Open Access Article
Open Access ArticleA study on a telo21 G-quadruplex DNA specific binding ligand: enhancing the molecular recognition ability via the amino group interactions†
Dongli Li‡
ab,
Jin-Qiang Hou‡c,
Wei Longc,
Yu-Jing Lubc,
Wing-Leung Wong *ab and
Kun Zhang*ab
*ab and
Kun Zhang*ab
aSchool of Chemical and Environmental Engineering, Wuyi University, Jiangmen 529020, P. R. China. E-mail: kzhang@gdut.edu.cn; Tel: +86-20-39322235
bInternational Healthcare Innovation Institute (Jiangmen), Jiangmen 529040, P. R. China. E-mail: wingleung@wyu.edu.cn; Tel: +86-0750-3299391
cInstitute of Natural Medicine and Green Chemistry, School of Chemical Engineering and Light Industry, Guangdong University of Technology, Guangzhou 510006, P. R. China
First published on 4th June 2018
Abstract
A symmetric ligand is synthesized composed of a core N-methylpyridinium scaffold and two para-substituted benzyl groups through a flexible ethylene bridge to form a novel three-ring-conjugated system. The ligand system was found to have only weak background fluorescent signal in aqueous or physiological conditions and exhibited strong fluorescent signal enhancement targeting at telo21 G-quadruplex structure rather than other types of nucleic acids. The comparison study with two terminal groups (–N(CH3)2 versus –SCH3) indicates that the stimulated signal enhancement of specific binding is probably attributed to the hydrogen-bonding interactions through the amino groups in the G-quartets. The docking result illuminates the experimental observation that the ligand system showed only weak fluorescent signals in aqueous or physiological conditions while exhibiting a strong fluorescent signal upon binding to the telo21 G-quadruplex structure (binding energy: −6.2 kcal mol−1).
Introduction
Fluorescent molecular signaling ligands or probes that can be utilized as effective bio-recognition, bio-labelling, and imaging agents in situ for target biomolecules such as amino acids, nucleic acids, peptides, proteins, and biomarkers of cancers, are of importance in many applications, particularly in the field of biological and biomedical sciences.1–6 In recent years, imaging technology developments in novel fluorophores with higher brightness and photo-stability have achieved a great improvement.7–9 There are a number of new materials including green fluorescent proteins, nanoparticles and quantum dots, and organic dyes active in the visible to the near infrared regions have been demonstrated as a non-invasive and sensitive tool for studying biological systems in vivo and for probing molecular dynamics in live cells with the fluorescent optical microscopy technique.10–12 However, the sensing or detection of a particular type of targeted biomolecule selectivity in their native environment is still challenging.13,14 In the nucleic acid research area, to discrimination of G-quadruplex (G4) structures from other type of DNA by visual methods is a big difficulty.15–17 For this reason, considerable efforts are in progress for the discovery of quadruplex specific binding ligands endowed with fluorescence properties.Diverse molecular design is recognized as a right strategy to search for G4-selective molecular fluorescent probes or ligands that display a strong enhancement upon binding with G4-DNA.18–20 Our recent study demonstrated that the small organic molecular ligand system is also able to achieve excellent G-quadruplex DNA selectivity with the induction of a strong fluorescent signal against other DNA tertiary structures.21 A crucial factor of the design reported is to build a π-conjugated co-planar two-ring system with a slightly rotatable ethylene bridge that increases the structural flexibility of the molecule (Fig. 1, L).22 The reported ligand L shows only limited signal discrimination ability although its signaling specificity towards G4-DNA is merit. Probably due to the ligand only has one amine group for interaction. To further advance the structure design, in the present study, a symmetrical ligand with N,N-dimethylaniline groups was constructed based on the π-conjugated co-planar three-ring system framework and investigated the significance of the presence of two amino groups in enhancing the interaction signaling through the comparison with a non-polar analog (methylthio group). The results indicated that the two amino groups provide extra binding affinity and selectivity towards telo21 G4-DNA, which is most probably attributed to the extra hydrogen bonds formed with the hydroxyl groups in groove structure of telo21.23 FRET melting experiments also support the amino groups granting an extra stabilization for the G-quadruplex structure of telomeric DNA.
Results and discussion
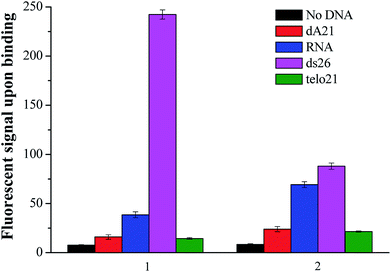
The new nucleic acid binding ligand targeting at human telo21 G-quadruplex DNA was synthesized by the piperidine-catalyzed Knoevenagel condensation reaction using 1,2,6-trimethylpyridin-1-ium iodide and para-substituted benzaldehyde, for examples p-(dimethylamino)benzaldehyde and p-(methylthio)benzaldehyde, as the reactants in n-butanol under reflux conditions. Pure compounds of 1 and 2 shown in Fig. 1 were obtained by re-crystallization from acetone with 70–85% yields.The spectroscopic properties of the nucleic acid binding ligands were listed in Table 2. The ligand with N-methylpyridinium scaffold as the core that is fused to form a three-ring-conjugated system exhibited absorption maxima at 402 nm and 450 nm and shows very weak emission signal in the range of 530–580 nm in aqueous buffer solution at pH 7.4. Interestingly, when addition of various nucleic acids such as those examined herein with the single-stand DNA: dA21, duplex DNA: ds26, G-quadruplex DNA: telo21, and RNA, the ligand was able to induce different degrees of fluorescent signal at yellow-green region (1: λem = 580 nm) as shown in Fig. 2. The difference in the fluorescent intensity observed indicates a recognition ability of the ligands towards different types of DNA structure. Particularly, ligand 1 upon binding human telo21 G-quadruplex DNA showing the highest emission signal while other types of DNA including RNA, dA21, and ds26 show very weak recognition response. However, its analog ligand 2 has no such distinctive recognition observation when tested with different nucleic acids under the same conditions. This could conclude that the side groups (dimethylamino versus methylthio) of the binding ligands take a significant effect on the fluorescent recognition of different type of nucleic acids structures. We speculated that the dimethylamino group is polar and is able to offer extra hydrogen bonds formation with the hydroxyl groups in groove structure of telo21 G4-DNA structure while the methylthio group has no such character. This explanation is further supported by the results obtained in the binding kinetics study and FRET melting experiments.
| Ligand | Side group | λAbs,max a(nm) | λex a(nm) | λem a(nm) | Φf b | K c(×105 M−1) |
|---|---|---|---|---|---|---|
| a Experiments were performed in 10 mM Tris–HCl buffer at pH 7.4.b Relative fluorescence quantum yield of probes upon addition of 3 μM telo21, standard of the relative fluorescence quantum yield is fluorescein (Φf = 0.85, in 1% NaOH).c Equilibrium binding constant between the ligand and telo21 DNA. | ||||||
| 1 | R = N(CH3)2 | 450 | 460 | 580 | 0.05 | 1.44 ± 0.35 |
| 2 | R = SCH3 | 402 | 402 | 530 | 0.02 | 1.18 ± 0.21 |
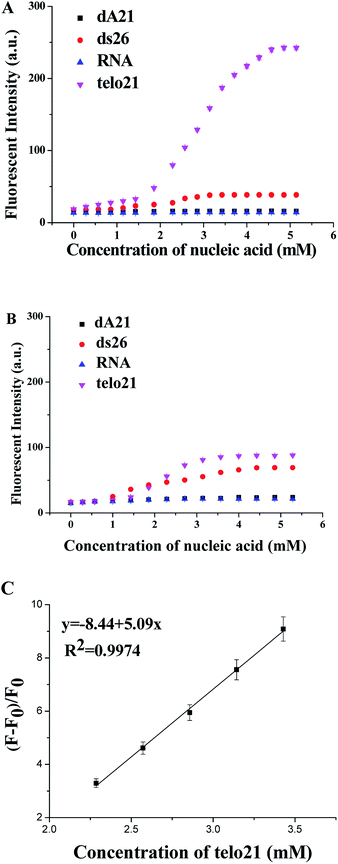
The molecular binding kinetics of the ligand with nucleic acids could be able to demonstrate the interaction difference due to dimethylamino and methylthio side group of the ligand. Fig. 3 showed the ligand binding assays at equilibrium, obviously the binding ligand with dimethylamino group showed much stronger recognition and interaction emission signal (2.5 times higher than methylthio group in the binding assay with G4-DNA telo21). Due to the three-ring-conjugated system is linked through the ethylene bridge that is flexible for free rotation, the increase in structural rigidity could enhance fluorescent signal upon bound with DNA molecule. The telo21 G-quadruplex structure took the advantage as its binding site (the G-quartets) is relatively planar and provides extra hydrogen bond interactions with the dimethylamino groups of 1. Previous studies also revealed that the binding affinities obtained with different amino-glycoside ligands for RNA often correlate with the number of amines present in the ligand.24,25 Therefore, it is possibly that the dimethylamino groups on the binding ligand could modulate the basicity of the amines and then leads to much stronger interactions with telo21 DNA. Both the fluorescent quantum yield upon binding with telo21 and the binding affinity of the ligand with dimethylamino groups is better than methylthio group (Table 1). The equilibrium binding constant for the dimethylamino groups is 1.44 × 105 M−1 while for the methylthio groups is significant lower (1.18 × 105 M−1). In addition, a good linear relationship (R2 = 0.9974) of the enhanced fluorescent signal of 1 with respect to telo21 DNA concentration in buffer solution (pH 7.4) was established (Fig. 3C) and the limit of detection (LOD) in the titration assay for telo21 DNA was estimated to be 5.8 nM.
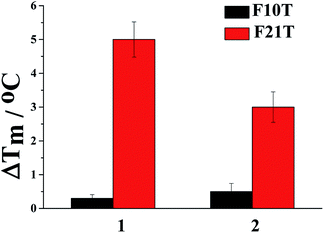
To obtain more information about the interaction property difference due to the side groups of ligand 1 and 2, the stabilization ability of the ligand bound with G-quadruplex (F21T) and duplex DNA (F10T) was investigated using fluorescence resonance energy transfer (FRET) melting assays. From Fig. 4, as indicated by the ΔTm values, both ligands provide much better stabilization ability to G-quadruplex DNA than duplex DNA. However, a larger ΔTm value increased by 5 °C was achieved with ligand 1 (at 2.0 μM) under the same conditions compared with 2. The result may suggest that the better molecular interactions of 1 with G-quadruplex (F21T) is a consequence of the dimethylamino group that provides extra hydrogen bond interactions while the non-polar methylthio group does not offer. In addition, ligand 1 could be a promising human telo21 G-quadruplex stabilizer.
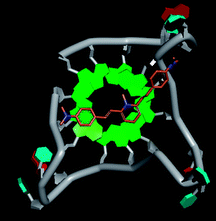
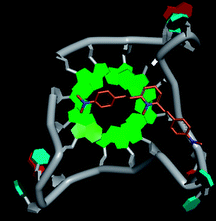
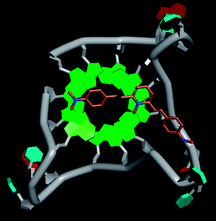
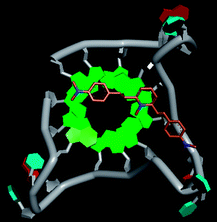
Molecular docking was performed to investigate the binding mode of compound 1 with telomeric G4. The crystal structure of human telomeric DNA G-quadruplex (4DA3)28 were used as a template and the result is depicted in Fig. 5. The compound is able to stack onto the G-quartets via pi–pi stacking interactions. The methylated pyridine ring of the compound carrying a positive charge was found to sit closely on the negatively charged carbonyl channel to enhance electrostatic interaction with G4. The three-ring system of 1 interacts with at least two guanines and the amino functional group of compound is likely to form water mediated H-bonds with G4 as evidenced by a number of crystal structures of ligand-G4 complex (e.g., PDB ID: 1L1H, 3CE5, 3CDM), where the ligands possesses amino functional groups. All those interactions with G-quartets constrain the π-conjugation system of 1 to a more planar conformation as compared to its unbound form, which is conformational flexible due to the ethylene bridge.22 The docking result explains the experimental observation that the ligand system showed weak fluorescent signal in aqueous or physiological conditions while exhibited strong fluorescent signal induction upon binding to telo21 G-quadruplex structure. Interestingly, our findings based on both experimental and docking results are in accord with to previously reported results that the introduction of a net positive charge onto the side chain (through the nitrogen atom of the terminal morpholino moiety) of a series of fluorenone carboxamides can improve G-quadruplex binding (docking study based on 2HY9, a NMR optimized DNA G-quadruplex model GRID MIFs, related to the telomeric sequence d[AG3(T2AG3)3]).29
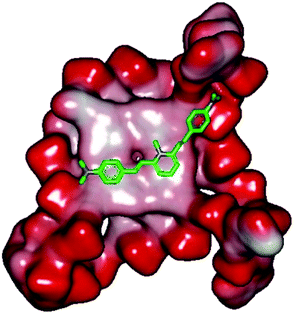 | ||
| Fig. 5 Molecular docking of 1 with a telomeric G4 (4DA3) to understand the binding mode. | ||
In addition, in the docking study based on the 1 to 1 stoichiometry ratio for the ligand interacts with telomeric G-quarduplex using autodock vina. The end stacking interaction with G-quartet is more likely the case because 1 is consisted of three aromatic rings connected by a conjugated system and the results show that 1 prefers to stack on 3′ end of the G-quadruplex. All these end stacking poses found from docking study are with 3′ end as listed in Table 2 for comparison.
Conclusions
In conclusion, a new symmetric ligand with a N-methylpyridinium scaffold as the molecular core fused p-(dimethylamino)benzyl groups through an ethylene bridge to form a three-ring-conjugated system exhibited very weak background emission in the yellow-green region under physiological conditions. The comparison studies demonstrated that the dimethylamino functional groups showed critical influence on the remarkable induction of fluorescent signal specific to telo21 G-quadruplex structure rather than other type of nucleic acids.Experimental section
General information
All commercial chemicals, reagents, and solvents were analytical grade and were used without further purification. Melting points (mp) measurements were performed using a SRS Opti Mel automated melting point instrument without correction. Mass spectra (MS) were recorded on Bruker amaZon SL mass spectrometer with an ESI or ACPI mass selective detector. 1H and 13C NMR spectra were recorded using TMS as the internal standard in CDCl3 or DMSO with a Bruker BioSpin GmbH spectrometer at 400 MHz and 100 MHz, respectively. All chemicals were purchased from commercial sources unless otherwise specified. All oligonucleotides used in this work were synthesized and purified by Shanghai Sangon Biotechnology Co., Ltd. (Shanghai, China), and their sequences were as follows: dA21 (single-stranded DNA): AAAAAAAAAAAAAAAAAAAAA; ds26 (duplex DNA): CAATCGGATCGAATTCGATCCGATTG; telo21 (G-quadruplex DNA): GGGTTAGGGTTAGGGTTAGGG; RNA (duplex): 16S- and 23S-Ribosomal from E. coli. The fluorescent titration measurements were repeated three times and the averaged values were reported.Synthesis of 1,2,6-trimethylpyridin-1-ium iodide
The reaction conditions were followed the reported literature.26 2,6-Dimethylpyridine (0.22 g, 2.05 mmol) and iodomethane (0.50 g, 3.52 mmol) were added into sulfolane (0.80 ml) at reflux conditions (50 °C) for 4 h. After cooling in an iced bath, the precipitate was filtrated, collected and rinsed with ethyl acetate. The solid was purified by using column chromatography to afford the compound in 95% yields. Mp 190–192 °C; 1H NMR (400 MHz, DMSO): δ 8.30 (t, J = 7.9 Hz, 1H), 7.90 (d, J = 7.9 Hz, 2H), 4.00 (s, 3H), 2.80 (s, 6H).General procedures for the synthesis of ligands 1 and 2
1,2,6-trimethylpyridin-1-ium iodide (0.53 g, 2.2 mmol) was reacted with 2.1 equivalents of p-(dimethylamino)benzaldehyde or p-(methylthio)benzaldehyde in 15 ml n-butanol using 0.3 ml 4-methylpiperidine as the base under reflux conditions for 2 h. After reaction, the mixture was cooled in an iced bath. The precipitate was filtrated and then washed twice with n-butanol. The collected solids were purified by re-crystallization in acetone to afford the pure compounds. The detailed characterization data were shown as follows:FRET assay
The FRET assay was performed as a high-throughput screen following previously published procedures.27 The labeled oligonucleotides F21T: 5′-FAM-d(GGG[TTAGGG]3)-TAMRA-3′ (donor fluorophore FAM is 6-carboxyfluorescein; acceptor fluorophore TAMRA is 6-carboxytetramethylrhodamine; HEG linker is [(–CH2–CH2–O–)6]) used as the FRET probes were diluted from stock to the correct concentration (0.4 μM) in buffer and then annealed by heating to 95 °C for 5 min, followed by cooling to room temperature. Samples were prepared by aliquoting 15 μL of the annealed F21T (at 2 × concentration, 0.4 μM) into LightCycler 96, followed by 15 μL of the ligand solution (at 2 × concentration, 0.2–2.0 μM) and further incubated for 1 h. Fluorescence melting curves were determined with a Roche LightCycler real-time PCR machine with excitation at 470 nm and detection at 530 nm. Fluorescence readings were taken at intervals of 3 °C over the range 37–93 °C, with a constant temperature being maintained for 300 s prior to each reading to ensure a stable value.Molecular docking study
The crystal structure of human telomeric DNA G-quadruplex (4DA3)28 were used as a template to perform docking study on compound 1. The 3D structures of the small molecules were sketched using DS viewer 3.5. Autodock Tools (ver. 1.5.6) was used to convert the structure files to pdbqt format.30 Docking calculation was performed using the AUTODOCK vina program.31 The dimensions of the active site box were chosen to be large enough to encompass the entire G4 structures. An exhaustiveness of 100 was used and other parameters were left as default.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We acknowledge the support received from the National Nature Science Foundation of China (21102021 and 81473082), Nature Science Foundation of Guangdong Province, China (2017A030313078), Science and Technology Program of Guangdong Province (2012B020306007), and the Department of Education Guangdong Province (No. 2016KCXTD005, 2017KSYS010).Notes and references
- A. T. Aron, K. M. Ramos-Torres, J. A. Cotruvo Jr and C. J. Chang, Acc. Chem. Res., 2015, 48(8), 2434–2442 CrossRef PubMed.
- M. C. Heffern, H. M. Park, H. Y. Au-Yeung, G. C. V. Bittner, C. M. Ackerman, A. Stahl and C. J. Chang, Proc. Natl. Acad. Sci., 2016, 113(50), 14219–14224 CrossRef PubMed.
- P. S. Zhang, H. Wang, Y. X. Hong, M. L. Yu, R. G. Zeng, Y. F. Long and J. Chen, Biosens. Bioelectron., 2018, 318–324 CrossRef PubMed.
- J. L. Kolanowski, F. Liu and E. J. New, Chem. Soc. Rev., 2018, 47(1), 195–208 RSC.
- M. J. Lohse, S. Nuber and C. Hoffmann, Pharmacol. Rev., 2012, 64(2), 299–336 CrossRef PubMed.
- R. Y. Tsien, Annu. Rev. Neurosci., 1989, 12, 227–253 CrossRef PubMed.
- T. Terai and T. Nagano, Pflügers Arch., 2013, 465(3), 347–359 CrossRef PubMed.
- M. Maekawa and G. D. Fairn, J. Cell Sci., 2014, 127, 4801–4812 CrossRef PubMed.
- X.-P. He and H. Tian, Chem, 2018, 4, 246–268 Search PubMed.
- X. Su, X. J. Xiao, C. Zhang and M. P. Zhao, Appl. Spectrosc., 2012, 66(11), 1249–1261 CrossRef PubMed.
- D. J. Stephens and V. J. Allan, Science, 2003, 300(5616), 82–86 CrossRef PubMed.
- A. Shivanandan, H. Deschout, M. Scarselli and A. Radenovic, FEBS Lett., 2014, 588, 3595–3602 CrossRef PubMed.
- S. Cox, Dev. Biol., 2015, 401, 175–181 CrossRef PubMed.
- S. Ray, J. R. Widom and N. G. Walter, Chem. Rev., 2018, 118 (8), 4120–4155 CrossRef PubMed.
- A. C. Bhasikuttan and J. Mohanty, Chem. Commun., 2015, 51, 7581–7597 RSC.
- A. Laguerre, K. Hukezalie, P. Winckler, F. Katranji, G. Chanteloup, M. Pirrotta, J.-M. Perrier-Cornet, J. M. Y. Wong and D. Monchaud, J. Am. Chem. Soc., 2015, 137, 8521–8525 CrossRef PubMed.
- Y. V. Suseela, N. Narayanaswamy, S. Pratihar and T. Govindaraju, Chem. Soc. Rev., 2018, 47, 1098–1131 RSC.
- M.-Q. Wang, Y. Wu, Z.-Y. Wang, Q.-Y. Chen, F.-Y. Xiao and Y.-C. Jiang, Dyes Pigm., 2017, 145, 1–6 CrossRef.
- B. R. Vummidi, J. Alzeer and N. W. Luedtke, ChemBioChem, 2013, 14(5), 540–558 CrossRef PubMed.
- L. Y. Zhang, J. Cheng, K. K. Ghosh, W. J. Chung, J. Yoo, W. Xu, W. Zhao, A. T. Phan and Y.-T. Chang, Sci. Rep., 2014, 4, 3776 CrossRef PubMed.
- A. Shivalingam, M. A. Izquierdo, A. L. Marois, A. Vyšniauskas, K. Suhling, M. K. Kuimova and R. Vilar, Nat. Commun., 2015, 6, 8178 CrossRef PubMed.
- N. Sun, C. Wang, M.-H. Xu, Y.-J. Lu, Y.-Y. Zheng and Y. Yan, Sens. Actuators, B, 2017, 250, 543–551 CrossRef.
- Y.-J. Lu, Q. Deng, J.-Q. Hou, D.-P. Hu, Z.-Y. Wang and K. Zhang, ACS Chem. Biol., 2016, 11, 1019–1029 CrossRef PubMed.
- C. H. Wong, M. Hendrix, E. S. Priestley and W. A. Greenberg, Chem. Biol., 1998, 5, 397–406 CrossRef PubMed.
- J. A. Means and J. V. Hines, Bioorg. Med. Chem. Lett., 2005, 15, 2169–2172 CrossRef PubMed.
- Y.-J. Lu, D.-P. Hu, Q. Deng, Z.-Y. Wang, B.-H. Huang, Y.-X. Fang, K. Zhang and W.-L. Wong, Analyst, 2015, 140, 5998–6004 RSC.
- X. Xie, A. Renvoise, A. Granzhan and M. P. Teulade-Fichou, New J. Chem., 2015, 39, 5931–5935 RSC.
- M. Micco, G. W. Collie, A. G. Dale, S. A. Ohnmacht, I. Pazitna, M. Gunaratnam, A. P. Reszka and S. Neidle, J. Med. Chem., 2013, 56, 2959–2974 CrossRef PubMed.
- S. Alcaro, A. Artese, J. N. Iley, S. Missailidis, F. Ortuso, L. Parrotta, R. Pasceri, F. Paduano, C. Sissi, F. Trapasso and M. G. Vigorita, ChemMedChem, 2010, 5, 575–583 CrossRef PubMed.
- G. Morris and R. Huey, J. Comput. Chem., 2009, 30, 2785–2791 CrossRef PubMed.
- S. Forli, R. Huey, M. E. Pique, M. F. Sanner, D. S. Goodsell and A. J. Olson, Nat. Protoc., 2016, 11, 905–9012 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra03833c |
| ‡ The authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2018 |