Open Access Article
Open Access ArticleA novel polysaccharide from Rhizoma panacis japonica exerts anti-inflammatory effects via STAT3 signal pathway
Qiu Li b,
Qun Lia,
Zhihui Haod,
Xucai Zheng*c and
Wei He*a
b,
Qun Lia,
Zhihui Haod,
Xucai Zheng*c and
Wei He*a
aDepartment of Immunology, School of Basic Medical Sciences, Anhui Medical University, Hefei, China. E-mail: weihe@ahmu.edu.cn
bState Key Laboratory of Quality Research in Chinese Medicine, Institute of Chinese Medical Sciences, University of Macau, Macau SAR, China
cDepartment of Head, Neck and Breast Surgery, Anhui Provincial Cancer Hospital, West Branch of Anhui Provincial Hospital, The First of University of Science and Technology of China, Hefei, China. E-mail: ahszlyyzxc@163.com
dAgricultural Bio-Pharmaceutical Laboratory, Qingdao Agricultural University, Qingdao, China. E-mail: abplab@126.com
First published on 23rd July 2018
Abstract
We report a novel water-soluble polysaccharide from Rhizoma panacis japonica, namely, PJ-1 with anti-inflammatory activity. We first obtained a polysaccharide from this herb by eluting with water. After fully characterizing PJ-1 with a series of chromatographic technologies, we evaluated its biological activity by stimulating Raw 264.7 cells to express inflammatory cytokines and found that PJ-1 effectively enhanced the production of anti-inflammatory factors such as IL-10. We further identified its anti-inflammatory effect in a mouse model of sepsis induced by a lipopolysaccharide. The treatment of PJ-1 evidently suppressed the secretion of major inflammatory cytokines and increased the survival rate of mice. Particularly, we verified the STAT3 signal pathway responsible for the anti-inflammatory activity. Thus, PJ-1, which has remarkable anti-inflammatory effect and mechanism of action, may be a promising candidate as an anti-inflammatory agent.
1 Introduction
Polysaccharides derived from natural herbs are commonly considered immunomodulatory, and they exert immunoregulatory effects in vitro and in vivo.1–4 Diverse functions of natural polysaccharides such as promoting macrophage phagocytosis,5 enhancing lymphocyte functions6 and boosting anti-tumor effects7 have been demonstrated. Among them, the interaction with receptors expressed by macrophages is a key molecular basis, which can evidently affect the functions of macrophages.8,9 For instance, dectin-1,10 mannose receptors (MR),11 and macrophage galactose-type lectin (MGL)12 specifically interplay with polysaccharides composed of mannose, glucose, and galactose, which triggers downstream signals and induces dynamic changes in cellular action. Inspired by these interactions, numerous researches have been carried out for developing polysaccharide-based macrophage-affinity therapeutic tools aimed at a series of therapeutic applications.9 As such, the screening of polysaccharides from herbs and inspection of their influences on macrophages are critical for the discovery of therapeutic agents in clinic.Rhizoma panacis japonica, a common herbal medicine, has attracted much attention due to its various pharmacological activities recorded by traditional oriental medicine. It can ‘stop bleeding’, ‘spread stasis’, ‘reduce swelling’, ‘relieve pain’, ‘stop expectorant’ and ‘relieve cough’ in clinic. Furthermore, its versatile biological activities including anti-tumor, anti-inflammatory, anti-viral, immunoregulatory and anti-fatigue activities are certified by modern pharmacology.13–17 Studies indicate that its polysaccharides may possess diverse biological activities such as anti-tumor effect17 and anti-oxidant activity.18,19 However, it remains unclear whether the polysaccharides can interact with macrophages to modulate their action. Studies on polysaccharides against macrophages illustrate clear immunoregulatory behavior and mechanism of action.
Therefore, in this study, we isolated a water-soluble polysaccharide via ion-exchange chromatography and gel column. Then, we evaluated the macrophage-modulating activities by stimulating secretion of the inflammatory cytokine. Furthermore, we observed the relative signal pathways and verified the biological activities of the polysaccharide in vitro and in vivo. Our findings may provide a promising prospect for developing new therapeutic agents based on this active polysaccharide.
2 Experimental
2.1 Materials and reagents
Rhizoma panacis japonica was purchased from China Pharmaceutical Corporation-Canton (Guangzhou, China), and third-party verification was performed at Anhui Medical University. DEAE-52Cellulose and Sephadex G-100 were purchased from GE Healthcare (Fairfield, Connecticut, USA). Standard monosaccharides including D-glucose, L-arabinose, D-mannose D-galactose, L-rhamnose, D-glucuronic acid and D-galacturonic acid were purchased from Sigma-Aldrich (USA). Phenol, sulfuric acid, TFA and PMP were purchased from Shanghai Aladdin Biochemical Polytron Technologies Inc. (Shanghai, China). Acetonitrile for HPLC was purchased from Merck KGaA (Darmstadt, Germany). All other chemicals and reagents used in this study were of analytical grade from Xi'an Chemical Co. (Xi'an, China). The water used was distilled water.Murine Raw 264.7 cells, purchased from the American Type Culture Collection (ATCC), were cultured in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (FBS), 100 U mL−1 penicillin and 100 mg mL−1 streptomycin in 5% CO2 at 37 °C. All cell-culture reagents were purchased from Gibco (USA). ELISA kits used for measurements of murine TNF-α, IL-1β and IL-10 were purchased from 4A Biotech Co., Ltd (Beijing, China).
2.2 Preparation of PJ-1 polysaccharide
We obtained PJ-1 polysaccharide according to our in-house protocol.20 Air-dried Rhizoma panacis japonica (500 g) was first extracted thrice with 95% ethanol for 2 h every time to remove fat-soluble components and organic molecules. Then, the dried residue was boiled with distilled water thrice for 3 h every time. The supernatant was collected, and a four-fold volume of ethanol was added and precipitated overnight at 4 °C. The precipitate was collected by centrifugation (4000 rpm, 20 min) at room temperature and then re-dissolved in distilled water. Proteins were removed by the Savage method (chloroform/n-butanol, 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v). The collected solution was subsequently dialyzed in a dialysis bag (MW 8000–14000 Da) and lyophilized to obtain crude polysaccharide. The crude polysaccharide was then applied to a DEAE-52 column (5 cm × 30 cm) eluted with distilled water (1000 mL) at a flow rate of 25 mL h−1. The fraction was collected and detected by the phenol-sulfuric acid method.21 This water fraction was further purified using a sephadex G-100 column (2 cm × 100 cm) eluted with distilled water at a flow rate of 12 mL h−1 to obtain water-soluble polysaccharide PJ-1.
1, v/v). The collected solution was subsequently dialyzed in a dialysis bag (MW 8000–14000 Da) and lyophilized to obtain crude polysaccharide. The crude polysaccharide was then applied to a DEAE-52 column (5 cm × 30 cm) eluted with distilled water (1000 mL) at a flow rate of 25 mL h−1. The fraction was collected and detected by the phenol-sulfuric acid method.21 This water fraction was further purified using a sephadex G-100 column (2 cm × 100 cm) eluted with distilled water at a flow rate of 12 mL h−1 to obtain water-soluble polysaccharide PJ-1.
2.3 Measurement of molecular weight
Each polysaccharide (2.0 mg) of PJ-1 was dissolved in water for homogeneity and molecular weight analysis using HPLC technology.22 The chromatographic conditions were as follows: Shimadzu LC20 with an RID-20 detector (Shimadzu, Japan) and with TSK-GEL G4000PWXL (TOSHO, Japan, 300 mm × 7.8 mm, i.d.) at 35 °C; 0.1 N NaNO3 and 0.06% NaN3 aqueous solutions were used as the mobile phase, and the flow rate was 0.6 mL min−1. Polyethyleneglycol standards with different molecular weights were used for the calibration curve.2.4 IR spectrum assay
FT-IR spectrum was obtained using a PerkinElmer Spectrum 100 FT-IR Spectrometer with KBr pellets (PerkinElmer, Waltham, USA). PJ-1 (3 mg) was mixed with KBr powder, and the IR spectrum was recorded in the frequency range of 4000–450 cm−1.2.5 Monosaccharide composition of PJ-1
The monosaccharide composition of PJ-1 was analyzed using the PMP pre-column derivatization method.20 Briefly, PJ-1 (4 mg) was completely hydrolyzed using 2 mL 2 M TFA in a sealed tube for 4 h at 110 °C. Then, the products were evaporated and washed with methanol twice and dried with nitrogen to remove TFA. Hydrolysate was dissolved in water and mixed with the same volume of sodium hydroxide. Then, 100 μL of 0.5 M methanolic solution of PMP was added to the mixed solution and incubated for 120 min at 70 °C. Afterwards, the solution was neutralized with 0.5 M hydrochloric acid, and chloroform (1 mL) was added; the solution was vortexed fully to generate layers. The organic layer was then removed. Finally, the solution was passed through a 0.45 μm membrane and analyzed by HPLC with a UV detector. It was eluted with a mixture of ammonium acetate buffer (pH 5.5) and acetonitrile (82![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 18 v/v for 0–25 min and 78
18 v/v for 0–25 min and 78![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 22 v/v for 25–50 min), and the results were obtained at 245 nm. A standard solution containing seven types of monosaccharides (mannose, rhamnose, glucuronic acid, galacturonic acid, glucose, galactose and arabinose) underwent the same protocol as described above.23
22 v/v for 25–50 min), and the results were obtained at 245 nm. A standard solution containing seven types of monosaccharides (mannose, rhamnose, glucuronic acid, galacturonic acid, glucose, galactose and arabinose) underwent the same protocol as described above.23
2.6 Cell viability
RAW 264.7 cells were cultured in 96-well plates (104 per well) in DMEM medium with 10% (v/v) FBS at 37 °C. After incubation for 24 h, the cells were treated with PJ-1 polysaccharide (50–200 μg mL−1) for 24, 48 and 72 h. Cell viability was measured by CCK-8 assay at different time points.2.7 Quantitative real-time PCR (Q-PCR)
Total RNA was obtained from the cells using TRIzol® Reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. The quantity and quality of total RNA were certified using Eppendorf Biophotometer Plus (Eppendorf AG, Hamburg, Germany). Q-PCR was carried out using LightCycler FastStar DNA Master SYBR Green I (Roche Diagnostics) following the protocol from the manufacturer. The fold change of each gene was normalized to that of β-actin.24 The primer sequences in this experiment are listed below (F: forward; R: reverse):iNOS: 5′-CCAAGCCCTCACCTACTTCC-3′ (F) 5′-CTCTGAGGGCTGACACAAGG-3′(R);
Arginase-1: 5′-CCAGAAGAATGGAAGAGTCAGTGT-3′ (F) 5′-GCAGATATGCAGGGAGTCACC-3′ (R);
β-Actin: 5′-TGCTGTCCCTGTATGCCTCT-3′ (F) and 5′-TTTGATGTCACGCACGATTT-3′ (R).
2.8 Western blotting
For Western blotting assay, cellular proteins were obtained by lysing cells in RIPA buffer containing 1 mM phenylmethylsulfonyl fluoride (PMSF). Equal amounts of samples were subjected to SDS-PAGE electrophoresis and then transferred to PVDF membranes (Bio-Rad, USA). After blocking with skimmed milk or bovine serum albumin (5%) for 1.5 h at room temperature with gentle shaking, the membranes were blotted with primary antibodies (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000) overnight at 4 °C and then exposed to secondary antibody for 2 hours at room temperature. The bands were visualized with SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific, USA). The corresponding antibodies including phosphor-STAT3, Total-STAT3 and GAPDH were obtained from cell signaling technology (Danvers, MA, USA).
1000) overnight at 4 °C and then exposed to secondary antibody for 2 hours at room temperature. The bands were visualized with SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific, USA). The corresponding antibodies including phosphor-STAT3, Total-STAT3 and GAPDH were obtained from cell signaling technology (Danvers, MA, USA).
2.9 The effect of PJ-1 on macrophages
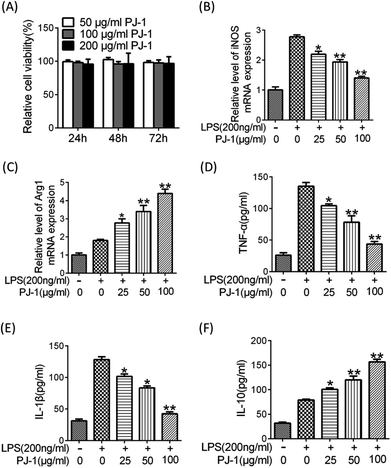
Raw 264.7 cells were cultured and pre-treated with PJ-1 at different concentrations (25–100 μg mL−1) for 1 h; then, LPS (200 ng mL−1) was added to different groups and incubated for 20 h at 37 °C. Afterwards, the concentrations of TNF-α, IL-1β, and IL-10 in cell-culture supernatant were measured using the corresponding ELISA kits (4A Biotech Co., Ltd, Beijing, China). The operation was strictly performed according to the instructions, and the plates were read at 450 nm using a microplate reader (Thermo Scientific). Furthermore, total RNA was extracted, and the level of RNA including Arg1 and iNOS genes was measured according to the above-mentioned procedure. Afterwards, under the same conditions, the cells were treated in the same manner with 10 μM Stattic (Sigma-Aldrich, St. Louis, Missouri, USA) as the inhibitor of the STAT3 signal pathway.2.10 The evaluation of PJ-1 in vivo
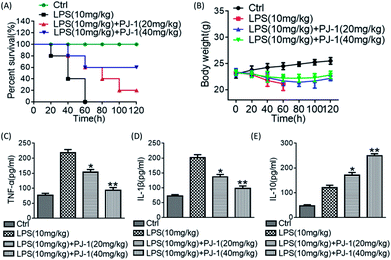
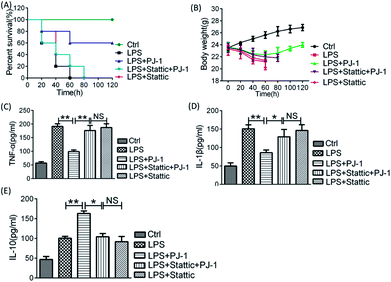
Male C57BL/6 mice (18–24 g) were supplied by the Experimental Animal Centre of Anhui Medical University (Hefei, China) and randomly divided into three groups (n = 10 per group) including control group (saline), LPS group (10 mg kg−1), and PJ-1 (20–40 mg kg−1) plus LPS group (10 mg kg−1). Particularly, PJ-1 polysaccharide was first executed by intraperitoneal injection and after 1 h, LPS injection was administered. After eight hours, the mice with different treatments were sacrificed, and blood serum was collected. The levels of TNF-α, IL-1β and IL-10 were quantified with commercial ELISA kits. The amounts and weights of surviving mice were counted at different time points. Meanwhile, the same operation was performed after inhibiting STAT3 activity with Stattic (1 mg kg−1), and the dose of PJ-1 was 20 mg kg−1 per mouse. Animal care and all experiments were performed according to the ‘Guidelines for the Care and Use of Laboratory Animals’ of Anhui Medical University.2.11 Statistics
Data are presented as mean ± standard error of the mean (SEM). Statistical analyses were performed using one-way ANOVA (GraphPad Prism 6), with * and ** standing for P < 0.05 and <0.01, respectively.3 Results and discussion
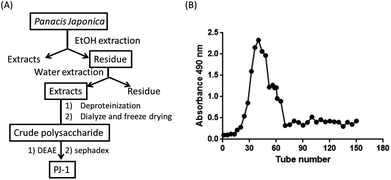
3.1 Purification of crude polysaccharide
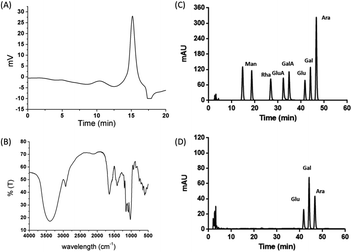
Crude polysaccharide was isolated, followed by removal of proteins using the Sevag method. A water-soluble fraction was obtained with a DEAE-52 column and elution using distilled water. The fraction was further purified using a Sephadex G-100 column with distilled water as the eluent. Finally, the polysaccharide, namely, PJ-1 was obtained (Fig. 1A and B).3.2 Chemical analysis of PJ-1
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.4
2.4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.6.
1.6.3.3 Biological activity of PJ-1 in vitro
3.4 Anti-inflammatory activity of PJ-1 in vivo
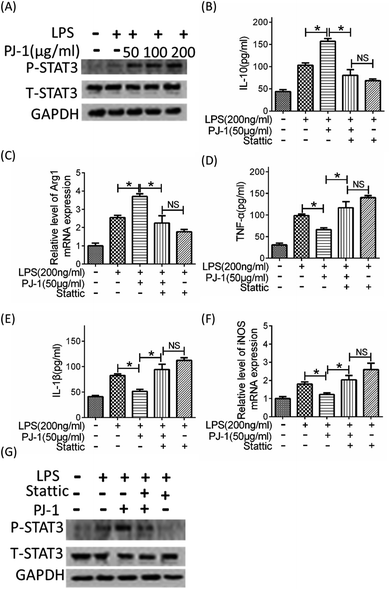
3.5 The anti-inflammatory effect of PJ-1 in vivo depends on STAT3 activation
We pre-treated LPS-challenged mice with Stattic to further investigate the role of STAT3 activation on the effect of PJ-1 on sepsis in vivo. Co-treatment with Stattic and PJ-1 could eliminate the therapeutic effect of PJ-1 including recovery of losses in body weight and enhancement of survival rates (Fig. 6A and B). Meanwhile, co-treatment with Stattic and PJ-1 eliminated the anti-inflammatory effect of PJ-1 on mice with sepsis (Fig. 6C–E). These results suggested that the anti-inflammatory effect of PJ-1 in vivo may be mediated by STAT3 activation.4 Conclusions
In this study, we obtained a water-soluble polysaccharide, PJ-1, which can modulate the function of macrophages. We characterized its molecular weight and monosaccharide composition and further validated the anti-inflammatory effect of this polysaccharide in vitro and in vivo. The results obtained indicated that PJ-1 could effectively suppress the secretion of major inflammatory cytokines. Moreover, we certified its mechanism of action with multiple experiments and confirmed that it exerted anti-inflammatory effect via the STAT3 signal pathway. Taken together, PJ-1, examined by a series of evaluations, has remarkable anti-inflammatory effect in vitro and in vivo. Therefore, we assume that PJ-1 has a promising prospect of becoming an outstanding candidate as an anti-inflammatory agent.Conflicts of interest
There are no conflicts to declare.Abbreviations
| PJ | Rhizoma panacis japonica |
| TFA | Trifluoroacetic acid |
| PMP | 1-Phenyl-3-methyl-5-pyrazolone |
| UV | Ultraviolet |
| IR | Infrared |
Acknowledgements
This work was supported by Scientific Research of BSKY (XJ201726) from Anhui Medical University, Anhui Institute of Translational Medicine Research Fund (2017zhyx10), and funding from the Qingdao Science and Technology Program (15-10-3-15-(41)-zch). All animal procedures were performed in accordance with the Guidelines for Care and Use of Laboratory Animals of Anhui Medical University and experiments were approved by the Animal Ethics Committee of Anhui Medical University.Notes and references
- Q. Li, Y. Feng, W. He, L. Wang, R. Wang, L. Dong and C. Wang, Carbohydr. Polym., 2017, 169, 304–314 CrossRef PubMed.
- Y.-Q. Du, Y. Liu and J.-H. Wang, Int. J. Biol. Macromol., 2015, 72, 1272–1276 CrossRef PubMed.
- Q. Fang, J.-F. Wang, X.-Q. Zha, S.-H. Cui, L. Cao and J.-P. Luo, Carbohydr. Polym., 2015, 134, 66–73 CrossRef PubMed.
- F. Kallel, D. Driss, F. Bouaziz, L. Belghith, S. Zouari-Ellouzi, A. Haddar, S. E. Chaabouni and R. Ghorbel, RSC Adv., 2015, 5, 6728–6741 RSC.
- X. Chu, X.-J. Liu, J.-M. Qiu, X.-L. Zeng, H.-R. Bao and J. Shu, Environ. Toxicol. Pharmacol., 2016, 48, 76–84 CrossRef PubMed.
- Q.-D. Xiang, Q. Yu, H. Wang, M.-M. Zhao, S.-Y. Liu, S.-P. Nie and M.-Y. Xie, J. Agric. Food Chem., 2017, 65, 5306–5315 CrossRef PubMed.
- A. Zong, Y. Liu, Y. Zhang, X. Song, Y. Shi, H. Cao, C. Liu, Y. Cheng, W. Jiang and F. Du, Carbohydr. Polym., 2015, 129, 50–54 CrossRef PubMed.
- S. S. Ferreira, C. P. Passos, P. Madureira, M. Vilanova and M. A. Coimbra, Carbohydr. Polym., 2015, 132, 378–396 CrossRef PubMed.
- S.-Z. Xie, R. Hao, X.-Q. Zha, L.-H. Pan, J. Liu and J.-P. Luo, Carbohydr. Polym., 2016, 146, 292–300 CrossRef PubMed.
- C. Deng, H. Fu, J. Shang, J. Chen and X. Xu, Int. J. Biol. Macromol., 2018, 109, 369–374 CrossRef PubMed.
- W.-J. Li, X.-F. Tang, X.-X. Shuai, C.-J. Jiang, X. Liu, L.-F. Wang, Y.-F. Yao, S.-P. Nie and M.-Y. Xie, J. Agric. Food Chem., 2017, 65, 348–357 CrossRef PubMed.
- I. G. Zizzari, P. Martufi, F. Battisti, H. Rahimi, S. Caponnetto, F. Bellati, M. Nuti, A. Rughetti and C. Napoletano, PLoS One, 2015, 10, e0132617 CrossRef PubMed.
- D. Xukun, M. Xue, C. Jian, Y. Wenzhong, L. You, Z. Biqun, J. Jie, C. Shuang and W. Fei, J. South-Cent. Univ. Natl., Nat. Sci. Ed., 2013, 1, 011 Search PubMed.
- T. Wang, Y. Dai, Y. Dun, C. Zhang, J. Wan, L. Deng, Z. Zhou, C. Liu and D. Yuan, Immunopharmacol. Immunotoxicol., 2014, 36, 404–411 CrossRef PubMed.
- X. Yang, R. Wang, S. Zhang, W. Zhu, J. Tang, J. Liu, P. Chen, D. Zhang, W. Ye and Y. Zheng, Carbohydr. Polym., 2014, 101, 386–391 CrossRef PubMed.
- B. Klimova and K. Kuca, Curr. Alzheimer Res., 2017, 14, 680–685 CrossRef PubMed.
- C. Chen, W. Wu, X. Xu, L. Zhang, Y. Liu and K. Wang, Carbohydr. Polym., 2014, 105, 308–316 CrossRef PubMed.
- X. Yang, R. Wang, S. Zhang, W. Zhu, J. Tang, J. Liu, P. Chen, D. Zhang, W. Ye and Y. Zheng, Carbohydr. Polym., 2014, 101, 386–391 CrossRef PubMed.
- H. Zhang, H. Zhao, X. Zhou, X. Yang, S. Shen, J. Wang, Z. Wang and L. Geng, RSC Adv., 2016, 6, 110706–110721 RSC.
- Q. Li, G. Guo, F. Meng, H. H. Wang, Y. Niu, Q. Zhang, J. Zhang, Y. Wang, L. Dong and C. Wang, ACS Macro Lett., 2016, 5, 617–621 CrossRef.
- M. Dubios, K. A. Gilles, J. K. Hamilton, P. A. Rebers and F. Smith, Anal. Chem., 1956, 28, 350–356 CrossRef.
- K. L. Cheong, D. T. Wu, J. Zhao and S. P. Li, J. Chromatogr. A, 2015, 1400, 98–106 CrossRef PubMed.
- F.-C. Meng, C. Yuan, X.-J. Huang, W.-J. Wang, L.-G. Lin, X.-T. Zhang, H.-Y. Jiao and Q.-W. Zhang, Phytochem. Lett., 2016, 15, 108–112 CrossRef.
- W. He, Y. Zhu, R. Mu, J. Xu, X. Zhang, C. Wang, Q. Li, Z. Huang, J. Zhang and Y. Pan, Biochem. Pharmacol., 2017, 145, 132–146 CrossRef PubMed.
- H. F. Zhang and R. Lai, Cancers, 2014, 6, 1408–1440 CrossRef PubMed.
- D. E. Levy and C. K. Lee, J. Clin. Invest., 2002, 109, 1143–1148 CrossRef PubMed.
- C. Qin, W. H. Fan, Q. Liu, K. Shang, M. Murugan, L. J. Wu, W. Wang and D. S. Tian, Stroke, 2017, 48, 3336–3346 CrossRef PubMed.
- H. S. Chung, B. S. Lee and J. Y. Ma, J. Evidence-Based Complementary Altern. Med., 2017, 2017, 4218468 Search PubMed.
- H. Zhao, Q. Shang, Z. Pan, Y. Bai, Z. Li, H. Zhang, Q. Zhang, C. Guo, L. Zhang and Q. Wang, Diabetes, 2018, 67, 235–247 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2018 |