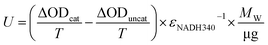Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Humanization and directed evolution of the selenium-containing scFv phage abzyme
Yan Xua,
Pengju Lia,
Jiaojiao Niea,
Qi Zhao b,
Shanshan Guana,
Ziyu Kuaia,
Yongbo Qiaoa,
Xiaoyu Jianga,
Ying Lia,
Wei Lia,
Yuhua Shia,
Wei Kongac and
Yaming Shan
b,
Shanshan Guana,
Ziyu Kuaia,
Yongbo Qiaoa,
Xiaoyu Jianga,
Ying Lia,
Wei Lia,
Yuhua Shia,
Wei Kongac and
Yaming Shan *ac
*ac
aNational Engineering Laboratory for AIDS Vaccine, School of Life Sciences, Jilin University, Changchun, Jilin, China. E-mail: shanym@jlu.edu.cn; Fax: +86 431 85167751; Tel: +86 431 89228979
bFaculty of Health Sciences, University of Macau, Macau, China
cKey Laboratory for Molecular Enzymology and Engineering, The Ministry of Education, School of Life Sciences, Jilin University, Changchun, Jilin, China
First published on 10th May 2018
Abstract
According to the binding site structure and the catalytic mechanism of the native glutathione peroxidase (GPX), three glutathione derivatives, GSH-S-DNP butyl ester (hapten Be), GSH-S-DNP hexyl ester (hapten He) and GSH-S-DNP hexamethylene ester (hapten Hme) were synthesized. By a four-round panning with a human synthetic scFv phage library against three haptens, the enrichment of the scFv phage particles with specific binding activity could be determined. Three phage particles were selected binding to each glutathione derivative, respectively. After a two-step chemical mutation to convert the serine residues of the scFv phage particles into selenocysteine residues, GPX activity could be observed and determined upto 3000 U μmol−1 in the selenium-containing scFv phage abzyme which was isolated by affinity capture against the hapten Be. Also the scFv phage abzymes elicited by different antigens displayed different catalytic activities. After a directed evolution by DNA shuffling to improve the affinity to the hapten Be, a secondary library with GPX activity was created in which the catalytic activity of the selenium-containing scFv phage abzyme could be increased 17%. This study might be helpful for new haptens or antigens design to optimize the abzymes with high binding activities and might also provide a novel scheme for GPX mimic candidates for drug development.
Introduction
Glutathione peroxidase (GPX) is a key peroxidase for protecting biomolecules from oxidative damage by eliminating hydrogen peroxide and lipid peroxide from the body and further blocking the injury of reactive oxygen free radicals in biological organisms. Selenocysteine is located in the active center of the GPX,1,2 which could also reflect the selenium level of the body. Furthermore, selenium, a structural part of a large group of selenoproteins is necessary for proper functioning of the body.3 In order to mimic the function of the GPX, numerous studies have been carried out. For example, when hemolyzates from erythrocytes of selenium-deficient rats were incubated in vitro in the presence of ascorbate or H2O2, added glutathione failed to protect the hemoglobin from oxidative damage. This occurred because the erythrocytes were practically devoid of glutathione-peroxidase activity.4 Extensively purified preparations of glutathione peroxidase contained a large part of the 75Se of erythrocytes labeled in vivo. Many of the nutritional effects of selenium can be explained by its role in glutathione peroxidase. Extensive research has been carried out to design and synthesize small organoselenium compounds as functional mimics of GPX. While the catalytic mechanism of the native enzyme itself is poorly understood, the synthetic mimics follow different catalytic pathways depending upon the structures and reactivities of various intermediates formed in the catalytic cycle.5 Therefore, GPX plays an important role in antioxidant drugs.In theory, humanized GPX should be one of the most effective anti-oxidation drugs.6 While as the limited source, the poor stability, and the large molecular weight, the application of the native GPX is limited.7 Furthermore the native GPX purification is not easy, which could not be conductive to the large-scale preparation.8
As a mimic enzyme, the antibody enzyme(abzyme) might possess the catalytic properties, the substrate specificities and the stereo-selectivities as the natural enzymes did. Also, it could be a shortcut that only a few weeks might be taken for an antibody affinity maturation as the incomparable diversity of the abzyme, while millions of years might be taken for a native enzyme evolution. Due to strong binding affinity and high specificity in antibodies against antigens, antibody-catalyzed reactions proceed in a highly regio- and stereo-selective manner, thus the development of new synthetic methods for organic synthesis provides a clear advantage. However, due to the nature of the binding of monoclonal antibody binding to its substrate, the substrate specificity of antibody catalytic reaction is very limited. Therefore, the practical application of catalytic antibodies in organic synthesis requires to expand its substrate specificity.9
The abzyme (especially the single-chain abzyme) may have a better stability, lower molecular weight, easier separation and purification process, which could be an ideal medicinal protein. In order to study the catalytic mechanism and develop novel pharmaceutical lead compounds, we have explored several mimic GPXs producing high activity according to the murine abzymes.10 Although some abzymes have been reported with efficiencies similar to that of corresponding natural enzymes,11 in general it has been difficult to obtain highly active catalysts using the standard approach. As an exogenous protein, the murine GPX enzyme could not be applied in the clinic. It is imperative to develop the humanized GPX abzyme.
The preparation of an abzyme depends on the hapten design. The structure of an ideal hapten candidate should be as close as possible to that of the transition-state analog in the actual reaction, at the same time, as far as possible be different from that of the product and the substrate, which could minimize the inhibition to introduce the active residues involved in the reaction. In vitro, there're usually a few approaches utilized to develop humanized hapten-specific antibodies: humanization of a murine antibody, biopanning of a human phage antibody library or a yeast display human scFv library, or eliciting antibodies with transgenic animals.
DNA shuffling is a new technique for the directed molecular evolution.12 By altering the original nucleotide sequence of a single gene or gene family, a brand new gene could be created and the expression product might display a new function. In recent years, the DNA shuffling technique has been widely applied with broad prospects and great application value in the area of biological engineering.
Phage display is a biotechnology technique that a DNA sequence of foreign proteins or peptides is inserted into the appropriate place of the structural gene of the phage coat protein. The foreign genes could be expressed with the coat protein. At the same time, the foreign proteins could be displayed onto the phage surface with the reassembly of the phage.13,14 Phage display technology has been widely applied in the study of protein–protein, protein–peptide, and protein–DNA interactions. Many remarkable advantages have been achieved by phage display technology, such as high flux elutriation, screening of simulant epitope, and easy purification of recombinant phage purification steps.15,16 Phage display technology has a great influence on life sciences, and allows users to select extremely rare ligands from the variant library based on user-defined selection criteria.17 A large number of studies have been used to find cell-based library selection approaches that many evidences have used to discover chemokine analogs, not only receptor antagonists but also variants with unusual effects on receptor signaling and trafficking.18 Thus it becomes a valuable tool for understanding the biology of chemokines.19
Phage antibody library technology is also a new genetic engineering antibody technique developed in recent years.20 In this technique, the gene fragments of the cloned antibodies in vitro were inserted into the phage vector, transfected into the engineering bacteria for the expression, and then the specific monoclonal phage antibody could be obtained by screening against the antigen or haptens. There are unique advantages for application of phage antibody library technology in the diagnosis and treatment of HIV and other viral infections and cancers.21,22
In this study, according to the binding site structure and the catalytic mechanism of the native glutathione peroxidase, three glutathione derivatives were synthesized as the haptens representing the maximum level of the basic glutathione structure and the hydrophobicity. After a four-round panning, phage particles recognized specifically by the haptens could be enriched from a human synthetic scFv phage library by regulating the concentration of the haptens and controlling the elution strength. It was expected that the binding sites of the antibodies recognized by the haptens could show strong affinities to the glutathione. Furthermore, due to the different modification of the synthesized haptens, the binding site structures of the antibodies might also be different. Then a two-step chemical mutation was employed to convert the serine residues of the scFv phage particles into the selenocysteine residues to prepare selenium-containing scFv phage abzymes for producing GPX activity.15 In order to further improve the activity of the GPX activity abzymes, DNA shuffling was utilized in the sequences of the enriched phage particles with potential GPX activities mimicking the somatic hypermutation occurred in vivo.
Comparing the GPX activities of these abzymes, the study might be helpful for new haptens or antigens design to optimize abzymes with high binding activities and might also provide a novel scheme to develop potential GPX mimic candidates for drug development.
Materials and methods
Human synthetic scFv phage library, E. coli TG1, Helper phage M13-K07, and Tg1 (pHEN2) were presented by MRC of Cambridge University.Synthesis of GSH-S-DNP
1.02 GSH was dissolved in 10 mL 1 mol L−1 NaOH solution with ice bath, 0.67 g dinitro chlorobenzene was dissolved in 10 mL ethanol. And then they are added to the GSH solution slowly, the mixture kept in 0 °C 10 minutes, mixed with dilute HCl to pH 4. The yellow crystal precipitation was observed, suction filtered, recrystallized with hot water to 70 °C drying. The final powder is GSH-S-DNP.Synthesis of GSH-S-DNP hexyl ester (hapten He) and GSH-S-DNP hexamethylene ester(hapten Hme)
0.21 g GSH-S-DNP was dissolved in 0.2 mL SOCl2, mixed with 2 mL n-hexylester for 3 hours, then additional 2 mL n-hexylester was added and kept at 90 °C for 1 hour. 20 mL ether and 10 mL petroleum ester were added. The yellow precipitation was separated out, frozen for 12 hours, filtered, washed and dried. The final powder is hapten He. The preparation method of hapten Hme is the same as that of hapten He.Separation and purification of haptens
As the synthetic derivatives containing the diester and the single ester, we separate them with Gf254 fluorescence silica gel. The liquid phase is chloroform/methanol (v/v = 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). The first peak at 280 nm was collected and dried. The products are the powder of diesters. The purities were above 95%.
1). The first peak at 280 nm was collected and dried. The products are the powder of diesters. The purities were above 95%.
Enzyme-Linked Immunosorbent Assay (ELISA) and data analysis
The binding activities of the enriched phage particles to the haptens were determined by ELISA. A 96-well plate (Jet Biofil, Guangzhou, China) coated with different haptens was employed for ELISA test. The binding activities of the enriched phage particles to M13-K07 and to the supernatant of TG1 containing pHEN2 were set as the positive control and negative control seperately. The blank is the screening buffer (PBS, 2×TY, TG1, 3% BSA). Optical density measurements were obtained at 450 nm.The catalytic activities (U) of the enriched scFv phage particles were calculated as the following equation:
MW/μg is the micromole of the sample. The micromole of the phage particles is used for calculating the catalytic activity, which is named phage catalytic activity; the unit is U μmol−1 phage. The catalytic activity of the pHEN phage particles absent of foreign protein expression is set as the background after converting the serine residues into the selenocysteine residues. The equation for the GPX catalytic activity of the selenium-containing scFv phage abzyme is:
KD value was obtained by the relationship between the number of the target protein displayed on the phage particle and the titer of the phage particles; the conversion equation is:
| KD phage-protein = KD phage/number of the displayed phage-protein |
After determined by ELISA, KD value of the phage particles was plotted by Graphpad Prism software version 5.0 (Graphpad Software Inc., San Diego, CA, USA).
DNA shuffling
The DNA mixture of the enriched phage particles with the GPX activity was used as the recombinant PCR template for the directed evolution.Results and discussion
The hapten design
According to the ability to elicit the murine scFv abzyme that is equivalent to the natural enzyme activity,10 GSH-S-DNP butyl ester was chosen as the hapten hoping to play the same role in the process of acquiring the humanized GPX mimic. And the hydrophobicities of the three haptens are increasing in order to explore the relationship between the hydrophobicity and the enzyme activity. The three haptens were designed to keep the molecular skeleton characteristics according to the GSH basic structure (Fig. 1). The structures might cause the induced antibodies recognizing the GSH binding sites, but avoid affecting the releasing of the substrate from the enzymatic reaction and further the reaction rate due to the strong specifically binding activity with the GSH. Therefore, it could be possible to obtain a structure of the antibody active site close to the native microenvironment of the GPX catalytic group.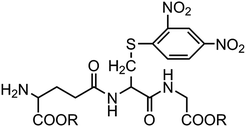 | ||
| Fig. 1 Structures of the haptens used for generating humanized abzymes with potential GPX activity. R: –CH2(CH2)2CH3 (hapten Be); –CH2(CH)4CH3 (hapten He); –C6H6 (hapten Hme). | ||
The enrichment of the specific scFv phage particles by affinity screening against haptens with human synthetic scFv phage library
Three haptens were screened against the human synthetic scFv phage library separately by biopanning.23 50 μg hapten dissolved in 1 mL coating buffer was coated on the Petri dish (Jet Biofil, Guangzhou, China) for the first round screening. And then the following quantity of the hapten was decreased as the increasing round of the screening. The ratio of the hapten amount to the phage particle number was about 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 for the 1st screening round, and was reduced gradually to 1
1 for the 1st screening round, and was reduced gradually to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 in the following screening rounds. The phage particles with specific binding activity could have been enriched according to the increased recovery ratios as shown in Tables 1–3.
50 in the following screening rounds. The phage particles with specific binding activity could have been enriched according to the increased recovery ratios as shown in Tables 1–3.
| Round | Hapten concentration (μg mL−1) | Input phage titer (cfu) | Output phage titer (cfu) | Recovery ratio |
|---|---|---|---|---|
| 1 | 50 | 1013 | 1.6 × 109 | 1.6 × 10−4 |
| 2 | 20 | 1013 | 2.4 × 106 | 2.4 × 10−7 |
| 3 | 10 | 1012 | 3.1 × 106 | 3.1 × 10−6 |
| 4 | 1 | 1012 | 1.5 × 106 | 1.5 × 10−6 |
| Round | Hapten concentration (μg mL−1) | Input phage titer (cfu) | Output phage titer (cfu) | Recovery ratio |
|---|---|---|---|---|
| 1 | 50 | 1013 | 7.7 × 109 | 7.7 × 10−4 |
| 2 | 20 | 1012 | 2.2 × 106 | 2.2 × 10−6 |
| 3 | 10 | 1012 | 6.8 × 106 | 6.8 × 10−6 |
| 4 | 1 | 1012 | 2.5 × 106 | 2.5 × 10−6 |
| Round | Hapten concentration (μg mL−1) | Input phage titer (cfu) | Output phage titer (cfu) | Recovery ratio |
|---|---|---|---|---|
| 1 | 50 | 1013 | 3.6 × 108 | 3.6 × 10−5 |
| 2 | 20 | 1012 | 6.5 × 106 | 6.5 × 10−6 |
| 3 | 10 | 1012 | 6.6 × 106 | 6.6 × 10−6 |
| 4 | 1 | 1012 | 1.2 × 106 | 1.2 × 10−6 |
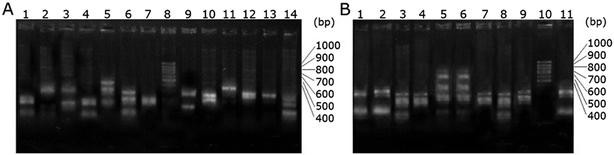
More than a dozen phage particles were picked and amplified from the enriched scFv phage libraries that were specifically binding to the respective haptens. The single strand DNAs were extracted and digested with HaeIII. The electrophoretogram of the digested enriched scFv-Be phage DNA is shown in Fig. 2B. The ratio of the screened particles with the similar digested DNA fingerprint patterns has increased obviously compared with that of the original scFv phage library (Fig. 2A). This further suggested the enrichment of the specific scFv phage particles.
Catalytic activity determination of the phage particles, Be8, He6 and Hme1 with specific binding activities
The phage particles, which were separately picked from each screened scFv phage library after a four-round panning, were recovered by a helper phage M13-K07. The specific binding activities of the recovered phage particles to the haptens were determined by ELISA (Tables 4–6).| Category | OD450 nm | Phage clone number | OD450 nm |
|---|---|---|---|
| Positive control | 1.621 | 1, 3, 4, 5, 6, 9, 10 | 0.1–0.5 |
| Negative control | 0.103 | 7, 11, 12, 13, 15 | 0.5–1.0 |
| Blank | ≤0.085 | 2, 8, 14 | 1.0–1.6 |
| Category | OD450 nm | Phage clone number | OD450 nm |
|---|---|---|---|
| Positive control | 1.841 | 2, 3, 4, 8, 11, 13 | 0.1–0.5 |
| Negative control | 0.160 | 1, 9, 10, 14, 15 | 0.5–1.0 |
| Blank | ≤0.061 | 6, 5, 7, 12 | 1.0–1.6 |
| Category | OD450 nm | Phage clone number | OD450 nm |
|---|---|---|---|
| Positive control | 1.506 | 3, 4, 8, 10, 11, 13, 15 | 0.1–0.5 |
| Negative control | 0.072 | 6, 2, 5, 7, 9, 12, 14 | 0.5–1.0 |
| Blank | ≤0.004 | 1 | 1.0–1.6 |
Phage particles, Be8, He6 and Hme1, which were the three most positive clones specifically binding to the glutathione derivatives, were purified and sequenced. After determined by ELISA, KDs of the phage particles were calculated by regression of Prism: KD Be8 = 14 μM; KD He6 = 12 μM; KD Hme1 = 12 μM. With the introduction of the selenocysteine residue into the scFv phage particles by the two-step chemical mutation, the scFv phage abzymes isolated by the affinity capture to the hapten Be could present the catalytic activities from 700 U μmol−1 to 3000 U μmol−1 similar to that of the native GPX (5780 U μmol−1) extracted from rabbit liver.24–26
Directed evolution of the scFv sequences
A secondary library with GPX activity was created by DNA shuffling. The phage particle affinities to the haptens were improved after the directed evolution13 as shown in Table 7. Phage particle B9 was selected for the activity analysis and the KD value was calculated after the chemical mutation. The correlative ELISA profile of the phage particle B9 is shown in Fig. 3 and the KD value is 13 μM by the regression calculation of Prism. The catalytic activity of the selenium-containing scFv phage abzyme could reach 3560 U μmol−1 according to the measurement of the phage particle activity.27,28| Category | OD450 nm | Phage clone number | OD450 nm |
|---|---|---|---|
| Positive control | 1.537 | 1, 3, 4, 5, 6, 8, 10, 11 | 0.1–0.5 |
| Negative control | 0.101 | 2, 7, 12, 13, 14, 15 | 0.5–1.0 |
| Blank | ≤0.061 | 9 | 1.0–1.4 |
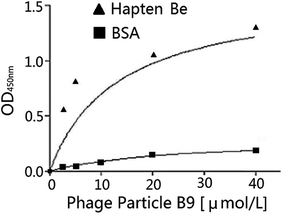 | ||
| Fig. 3 Binding activity of the scFv phage particle Be9 after the directed evolution to the immobilized hapten Be7. | ||
Conclusion
According to the binding site structure and the catalytic mechanism of native glutathione peroxidase, three glutathione derivatives were synthesized in this study. A four-round screening was performed using the MRC human synthetic scFv phage library by affinity capture to the respective haptens. The results of the DNA fingerprint analysis and ELISA indicated the enrichment of the phage particles with the specificity to the hapten respectively. After the introduction of the selenocysteine residues into the enriched scFv phage particles, the chemical mutated phage abzymes could give the catalytic activities ranging from 700 U μmol−1 to 3000 U μmol−1. After the directed evolution of these scFv phage sequences by DNA shuffling to improve the affinity to the hapten Be, the catalytic activity of the selenium-containing scFv phage abzymes could be improved to 3560 U μmol−1 similar to that of the native GPX. In short, after bio-panning, a secondary library with GPX activity has been created in which the catalytic activity of the positive phage abzymes could be increased 17% compared with that of either native phage abzymes. The study suggests a novel scheme to develop potential humanized GPX mimic candidates for drug development and might be helpful for new haptens or antigens design to optimize abzymes with high binding and catalytic activities.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The current work was supported by the National Natural Science Foundation of China (Grant no. 31770996), Industrial Technology Research and Development Projects of Jilin Province Development and Reform Commission (Grant no. 2014Y081), the Science and Technology Development Fund of Macau (FDCT/131/2016/A3), and Science and Technology Enterprise Technology Innovation Fund by Science and Technology Department of Jiangsu Province (Grant no. BC2015065).References
- F. Li, J. Liu and S. Rozovsky, Glutathione peroxidase's reaction intermediate selenenic acid is stabilized by the protein microenvironment, Free Radical Biol. Med., 2014, 76, 127–135 CrossRef CAS PubMed.
- L. Dery, P. S. Reddy, S. Dery, R. Mousa, O. Ktorza, A. Talhami and N. Metanis, Accessing human selenoproteins through chemical protein synthesis, Chem. Sci., 2017, 8(3), 1922–1926 RSC.
- M. Wolonciej, E. Milewska and W. Roszkowska-Jakimiec, Trace elements as an activator of antioxidant enzymes, Postepy Hig. Med. Dosw., 2016, 70(0), 1483–1498 CrossRef PubMed.
- J. T. Rotruck, A. L. Pope, H. E. Ganther, A. B. Swanson, D. G. Hafeman and W. G. Hoekstra, Selenium: biochemical role as a component of glutathione peroxidase, Science, 1973, 179(4073), 588–590 CAS.
- D. Bhowmick and G. Mugesh, Insights into the catalytic mechanism of synthetic glutathione peroxidase mimetics, Org. Biomol. Chem., 2015, 13(41), 10262–10272 CAS.
- L. Alpsoy, H. Sahin and S. Karaman, Anti-oxidative and anti-genotoxic effects of methanolic extract of Mentha pulegium on human lymphocyte culture, Toxicol. Ind. Health, 2011, 27, 647–654 CrossRef CAS PubMed.
- M. Jaskulak, A. Rorat, A. Grobelak and M. Kacprzak, Antioxidative enzymes and expression of rbcL gene as tools to monitor heavy metal-related stress in plants, J. Environ. Manage., 2018, 218, 71–78 CrossRef CAS PubMed.
- H. Lemmerhirt, S. Behnisch, A. Bodtke, C. H. Lillig, L. Pazderova, J. Kasparkova, V. Brabec and P. J. Bednarski, Effects of cytotoxic cis- and trans-diamine monochlorido platinum(II) complexes on selenium-dependent redox enzymes and DNA, J. Inorg. Biochem., 2018, 178, 94–105 CrossRef CAS PubMed.
- N. Miyamoto, M. Yoshimura, Y. Okubo, K. Suzuki-Nagata, T. Tsumuraya, N. Ito and I. Fujii, Structural basis of the broad substrate tolerance of the antibody 7B9-catalyzed hydrolysis of p-nitrobenzyl esters, Bioorg. Med. Chem., 2018, 26, 1412–1417 CrossRef CAS PubMed.
- R. Huo, J. Wei, J. Xu, S. Lv, Q. Zheng, F. Yan, J. Su, J. Fan, J. Li, Y. Duan, Y. Yu, F. Jin, W. Sun, Y. Shi, D. Cong, W. Li, G. Yan and G. Luo, Human catalytic antibody Se-scFv-B3 with high glutathione peroxidase activity, J. Mol. Recognit., 2008, 21(5), 324–329 CrossRef PubMed.
- K. Wang, P. Gong, L. Liu, S. Gong, J. Liu, J. Shen and G. Luo, Effect of 2-TeCD on the expression of adhesion molecules in human umbilical vein endothelial cells under the stimulation of tumor necrosis factor-alpha, Int. Immunopharmacol., 2009, 9(9), 1087–1091 CrossRef CAS PubMed.
- H. Li, X. Chu, B. Peng and X. X. Peng, DNA shuffling approach for recombinant polyvalent OmpAs against V. alginolyticus and E. tarda infections, Fish Shellfish Immunol., 2016, 58, 508–513 CrossRef CAS PubMed.
- E. M. Driggers and P. G. Schultz, Catalytic antibodies, Adv. Protein Chem., 1996, 49, 261–287 CrossRef CAS PubMed.
- W. P. Stemmer, Rapid evolution of a protein in vitro by DNA shuffling, Nature, 1994, 370(6488), 389–391 CrossRef CAS PubMed.
- R. Pasqualini and E. Ruoslahti, Organ targeting in vivo using phage display peptide libraries, Nature, 1996, 380(6572), 364–366 CrossRef CAS PubMed.
- J. Wang, J. Song, S. Zhou, Y. Fu, J. A. Bailey and C. Shen, Screening and identification of RhD antigen mimic epitopes from a phage display random peptide library for the serodiagnosis of haemolytic disease of the foetus and newborn, Blood Transfus., 2018, 16, 1–7 Search PubMed.
- A. Mechaly, R. Alcalay, T. Noy-Porat, E. Epstein, Y. Gal and O. Mazor, Novel Phage Display-Derived Anti-Abrin Antibodies Confer Post-Exposure Protection against Abrin Intoxication, Toxins, 2018, 10(2), 80 CrossRef PubMed.
- V. Flores, O. Sepulveda-Robles, A. Cazares, L. Kameyama and G. Guarneros, Comparative genomic analysis of Pseudomonas aeruginosa phage PaMx25 reveals a novel siphovirus group related to phages infecting hosts of different taxonomic classes, Arch. Virol., 2017, 162(8), 2345–2355 CrossRef CAS PubMed.
- M. Gorelik, S. Orlicky, M. A. Sartori, X. Tang, E. Marcon, I. Kurinov, J. F. Greenblatt, M. Tyers, J. Moffat, F. Sicheri and S. S. Sidhu, Inhibition of SCF ubiquitin ligases by engineered ubiquitin variants that target the Cul1 binding site on the Skp1-F-box interface, Proc. Natl. Acad. Sci. U. S. A., 2016, 113(13), 3527–3532 CrossRef CAS PubMed.
- K. Dorgham, F. Cerini, H. Gaertner, A. Melotti, I. Rossitto-Borlat, G. Gorochov and O. Hartley, Generating Chemokine Analogs with Enhanced Pharmacological Properties Using Phage Display, Methods Enzymol., 2016, 570, 47–72 Search PubMed.
- J. D. Marks, H. R. Hoogenboom, T. P. Bonnert, J. McCafferty, A. D. Griffiths and G. Winter, By-passing immunization: Human antibodies from V-gene libraries displayed on phage, J. Mol. Biol., 1991, 222(3), 581–597 CrossRef CAS PubMed.
- L. M. Watanabe, F. Barbosa Junior, A. A. Jordao and A. M. Navarro, Influence of HIV infection and the use of antiretroviral therapy on selenium and selenomethionine concentrations and antioxidant protection, Nutrition, 2016, 32(11–12), 1238–1242 CrossRef CAS PubMed.
- B. C. Saunders, Glutathione. Its reaction with alkali and some N and S derivatives, Biochem. J., 1934, 28(6), 1977–1981 CrossRef CAS PubMed.
- J. K. Scott and G. P. Smith, Searching for peptide ligands with an epitope library, Science, 1990, 249(4967), 386–390 CAS.
- M. M. Mader and P. A. Bartlett, Binding Energy and Catalysis: The Implications for Transition-State Analogs and Catalytic Antibodies, Chem. Rev., 1997, 97(5), 1281–1302 CrossRef CAS PubMed.
- X. Ren, S. Gao, D. You, H. Huang, Z. Liu, Y. Mu, J. Liu, Y. Zhang, G. Yan, G. Luo, T. Yang and J. Shen, Cloning and expression of a single-chain catalytic antibody that acts as a glutathione peroxidase mimic with high catalytic efficiency, Biochem. J., 2001, 359, 369–374 CrossRef CAS PubMed.
- R. Pasqualini and E. Ruoslahti, Tissue targeting with phage peptide libraries, Mol. Psychiatry, 1996, 1(6), 423 CAS.
- H. Zhao, L. Giver, Z. Shao, J. A. Affholter and F. H. Arnold, Molecular evolution by staggered extension process (StEP) in vitro recombination, Nat. Biotechnol., 1998, 16(3), 258–261 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2018 |