxide to prepare 100 mM storage solution. Ibuprofen (purity 99%) was obtained from Sinopharm Chemical Reagent Co., Ltd (Shanghai, China). Phenylbutazone (purity ≥ 98%) was obtained from Shanghai Aladdin biological technology Co., Ltd (Shanghai, China). BSA (purity > 96%) was bought from Gen-View Scientific INC (USA). BSA solution (1 × 10−4 mol L−1) was dissolved in 0.01 mol L−1 PBS. The stock solutions of phenylbutazone (6 × 10−3 mol L−1) and ibuprofen (6 × 10−3 mol L−1) were dissolved in ethanol. All storage solutions were preserved in dark at 4 °C.
Fluorescence spectroscopy experiments
At three temperatures (290, 298 and 310 K), the fluorescence spectroscopy experiments were performed by using the Hitachi F-7000 type Fluorescence Spectrophotometer (Hitachi High-Technologies Co., Ltd., Tokyo, Japan) under 5/5 nm slit widths. The scanning wavelength range was 200–500 nm. The excitation wavelength was set at 280 nm.9 In this research, considering the inner filter effect (IFE), so all fluorescence intensities were corrected according to some references.10–13.
Synchronous fluorescence spectroscopy experiments
At 298 K, synchronous fluorescence spectrum experiments were conducted. The wavelength intervals (Δλ = λem − λex) of 15 nm and 60 nm were respectively set. A 200–500 nm scanning wavelength range was set.14
Circular dichroism (CD) spectroscopy experiments
CD spectroscopy experiments were carried out by using JASCO J-810 Spectrophotometer (Japan Spectroscopic Company, Tokyo, Japan) with a 1 mm quartz cell at 298 K. The scan rate was 100 nm min−1 and the scanning range was 190–250 nm with an interval of 1 nm.15
UV-visible absorption spectrum experiments
At 298 K, UV-visible absorption spectrum experiments were conducted by utilizing UV-2700 spectrophotometer (Shimadzu Co., Kyoto, Japan). A 190–500 nm scanning wavelength range with an interval of 1 nm was set.16
Molecular docking
The 3D structure of scoparone was obtained from chemspider (http://www.chemspider.com). Then, it was converted into mol2 format by OpenBabel-2.3.2. The crystal structure of BSA (PDB ID: 4F5S) was obtained from protein data bank (http://www.rcsb.org/pdb/home/home.do). All water molecules were removed from the protein. All hydrogen atoms were added, and the AD4 atom type and Gaisteiger charges were assigned to the receptor protein. Molecular docking simulation was conducted by Autodock tools 1.5.6 to obtain the probable binding mode of scoparone with BSA. The docking site for scoparone on BSA was regarded as the active site with grid box size of 60 × 60 × 60, spacing of 0.375 Å, and grid centre of −10.586, 33.853, and 102.903. The running times of genetic algorithm was set at 100. All other parameters were set at the default. The minimum binding free energy conformation of scoparone–BSA complex was visualized by utilizing PyMol 1.8.2.0, Discovery Studio version 2.5 and Ligplot+ 1.4.5.
Results and discussion
Effect of scoparone on fluorescence spectra of BSA
The fluorescence spectrum of proteins can reflect the species, structure, microenvironment and distribution of the fluorescent chromophore groups in protein molecules. Protein molecules can emit strong endogenous fluorescence due to amino acid residues, such as tryptophan, tyrosine and phenylalanine.17 When excited at 280 nm, protein fluorescence is mainly caused by tryptophan, because the fluorescence of tyrosine is practically totally quenched if it is ionized or near a carboxyl group, an amino group, or a tryptophan residue and phenylalanine has a very low quantum yield.18,19 The fluorescence spectra about the interaction of scoparone with BSA at different temperatures were shown in Fig. 1. When excited by exciting light with the wavelength of 280 nm, the maximum emission peak of BSA appeared at near 340 nm. When the concentration of BSA solution (1 × 10−5 mol L−1) was fixed, the endogenous fluorescence intensity of BSA decreased with the increase of scoparone concentrations. These results indicated that the interaction between scoparone and BSA occurred.20 Sun et al. proved that isoalantolactone could interact with HSA to quench the endogenous fluorescence of HSA, which was very similar to our experimental results.21
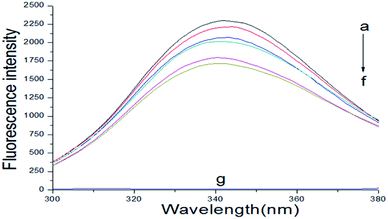 |
| | Fig. 1 The fluorescence emission spectrum of BSA with or without various concentrations of scoparone (a–f) was measured at 298 K. The concentrations of scoparone were from 0 to 1.65 × 10−5 mol L−1 with a concentration gradient of 0.33 × 10−5 mol L−1. Curve g represented the fluorescence emission spectrum of 1 × 10−5 mol L−1 scoparone. CBSA = 1 × 10−5 mol L−1. | |
Fluorescence quenching can decrease the fluorescence intensity by the excited molecules losing their energy, which generally induced by some molecular interactions, including excited-state reactions, collisional quenching, ground-state complex formation, molecular rearrangements and energy transfer.22 The quenching effect of fluorescence can usually be categorized into dynamic quenching and static quenching according to different quenching mechanisms. Static quenching can form a new complex between quenchers and fluorophores, and the quenching constant decreases with increasing temperature. In contrast, for dynamic quenching, the quenching constant increases by increasing the collisional encounters that can be increased with increasing temperature.23,24 In the dynamic quenching process, the interaction between the fluorophores and quenchers can be described by the Stern–Volmer equation:25
| |
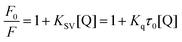 | (1) |
where
F and
F0 represent the fluorescence intensities of BSA with or without the quencher (scoparone) respectively, [Q] means the concentration of scoparone,
Kq is the fluorescence quenching rate constant of protein,
Ksv is the quenching constant,
τ0 represents the mean fluorescence lifetime of protein without quencher (
τ0 is equal to 10
−8 s).
26
A good linear relationship was found in Stern–Volmer plots at three different temperatures in Fig. 2A. The quenching constant of the intrinsic fluorescence of BSA quenched by scoparone was decreased with the increasing temperatures. Moreover, the quenching rate constants (Kq) of BSA fluorescence were much greater than 2.0 × 1010 L mol−1 s−1 as shown in Table 1. Based on these results, it could be seen that the fluorescence quenching effect of scoparone to BSA was due to the static quenching caused by scoparone binding to BSA.27
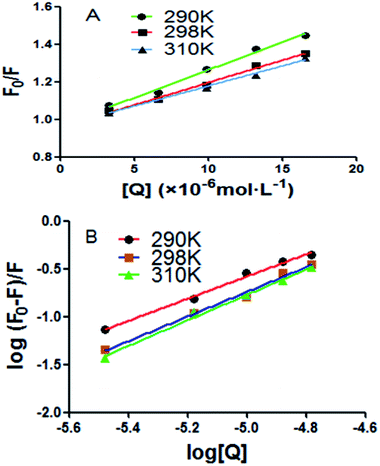 |
| | Fig. 2 (A) Stern–Volmer plots of BSA quenched by scoparone at three temperatures. (B) Modified Stern–Volmer plots of BSA quenched by scoparone at three temperatures. | |
Table 1 Stern–Volmer equation constants of BSA quenched by scoparone at three temperaturesa
| T (K) |
Ksv (×104 mol L−1) |
kq (×1012 L mol−1 s−1) |
r2 |
| r2 is the correlation coefficient. |
| 290 |
2.96 |
2.96 |
0.9904 |
| 298 |
2.39 |
2.39 |
0.9927 |
| 310 |
2.14 |
2.14 |
0.9933 |
For static quenching, the relationship between fluorescence intensity and quencher concentration are according with Line weaver–Burk double logarithmic equation:28
| |
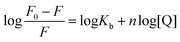 | (2) |
where [Q],
F and
F0 are identical with
eqn (1),
n denotes the number of binding sites and
Kb means the binding constant.
The log[(F0 − F)/F] against lg[Q] at three temperatures were plotted as shown in Fig. 2B. From the slope and intercept of the plot, the binding constants (Kb) and number of binding sites (n) between BSA and scoparone could be obtained. As presented in Table 2, the values of n were approximately equal to 1. In addition, the values of Kb were in the magnitude of 105 mol L−1, which meant that scoparone had a high affinity binding on BSA.29 It is generally accepted that strong binding could decrease the concentration of free drug in plasma. However, weak binding could lead to a short life time or poor distribution. Due to scoparone having a strong affinity binding on BSA. It could be inferred that scoparone was slowly released in bloodstream and played a lasting pharmaceutical effect in vivo.2
Table 2 The thermodynamic parameters and binding constants of BSA quenched by scoparone at three temperaturesa
| T (K) |
Kb (×105 mol L−1) |
n |
r2 |
ΔH (kJ mol−1) |
ΔG (kJ mol−1) |
ΔS (J mol−1 K−1) |
| r2 is the correlation coefficient. |
| 290 |
2.31 |
1.19 |
0.9918 |
39.93 |
−29.85 |
240.64 |
| 298 |
4.39 |
1.28 |
0.9912 |
|
−31.78 |
|
| 310 |
6.79 |
1.32 |
0.9922 |
|
−34.67 |
|
The binding forces between drug molecules and proteins mainly include hydrophobic interaction, van der Waals force, electrostatic force, and hydrogen bonding.30 Ross et al. found some binding rules of small molecules and biomacromolecules, mainly including: when ΔH < 0 and ΔS < 0, the hydrogen bond and van der Waals force are the main forces. When ΔH ≈ 0 and ΔS > 0, the electrostatic force dominates the interaction. When ΔH > 0 and ΔS > 0, the hydrophobic force plays the major role in interaction.31 When the temperature does not change much, the enthalpy change (ΔH) of the reaction can be seen as a constant. According to the relative magnitudes of the thermodynamic parameters of enthalpy change (ΔH) and entropy change (ΔS) before and after the reaction, the types of the main interaction forces between drug molecules and proteins can be acquired.32 The relationship of thermodynamic parameters is as follows:33
| |
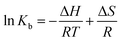 | (3) |
where
R represents the gas constant,
T represents the experimental temperature and
Kb means the binding constant. The values of Δ
H and Δ
S can be obtained
via the slope and intercept of curve of ln
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Kb versus
Kb versus 1/
T as shown in
Fig. 3A. The values of Δ
G can be obtained with
eqn (4).
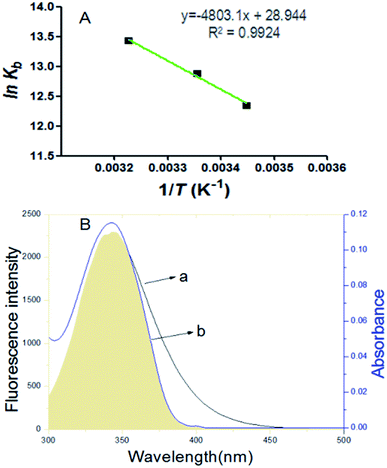 |
| | Fig. 3 (A) Van't Hoff plots of BSA quenched by scoparone at three temperatures. (B) At 298 K, the overlaps of fluorescence spectra of BSA (a) with the UV-visible absorption spectra of scoparone (b). The concentrations of BSA and scoparone were 1 × 10−5 mol L−1. | |
The values of ΔG, ΔH and ΔS were calculated as presented in Table 2. The negative value of ΔG and positive values of ΔH and ΔS revealed that the interaction of scoparone with BSA was a spontaneous reaction mainly driven by hydrophobic interaction.15
Binding distance between scoparone and BSA
Förster's theory of dipole–dipole non-radiative energy transfer states that when the energy donor is close enough with energy receptor and the distance is within 7 nm, the excited energy donor molecules will undergo non-radiative energy transfer with neighboring energy receptor molecules.9 The energy transfer efficiency (E) and binding distance (r) between energy donor molecules (BSA) and energy receptor molecules (scoparone) can be determined. The value of E can be obtained by eqn (5):17| |
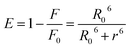 | (5) |
where F and F0 are identical with eqn (1), R0 denotes the critical distance when E is 50%. The value of R0 can be calculated with eqn (6):18| | |
R06 = 8.8 × 10−25 k2 N−4 Φ J(λ)
| (6) |
where Φ represents the fluorescence quantum yield of BSA, Φ is equal to 0.118, k2 denotes the spatial orientation factor of the dipole, k2 is equal to 2/3, N represents the average refractive index of the medium, N is equal to 1.336, and J(λ) represents the spectral overlap integral between the absorption spectrum of scoparone and the emission spectrum of BSA. It can be computed with eqn (7):20| |
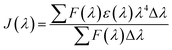 | (7) |
where ε(λ) and F(λ) denote the molar extinction coefficient of scoparone and fluorescence intensity of BSA at wavelength λ, respectively. As shown in Fig. 3B, the fluorescence emission spectrum of BSA partially overlapped with the UV-vis absorption spectrum of scoparone. The value of J(λ) was 7.32 × 10−14 cm3 L−1 mol−1. Furthermore, the values of E, R0 and r respectively were 11%, 3.42 nm and 4.81 nm. These experimental results revealed that the energy transfer from BSA to scoparone arose with a high possibility due to 0.5R0 < r < 1.5R0 and r < 7 nm.34
Synchronous fluorescence spectra
In the synchronous fluorescence spectrum analysis, Δλ = 15 nm shows the fluorescence spectrum characteristic of tyrosine residues, and Δλ = 60 nm shows the fluorescence spectrum characteristic of tryptophan residues.35 The maximum emission wavelength of tyrosine and tryptophan residues in proteins is related to the polarity of the microenvironment in which these two residues are located. Therefore, the changes in the conformation of the proteins can be determined by observing the change of the maximum emission wavelength.36 The BSA–scoparone system's synchronous fluorescence spectrum can be obtained by fixing the BSA concentration and increasing the scoparone concentrations gradually. As shown in Fig. 4, the fluorescence intensity of tryptophan and tyrosine residues in BSA was significantly reduced with the increasing concentrations of scoparone. However, the pitch of quenching of Trp residues was more obvious than Tyr residues, which indicated that the binding site of scoparone on BSA was mainly focused on Trp nearby.19 Furthermore, the maximum λem of Tyr residues almost did not change and a slight blue shift (2 nm) was found in the maximum λem of Trp residues. These results confirmed that there was almost no change in the microenvironment around Tyr residues but had slight change in the microenvironment around Trp residues, which meant that BSA conformation was altered slightly. Lou et al. conducted a study of the interaction between fungicide boscalid and BSA by synchronous fluorescence spectra, proving that fungicide boscalid almost did not change in the microenvironment around Tyr residues but made slight change in the microenvironment around Trp residues, which made our research results more convincing.22
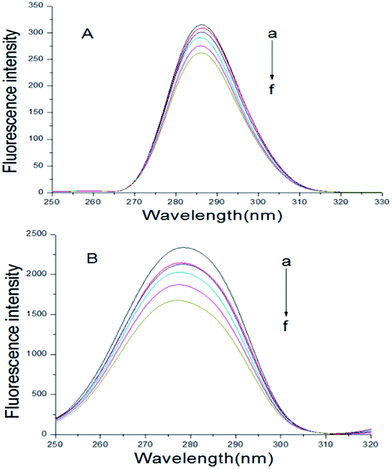 |
| | Fig. 4 At 298 K, synchronous fluorescence spectrum of BSA with or without scoparone (a–f) were measured. The concentrations of scoparone differed from 0 to 1.65 × 10−5 mol L−1 with a concentration gradient of 0.33 × 10−5 mol L−1. CBSA = 1 × 10−5 mol L−1, (A) Δλ = 15 nm, (B) Δλ = 60 nm. | |
Circular dichroism spectroscopy
To further examine the changes in the conformation of BSA induced by scoparone, circular dichroism spectroscopy was used to study the interaction between scoparone and BSA. As shown in Fig. 5A, the circular dichroism spectra of BSA showed two negative peaks at near 208 nm and 222 nm, which were the characteristic peaks of the α-helix structure in BSA. The α-helix content was calculated by the following two formulas:3| |
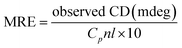 | (8) |
| |
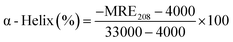 | (9) |
where MRE is the residue ellipticity, Cp represents the molar concentration of BSA, l denotes the path length and n means the number of amino acid residues (n = 583, for BSA). 4000 represents the MRE value of the β-form and random coil conformation cross at 208 nm. 33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 represent the MRE value of pure α-helix at 208 nm. MRE208 denotes the observed MRE value at 208 nm.
000 represent the MRE value of pure α-helix at 208 nm. MRE208 denotes the observed MRE value at 208 nm.
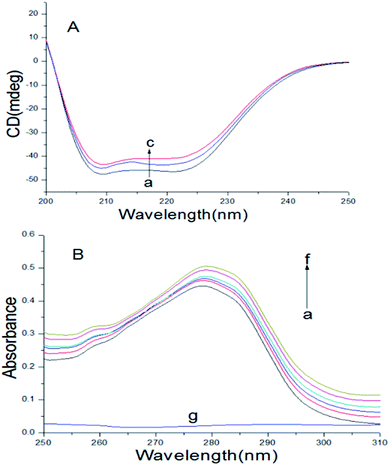 |
| | Fig. 5 (A) At 298 K, CD spectra of BSA with or without scoparone (a–c) were obtained. The concentrations of scoparone were from 0 to 1 × 10−5 mol L−1 with a concentration gradient of 0.5 × 10−5 mol L−1, CBSA = 5 × 10−6 mol L−1. (B) At 298 K, the UV-vis absorption spectrum of BSA with or without scoparone (a–f) were obtained. The concentrations of scoparone were from 0 to 1.65 × 10−5 mol L−1 with a concentration gradient of 0.33 × 10−5 mol L−1. Curve g represented the spectrum of 0.33 × 10−5 mol L−1 scoparone, CBSA = 1 × 10−5 mol L−1. | |
When the concentration of scoparone was gradually increased, the intensity of negative peaks of BSA at 208 and 222 nm in the circular dichroism spectra reduced, but the peak shape remained basically unchanged. In addition, by eqn (8) and (9), the calculated content of α-helix structure in BSA decreased from 45.26% to 42.14%. These results proved that the conformation of BSA was slightly changed by interaction with scoparone, which was consistent with the results of synchronous fluorescence spectroscopy. Raza et al. conducted a study of the interaction between amphotericin B and BSA by circular dichroism spectra. The results revealed that with the increase of amphotericin B concentrations, the α-helix content of BSA decreased from 45.22% to 41.75%, which also showed that the conformation of BSA was slightly changed.30
UV-vis absorption spectra
UV-vis absorption spectroscopy can usually be applied to research the protein conformational changes and complex formation between proteins and drugs.28 As shown in Fig. 5B, there was a absorption peak at near 280 nm, because of the π → π* transition of some aromatic amino acids of BSA, including Phe, Tyr and Trp.25 The absorption intensity of peak at about 280 nm increased with the increase of scoparone concentrations, revealing that a new complex of scoparone–BSA was formed. In addition, the changes of absorption spectrum of BSA further verified that the quenching effect of scoparone to BSA was static quenching. Bardhan et al. investigated the interaction of aurintricarboxylic acid with bovine serum albumin by UV-vis absorption spectroscopy, proving that absorption spectrum of BSA gradually increased with increase of aurintricarboxylic acid concentrations, which was not much different from our research.3
Binding site studies
Many studies have shown that most of the drug molecules can be located in the hydrophobic cavities of subdomain IIA and subdomain IIIA, namely site I and site II.18 Related research results have proved that phenylbutazone and ibuprofen could be used for site I and site II probes respectively.17,19 So these chemicals were used to study the binding sites of scoparone on BSA. The results of the experiment showed that the value of Kb was 4.53 × 102 mol L−1 in the presence of phenylbutazone and the correlation coefficient was 0.9905. The value of Kb was 4.73 × 105 mol L−1 in the presence of ibuprofen and the correlation coefficient was 0.9867. For scoparone–BSA complex, the value of Kb decreased obviously in the presence of phenylbutazone as shown in Fig. 6. These results revealed that scoparone was more likely to be located in site I of BSA.29
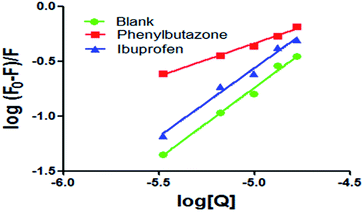 |
| | Fig. 6 Modified Stern–Volmer plots of co-solution of BSA and site markers quenched by scoparone at 298 K. | |
Molecular docking studies
Molecular docking technique is an important tool for understanding the ligand–protein interactions, which can further substantiate the experimental data.4 The docking results with the lowest binding free energy (−4.95 kcal mol−1) were shown in Fig. 7, which is basically consistent with ΔG at 298 K. Scoparone was located in the hydrophobic cavity of IIA of BSA (site I), which was consistent with site marker competitive experimental results. Furthermore, ΔE1 (intermolecular interaction energy, which is a sum of van der Waals energy, hydrogen bonding energy, desolvation free energy and electrostatic energy), ΔE2 (the sum of van der Waals energy, hydrogen bonding energy and desolvation free energy) and ΔE3 (electrostatic energy) were obtained. ΔE1 was equal to −5.55 kcal mol−1. ΔE2 was equal to −5.24 kcal mol−1 and ΔE3 was equal to −0.3 kcal mol−1. ΔE2 was markedly more negative than ΔE3. Therefore, based on these molecular docking information and the results of thermodynamic parameters, it could be speculated that hydrophobic interaction was the main forces for scoparone binding to BSA.
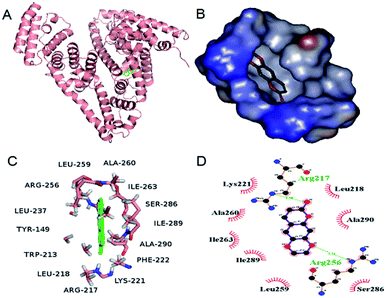 |
| | Fig. 7 (A) Scoparone docked in the site I of BSA. (B) Scoparone located in the hydrophobic cavity of site I. (C) Scoparone was surrounded by amino acid residues of BSA in 4 Å. (D) The partial forces between scoparone and some amino acids of BSA. | |
Conclusions
In the present work, the interaction between scoparone and BSA under simulated physiological conditions was studied by fluorescence spectroscopy, circular dichroism spectroscopy, UV-vis absorption spectroscopy and molecular simulation techniques. The results of fluorescence spectroscopy indicated that scoparone quenched the endogenous fluorescence of BSA and the quenching mechanism was static quenching. The binding between scoparone and BSA was a spontaneous reaction mainly driven by hydrophobic interaction. The results of molecular docking and the site marker experiment revealed that scoparone was more likely to be located in site I of BSA. The results of synchronous fluorescence and circular dichroism spectroscopy confirmed that scoparone induced the conformational changes of BSA slightly. This study would be very useful for us to explain the pharmacokinetics of scoparone and the pharmacological mechanism. The efficacy and metabolism of scoparone in the human body will be further studied in future studies.
Conflicts of interest
There are no conflicts to declare.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (No. 31770017), Cultivation Plan for Youth Agricultural Science and Technology Innovative Talents of Liaoning Province (No. 2015013), Project Supported by Scientific Research Fund of Liaoning Provincial Education Department (No. LQN201714), Startup Foundation for Doctors of Liaoning Province (No. 20170520258).
References
- M. Raza, A. Ahmad, F. Yue, Z. Khan, Y. Jiang, Y. Wei, S. Raza, W. W. He, F. U. Khan and Y. Q. Peng, J. Photochem. Photobiol., B, 2017, 170, 6–15 CrossRef PubMed.
- Q. Wang, C. R. Huang, M. Jiang, Y. Y. Zhu, J. Wang, J. Chen and J. H. Shi, Spectrochim. Acta, Part A, 2016, 156, 155–163 CrossRef PubMed.
- M. Bardhan, J. Chowdhury and T. Ganguly, J. Photochem. Photobiol., B, 2011, 102, 11–19 CrossRef PubMed.
- M. Manjushree and H. D. Revanasiddappa, J. Mol. Liq., 2017, 237, 23–37 CrossRef.
- S. Minic, D. Stanic-Vucinic, M. Radomirovic, M. Radibratovic, M. Milcic, M. Nikolic and T. C. Velickovic, Food Chem., 2018, 239, 1090–1099 CrossRef PubMed.
- Y. Y. Lou, K. L. Zhou, D. Q. Pan, J. L. Shen and J. H. Shi, J. Photochem. Photobiol., B, 2017, 167, 158–167 CrossRef PubMed.
- Y. Q. Wang, M. Wang, B. P. Chen and J. Shi, Eur. J. Pharmacol., 2017, 815, 376–380 CrossRef PubMed.
- X. Liu and X. X. Zhao, Biomed. Pharmacother., 2017, 93, 57–61 CrossRef PubMed.
- M. S. Nair, J. Photochem. Photobiol., B, 2015, 149, 58–67 CrossRef PubMed.
- G. W. Zhang, Y. T. Hu and J. H. Pan, J. Solution Chem., 2014, 43, 727–745 CrossRef.
- Y. P. Wang, G. W. Zhang and L. H. Wang, Pestic. Biochem. Physiol., 2014, 108, 66–73 CrossRef PubMed.
- G. W. Zhang and Y. D. Ma, Food Chem., 2013, 136, 442–449 CrossRef PubMed.
- G. W. Zhang, Y. D. Ma, L. Wang, Y. P. Zhang and J. Zhou, Food Chem., 2012, 133, 264–270 CrossRef PubMed.
- Y. Y. Lou, K. L. Zhou, J. H. Shi and D. Q. Pan, J. Photochem. Photobiol., B, 2017, 173, 58–67 CrossRef PubMed.
- J. J. Ao, L. Gao, T. Yuan and G. F. Jiang, Chemosphere, 2015, 119, 590–600 CrossRef PubMed.
- G. F. Shen, T. T. Liu, Q. Wang, M. Jiang and J. H. Shi, J. Photochem. Photobiol., B, 2015, 153, 380–390 CrossRef PubMed.
- J. H. Shi, D. Q. Pan, M. Jiang, T. T. Liu and Q. Wang, J. Photochem. Photobiol., B, 2016, 164, 103–111 CrossRef PubMed.
- J. H. Shi, D. Q. Pan, X. X. Wang, T. T. Liu, M. Jiang and Q. Wang, J. Photochem. Photobiol., B, 2016, 162, 14–23 CrossRef PubMed.
- J. H. Shi, K. L. Zhou, Y. Y. Lou and D. Q. Pan, Spectrochim. Acta, Part A, 2018, 188, 362–371 CrossRef PubMed.
- L. Fu, Y. Q. Sun, L. N. Ding, Y. Y. Wang, Z. Gao, Z. Wu, S. M. Wang, W. Li and Y. F. Bi, Food Chem., 2016, 203, 150–157 CrossRef PubMed.
- Y. Sun, C. M. Hu, G. B. Li, S. Y. Yang, L. S. Liu, C. L. Tan, S. Wei, X. Y. Hu, Y. Y. Zhao and J. Fan, Indian J. Chem., 2011, 50, 1477–1484 Search PubMed.
- Y. Y. Lou, K. L. Zhou, J. H. Shi and D. Q. Pan, J. Photochem. Photobiol., B, 2017, 173, 589–597 CrossRef PubMed.
- A. J. Esfahlan and V. P. Azar, Food Chem., 2016, 202, 426–431 CrossRef PubMed.
- K. B. Uppuluri, K. B. Ayaz Ahmed, A. Jothi and A. Veerappan, J. Mol. Liq., 2016, 219, 602–607 CrossRef.
- B. Tang, Y. M. Huang, X. L. Ma, X. X. Liao, Q. Wang, X. N. Xiong and H. Li, Food Chem., 2016, 212, 434–442 CrossRef PubMed.
- V. Anbazhagan and R. Renganathan, J. Lumin., 2008, 128, 1454–1458 CrossRef.
- L. Xu, Y. X. Hu, J. Li, Y. F. Liu, L. Zhang, H. X. Ai and H. S. Liu, J. Photochem. Photobiol., B, 2017, 173, 187–195 CrossRef PubMed.
- M. Kumari, J. K. Maurya, U. K. Singh, A. B. Khan, M. Ali, P. Singh and R. Patel, Spectrochim. Acta, Part A, 2014, 124, 349–356 CrossRef PubMed.
- J. H. Shi, J. Chen, J. Wang, Y. Y. Zhu and Q. Wang, Spectrochim. Acta, Part A, 2015, 149, 630–637 CrossRef PubMed.
- M. Raza, A. Ahmad, F. Yue, Z. Khan, Y. Jiang, Y. Wei, S. Raza, W. W. He, F. U. Khan and Y. Q. Peng, J. Photochem. Photobiol., B, 2017, 170, 6–15 CrossRef PubMed.
- R. Q. Wang, Z. G. Li, L. L. Yang, T. Ren, L. J. Zhang and R. Y. Wang, J. Mol. Liq., 2015, 211, 584–590 CrossRef.
- M. Sarkar, S. S. Paul and K. K. Mukherjea, J. Lumin., 2013, 142, 220–230 CrossRef.
- B. Tang, Y. M. Huang, H. Q. Yang, P. X. Tang and H. Li, J. Photochem. Photobiol., B, 2016, 165, 24–33 CrossRef PubMed.
- S. Yasmeen and Riyazuddeen, Thermochim. Acta, 2017, 655, 76–86 CrossRef.
- H. B. Shen, Z. Q. Gu, K. Jian and J. Qi, J. Pharm. Biomed. Anal., 2013, 75, 86–93 CrossRef PubMed.
- S. Roy, R. K. Nandi, S. Ganai, K. C. Majumdar and T. K. Das, J. Pharm. Anal., 2017, 7, 19–26 CrossRef PubMed.
Footnote |
| † These authors contributed equally to this work. |
|
| This journal is © The Royal Society of Chemistry 2018 |
Click here to see how this site uses Cookies. View our privacy policy here.  Open Access Article
Open Access Article ,
Yonglin He†,
Dan Liu,
Yin He,
Xiao Hou,
Ye Cheng and
Jianli Liu*
,
Yonglin He†,
Dan Liu,
Yin He,
Xiao Hou,
Ye Cheng and
Jianli Liu*
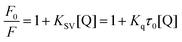

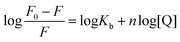
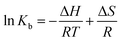
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Kb versus 1/T as shown in Fig. 3A. The values of ΔG can be obtained with eqn (4).
Kb versus 1/T as shown in Fig. 3A. The values of ΔG can be obtained with eqn (4).
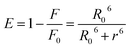
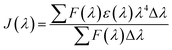
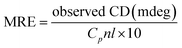
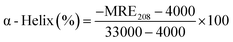
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 represent the MRE value of pure α-helix at 208 nm. MRE208 denotes the observed MRE value at 208 nm.
000 represent the MRE value of pure α-helix at 208 nm. MRE208 denotes the observed MRE value at 208 nm.






