Open Access Article
Open Access ArticleTumor suppressor miR-449a inhibits the development of gastric cancer via down-regulation of SGPL1
Qian Chena,
Zhen Yanga,
Gaofeng Pana,
Hongjian Dinga,
Daowen Jianga,
Jianfang Huang*b and
Weiyan Liu *a
*a
aDepartment of General Surgery, Minhang Hospital, Fudan University, No. 170, Xinsong Road, Minhang District, Shanghai 201199, China. E-mail: weiyanliu28@163.com
bDepartment of Infection Diseases, The Fifth People's Hospital Affiliated to Fudan University, Shanghai 201199, China. E-mail: drjianfanghuan@163.com
First published on 19th July 2018
Abstract
MicroRNAs (miRNAs) are small noncoding RNAs that are known to participate in the regulation of many physiological and pathological processes, which can indirectly influence the development of malignant behaviors. Numerous studies have demonstrated that miR-449a plays important roles in human carcinogenesis. However, its precise functional and regulatory roles remain unclear. In this study, we mainly explored the functional role of miR-449a in gastric cancer (GC). The expression levels of miR-449a in 98 cases of GC tissues and cell lines were determined by qRT-PCR. The possible mechanisms of miR-449a in GC cells were explored by fluorescence reporter assay. miR-449a expression was significantly lower in GC tissues compared to matched para-carcinoma tissues and was associated with tumor differentiation. Furthermore, in vitro knockdown of miR-449a by siRNA significantly inhibited MKN-28 cell proliferation, migration and invasion as well as tumorigenesis via inducing G0/G1 arrest of GC cells. In addition, we identified SGPL1 as a target of miR-449a and demonstrated that miR-449a regulated SGPL1 expression via binding its 3′-UTR region. The experiments indicated that miR-449a functions as a novel tumor suppressor in GC and its anti-oncogenic activity may involve its inhibition of the target gene SGPL1. These findings suggested that miR-449a may be a promising candidate for the development of antitumor drugs targeting GC.
Background
Gastric cancer (GC) is the fifth most common cancer in the world and also the third leading cause of cancer-related death.1,2 However, the pathogenesis of GC is not completely elucidated. At present, it has been considered that the occurrence of GC is closely related to Helicobacter pylori infection, gene mutation, diet, and other risk factors.3–5 Despite much progress in the treatment of GC, the overall 5 year survival rate of patients with GC is still less than 30%.6 Heterogeneity and a lack of effective therapeutic targets are the two major obstacles to the precise treatment of GC.7,8 Therefore, it is urgent to seek new diagnostic and prognostic markers for the targeted therapy of GC.MiRNAs are a class of non-coding small molecule RNA with evolutionary conservation, which can regulate gene expression by transcriptional suppression or degradation of target mRNA transcript.9–11 Each miRNA has the potential to regulate a number of target genes, and then modulate the expression of thousands of mRNAs.12 MiRNAs are involved in a variety of biological processes, including tumorigenesis. Some miRNAs are tumor suppressors, while others are carcinogens that promote the development and progression of tumors.13
Previous studies have suggested that miR-449a is a tumor suppressor in various cancers, such as hepatocellular carcinoma and lung cancer.14,15 It has been reported that aberrant expression of specific miRNAs in GC is closely associated with the progression and prognosis of GC.16 Yao et al. have reported that the expression of miR-449a was significantly downregulated in GC cell lines and GC tissues.17 Other studies have also indicated a tumor-suppressive function of miR449a in GC (Hu et al., 2014; Li et al., 2014; Li et al., 2015). However, the function and mechanism of miR449a involved in GC progression are not fully understood. In the current study, the expression of miR-449a in gastric carcinoma and the relationship between miR-449a expression and clinicopathological parameters of GC patients were analyzed. The effects of miR-449a overexpression or knockdown on the growth, migration, invasion and tumorigenesis of GC cells were also investigated. Moreover, we identified SGPL1 as a potential target gene of miR-449a. Our data suggest that miR-449 may suppress GC tumorigenesis and metastasis via by directly targeting SGPL1 gene.
Methods
Cell lines
Human GC cell lines SNU-1, SNU-638, BGC-823, SGC-7901, KATOIII and normal gastric epithelial cell line (GES-1) were purchased from American Type Culture Collection (ATCC). MKN-28, HGC-27 and MGC-803 cell lines were donated by Shanghai Cancer Institute and Renji Hospital Affiliated to Shanghai Jiaotong University School of Medicine. The cell lines were cultured in RPMI1640 medium (HyClone) containing 10% fetal bovine serum (FBS, GIBCO), 2 mM L-glutamine, 100U ml−1 penicillin and 100 μg ml−1 streptomycin. The cells were incubated in an atmosphere of 5% CO2 at 37 °C.Sample collection
The primary cancer tissues and the paired adjacent non-cancer tissues were obtained from 86 GC patients who received radical gastrectomy in Minhang Hospital between January 2012 and July 2013. Tumor tissues were immediately snap-frozen in liquid nitrogen after excision during the surgical procedure and then stored at −80 °C for cryopreservation. Clinicopathological data of patients were reviewed and TNM staging was conducted based on the standard of the American Joint Cancer. All samples were confirmed by pathological examination. This work was approved by the Clinical Research and Ethics Committee at Minhang Hospital and all patients were informed consent.RNA extraction and qRT-PCR
According to the instructions, total RNA was extracted from the cell lines and tissue samples using TRIzol reagent (Life Technologies, Inc., CA, USA). The quantification of miRNA was analyzed using TaqMan miRNA Reverse Transcription Kit and TaqMan MicroRNA Assay Kit (GenePharma, Shanghai, China). The relative expression of miR-449a in tumor tissues and paired non-tumor tissues was calculated using the 2−ΔΔCt method. The primer sequences of SGPL1 were 5′-CACAAAGCAGACGGGCAGACCC-3′ (forward) and 5′-GCTTATCGTCGGCTTGTTGCT-3′ (reverse). The primer sequences of c-MYC were 5′-TCCTTCTACTCCCCCATCACGA-3′ (forward) and 5′-TTCCT CAGTTCTTCCTCTTTCCTCA-3′ (reverse). The primer sequences of GAPDH were 5′-GGACCTGGCCGACCTTCTAG-3′ (forward) and 5′-GTAGCAGCGATGCCCCTTGA-3′ (reverse). The expression of miR-449a was normalized to GAPDH mRNA. PCR for each sample was repeated three times.Transient transfection
Hsa-miR-449a mimics (miR-449a), negative controls (miR-NC), hsa-miR-449a inhibitor (anti-miR-449a) and inhibitor negative control oligonucleotides (anti-miR-NC) were purchased from GenePharma Company. Cells at the logarithmic growth phase were digested with trypsin and inoculated into 6-well plates. When cells reached 50% confluence on the second day, cells were transfected with oligonucleotides (with a final concentration of 100 nM) using Lipofectamine™ 2000 reagent (Invitrogen) according to the manufacturer's instructions. After transfection for 48 h, cells were harvested for further experiments.ELISA
After transfection for 48 h, ELISA kit (Cusabio, Wuhan, China) was used to detect SGPL1 level. The culture supernatant was collected. SGPL1 level was detected in both culture supernatant and tumor tissues by ELISA according to the manufacturer's instructions.Cell proliferation assay
MKN-28 cell proliferation was determined using the Cell Counting Kit-8 assay kit (Promega, Madison, Wisconsin, USA) and water soluble tetrazolium salt colorimetric method (MTT), according to the manufacturer's instructions. Cells were seeded at a density of 2 × 103 per well in 96-well plates 24 h after transfection. Cell proliferation assay was performed at each 24 h for 5 days. The absorbance was read at 490 nm using a GloMax Microplate Absorbance Reader (Promega). All the experiments were repeated three times.Cell migration and invasion assay
The capability of cell migration was examined by transwell migration assay and invasion assay using Transwell® Microcell and 8.0 μm polycarbonate microporous membrane (Corning, NY, USA). MKN-28 cells were seeded and cultured in serum-free medium for 24 h after transfection with miR-449a mimic, inhibitor or the proper negative control (100 nM) for 24 h. Then 3 × 104 cells in 200 μl serum-free medium were added into the upper chamber and 500 μl medium containing 10% FBS was added to the lower chamber as chemokine, and cultured for 36 h at 37 °C. A cotton swab was used to gently scrape the non-metastatic cells from the surface of the polycarbonate microporous membrane. Cells transferred to the bottom of the membrane were stained with stain reagent for 20 min, and observed under the microscope. According to the instructions, cell invasion assay was carried out using 6.5 mm Transwell® Microcell and 8.0 μm polycarbonate microporous membrane (Corning), and extracellular matrix was simulated by Matrigel gel (BD Biosciences, USA). MKN-28 cells in 200 μl serum-free medium were added into the upper chamber coated with Matrigel gel, and 500 μl medium containing 10% FBS was added to the lower chamber as chemokine, and incubated for 48 h at 37 °C. A cotton swab was used to gently scrape the non-invasive cells. The invasive cells were collected, fixed, stained with 0.04% crystal violet, and counted as described above.Construction of 3′UTR of SGPL1 reporter gene system
RT-PCR was used to synthesize 600 bp wild type SGPL1-3 ′UTR containing the putative miR-449a binding site (wild-type) and mutant SGPL1-3′ UTR sequence (mutant). The sequence of Luc-SGPL1-3′UTR (wild-type): 5′-ggactagtagcaagaatcaagattctgctaact-3′ (forward) and 5′-cgacgcgttgggtgtatttcttggtcttatttc-3′ (reverse). Mutant SGPL1-3′UTR contained mutations of three discontinuous nucleotides in the miR-449a target site. The sequence of Luc-SGPL1-mut 3′UTR (mutant): 5′-gaggttgctgagatgcagttcagtaatggtgaatgtggaa-3′ (forward) and 5′-ttccacattcaccattactgaactggaacacagcaacctc-3′ (reverse). After digestion with SpeI and MluI, wild type and mutant SGPL1-3′UTR fragment was cloned into SpeI and MluI loci of pMIR luciferase vector (Applied Biosystems), named pMIR/SGPL1 and pMIR/SGPL1/mut, respectively. The accuracy of insertion was verified by sequencing. MKN-28 cells were seeded in 24-well plates and cotransfected with pMIR/SGPL1 or pMIR/SGPL1/mut, 2 ng pRL-TK vector, and 60 pmol miR-449a mimics, inhibitor or control sequence (Promega, Madison, Wisconsin, USA) using Lipofectamine™ 2000 (Invitrogen). Cells were collected 48 h after transfection. The activity of firefly luciferase and Renilla luciferase was measured using the Dual-Luciferase Reporter Assay System (Promega) following the manufacturer's instructions. The firefly luciferase activity was normalized to Renilla luciferase activity.Immunohistochemistry
4 μm paraffin sections were baked at 68 °C for 1 h. After dewaxing and rehydration, the sections were heated in the boiling 10 mmol l−1 citrate buffer (pH6.0) for 10 min for antigen recovery. Methanol containing 0.3% H2O2 was added to remove endogenous peroxidase. The sections were blocked with PBS containing 2% bovine serum albumin (BSA) for 30 min, and then incubated with the mouse-anti-human SGPL1 monoclonal antibody (Abcam, Hongkong, concentration 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 150), followed by peroxidase/DAB and rabbit-anti-mouse secondary antibody (Dako, Denmark). DakoREAL™EnVision™ detection system was used to detect immune complexes.
150), followed by peroxidase/DAB and rabbit-anti-mouse secondary antibody (Dako, Denmark). DakoREAL™EnVision™ detection system was used to detect immune complexes.
Stable retroviral transfection
The genomic region containing miR-449a primary transcript was cloned to the EcoRI-XhoI site of the modified pMSCV-GW-RfA-PGK-EGFP retroviral vector. The negative control vector didn't contain the insertion sequence. The plasmid mixture of 10 μg miR-449a, 10 μg gag/pol vector and 10 μg VSVG vector were co-transfected into 293T cells using FuGENE6 transfection reagent (Roche, Basel, Switzerland). Plasmid/medium/FuGENE6 mixture was dropped to 293T cells. Cell supernatants were collected twice a day for 2 days. MKN-28 cells were infected after 8 μg ml−1 polyamines was added to 6-well plate. The virus supernatant was removed after 24 h. Positive cells were screened according to the expression of GFP using fluorescence activated cell sorting (FACS), named as MKN-28-miR-449a or MKN-28-vector, and the expression of miR-449a was verified by qRT-PCR.Tumor xenograft model
100 μl cells (2 × 106) transfected with MKN-28-miR-449a or MKN-28-vector were subcutaneously inoculated to 4 week-old male nude mice (Institute of zoology, Chinese Academy of Sciences, Shanghai, China). The size of the tumor was calculated as (long × width2)/2.13 After 5 weeks, nude mice in both groups were sacrificed. The proliferation index (PI) was estimated by the mean percentage of cell nuclear antigen Ki-67 positive cells in every 1000 cells.Statistic analysis
Statistical significance was assessed by Student's t-test using SPSS 22.0 statistical software (SPSS, Inc., Chicago, IL, USA). Data were presented as the mean ± S.D. from at least three independent experiments. A P value less than 0.05 was considered statistically significant.Results
Overexpression of miR-449a inhibits the proliferation of GC cells
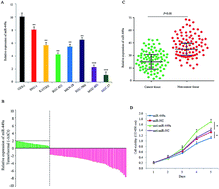
We first employed qRT-PCR to detect miR-449a levels in 7 GC cells, which are from the primary tumor, malignant ascites or metastatic sites. The result showed that miR-449a expression was downregulated in all 7 GC cell lines as compared with the immortalized normal human gastric epithelial cells GES-1 (Fig. 1A). Furthermore, we examined the expression of miR-449a in 86 GC tissues and paired non-cancerous tissues by qRT-PCR (Fig. 1B). Compared with paired non-cancerous tissues, the expression levels of miR-449a were lower in tumor tissues (Fig. 1C). These results indicated that miR-449a was down-regulated in GC. We subsequently analyzed the association between miR-449a expression and clinicopathological features of GC patients. Clinicopathological features of 86 GC patients are shown in Table 1. The tumor differentiation level in the group of low miR-449a expression was significantly decreased (P = 0.028), but no significant correlation was found between miR-449a expression and age, gender, tumor location, local invasion depth, lymph node metastasis, and TNM stage. In order to explore the role of miR-449a in the occurrence and progression of GC, we transfected miR-449a mimics (miR-449a group) or miR-449a inhibitor (anti-miR-449a group) into MKN-28 cells, respectively. Compared with miR-NC transfection, miR-449a overexpression inhibited the growth of MKN-28 cells (P < 0.05), while compared with anti-miR-NC transfection, anti-miR-449a increased the proliferative activity of MKN-28 cells (P < 0.05, Fig. 1D).| Clinicopathologic parameters | miR-449a expression | P value | |
|---|---|---|---|
| High (n = 39) | Low (n = 47) | ||
| Gender | |||
| Male | 23 | 29 | 0.791 |
| Female | 16 | 18 | |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| Age (years) | |||
| <60 | 20 | 24 | 0.942 |
| ≥60 | 19 | 23 | |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| Tumor location | |||
| Distal third | 11 | 11 | 0.310 |
| Middle third | 8 | 18 | |
| Proximal third | 20 | 18 | |
| WHO classification | |||
| Adenocarcinoma | 33 | 39 | 0.206 |
| Signet-ring cell carcinoma | 1 | 7 | |
| Mucinous adenocarcinoma | 5 | 1 | |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| Differentiation | |||
| Well/moderately | 20 | 11 | 0.028 |
| Poorly | 19 | 36 | |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| Borrmann classification | |||
| I/II | 15 | 17 | 0.932 |
| III/IV | 24 | 30 | |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| Local invasion | |||
| T1/T2 | 2 | 10 | 0.095 |
| T3/T4 | 37 | 37 | |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| Lymph node metastasis | |||
| No | 8 | 6 | 0.547 |
| Yes | 31 | 41 | |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| Distant metastasis | |||
| No | 38 | 43 | 0.851 |
| Yes | 1 | 4 | |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| TNM stage | |||
| I/II | 9 | 11 | 0.949 |
| III/IV | 30 | 36 | |
Overexpression of miR-449a inhibits the migration and invasion of GC cells
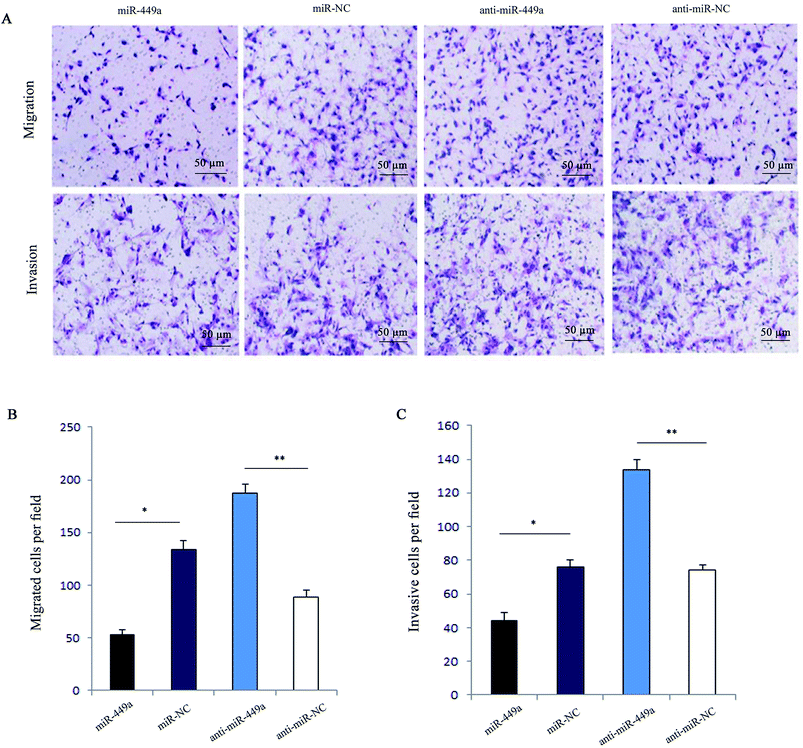
Next, we analyzed the effect of miR-449a on migration and invasion of GC cells. The results of cell migration and invasion assays showed that overexpression of miR-449a inhibited tumor cell migration and invasion (both P < 0.05, Fig. 2A and B). In contrast, miR-449a knockdown significantly promoted cell migration and invasion (both P < 0.01, Fig. 2A and C).miR-449a promotes the apoptosis of GC cells
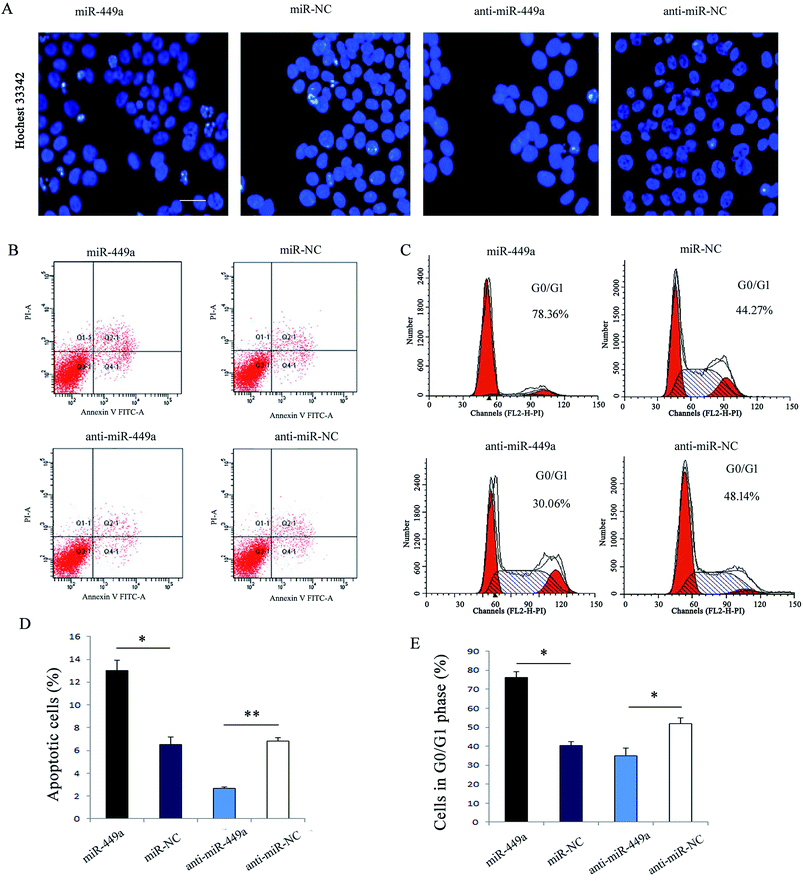
Given the important role of apoptosis involved in cell growth, we first analyzed the effect of miR-449a on GC cell apoptosis by flow cytometry. Hoechst33342 staining which indicates chromatin condensation and nuclear fragmentation showed that there were more cells in miR-449a group showing typical morphological apoptotic characteristics as compared with miR-NC group (Fig. 3A), but there was no significant difference between the anti-miR-449a group and anti-miR-NC group (Fig. 3A). Consistent with this results, flow cytometry results showed that the apoptosis rate in miR-449a group was significantly increased compared with miR-NC group (15.03% ± 1.27% vs. 6.42% ± 0.58%, P < 0.01). In contrast, the apoptosis rate in anti-miR-449a group was significantly decreased compared to the control group (3.91% ± 0.27% vs. 6.46% ± 0.64%; P < 0.05) (Fig. 3B and D).miR-449a induces cell cycle arrest in G0/G1 phase in GC cells
To further understand the mechanism of miR-449a-mediated inhibition in GC proliferation, we performed cell cycle analysis (Fig. 3C and E). The results showed that the cell population in G0/G1 phase was significantly higher in miR-449a group than in miR-NC group (75.35% ± 3.81% vs. 41.39% ± 3.84%, P < 0.01). In contrast, miR-449a knockdown significantly reduced the cell population in G0/G1 phase as compared with anti-miR-NC group (P < 0.05).SGPL1 is a direct target of miR-449a
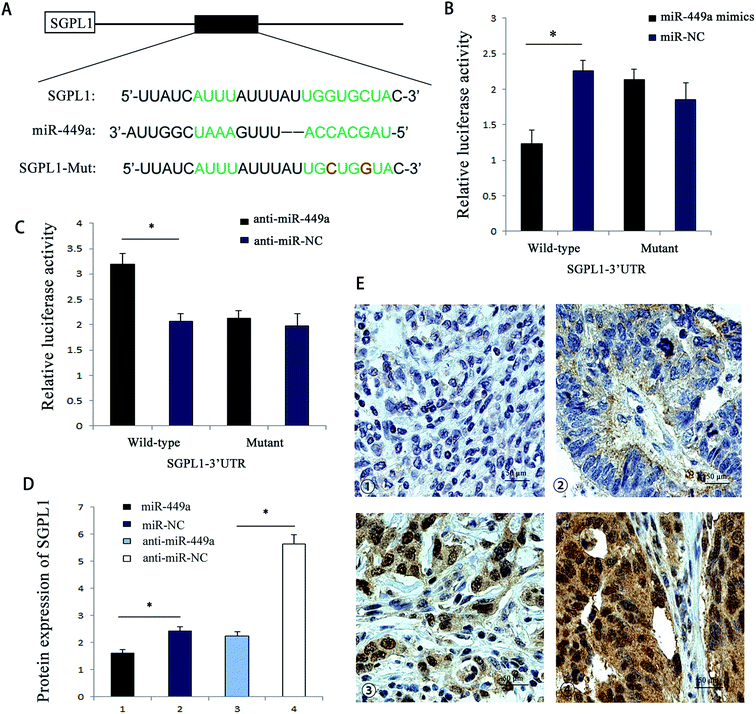
In order to further reveal the mechanism of miR-449a in GC, we employed bioinformatics to search for miR-449a candidate target genes, including TargetScan, miRBaseTarget, and StarBase. Of all the predicted target genes, SGPL1 was one of the target genes of miR-449a. We also found that the 3′UTR region of SGPL1 had a potential miR-449a binding site (Fig. 4A). To determine whether the 3′-UTR of SGPL1 mRNA was the functional target of miR-449a, we performed the luciferase reporter assay. We first determined the luciferase activity of MKN-28 cells co-transfected with miR-449a (or miR-NC) and Luc-SGPL1-3′UTR plasmid or Luc-SGPL1-mut 3′UTR plasmid (containing the putative mutant miR-449a binding site), and the pRL-TK plasmid containing Renilla luciferase gene was used as an internal control. The results showed that miR-449a decreased the luciferase activity of Luc-SGPL1-3′UTR (P < 0.05), but had no significant effect on Luc-SGPL1-mut 3′UTR (P > 0.05) (Fig. 4B). In contrast, anti-miR-449a increased the luciferase activity of Luc-SGPL1-3′UTR (P < 0.05), but had no significant effect on Luc-SGPL1-mut 3′UTR (P > 0.05) (Fig. 4C). These results indicated that SGPL1 is a potential target of miR-449a.To further confirm that miR-449a targets SGPL1, we analyzed the effect of miR-449a on SGPL1 protein expression. ELISA results showed that compared with miR-NC group, SGPL1 protein expression in miR-449a group was significantly inhibited, while SGPL1 protein expression in the anti-miR-449a group was increased as compared with anti-miR-NC group (Fig. 4D). Previous studies have shown that SGPL1 was related to the proliferation and migration of GC cells. We then conducted immunohistochemistry for SGPL1 expression in GC tissues. The results showed that SGPL1 was up-regulated in GC tissues. Representative images of all intensities are depicted in Fig. 4E. To further evaluate the clinical relevance of the above results, we analyzed the correlation between SGPL1 and miR-449a in GC tissues. The results showed that the expression pattern of SGPL1 and miR-449a in GC tissues was opposite (P = 0.032, Table 2). These results suggested that miR-449a could regulate the protein expression of SGPL1 in GC.
| miR-449a expression | P value | ||
|---|---|---|---|
| Low | High | ||
| a P < 0.05 was considered statistically significant. | |||
| SGPL1 (IHC) | |||
| Negative | 12 | 22 | 0.032 |
| Positive | 35 | 17 | |
miR-449a inhibits the formation of tumor in vivo
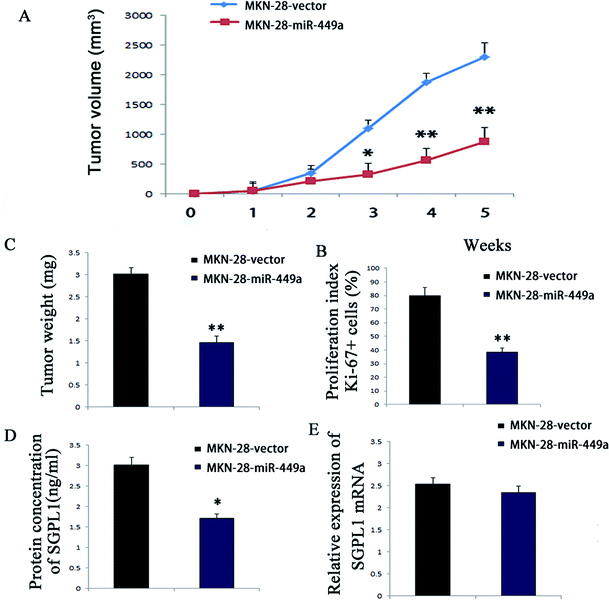
Since miR-449a promoted the proliferation of GC cells in vitro, we further investigated whether miR-449a could affect the formation of GC in vivo. According to the method described above, stable MKN-28-miR-449a and MKN-28-vector cell lines were obtained by retrovirus mediation. Cells transfected with MKN-28-miR-449a or MKN-28-vector were subcutaneously injected to 4 week-old male nude mice and tumor formation was monitored. Compared with the MKN-28-vector group, the tumor growth rate in the MKN-28-miR-449a group was decreased (Fig. 5A). On the 28th day, the average tumor volume of nude mice inoculated with MKN-28-miR-449a cells was significantly lower than that of nude mice inoculated with MKN-28-vector cells (403.57 ± 69.14 mm3 vs. 1085.36 ± 90.53 mm3) (Fig. 5B and C, P < 0.05).To determine whether the tumor growth inhibition induced by MKN-28-miR-449a cells was partly associated with proliferation inhibition, we carried out immunohistochemistry. Ki-67 staining showed that the tumor volume of mice injected with MKN-28-miR-449a cells decreased, which may be partly due to the decline of proliferation induced by miR-449a overexpression. Compared to MKN-28-vector group, the proportion of Ki-67 positive cells in the MKN-28-miR-449a group decreased significantly (38.7% ± 3.6% vs. 83.1% ± 6.2%) (Fig. 5D, P < 0.01). ELISA results showed that compared to the MKN-28-vector group, SGPL1 expression in MKN-28-miR-449a group decreased significantly (1.97 ± 0.25 ng ml−1 vs. 3.61 ± 0.38 ng ml−1, P < 0.05, Fig. 5E). However, there was no significant difference in mRNA expression level of SGPL1 between the two groups (Fig. 5F). Together, these results suggested that miR-449a could inhibit the tumorigenicity of MKN-28 cells in vivo.
Discussion
More and more evidence has proved that miRNAs play an important role in GC as tumor suppressors or carcinogenic factors.18,19 Many miRNAs are supposed to provide potential therapeutic opportunities as they can target plenty of genes in different signaling pathways.20 Our results showed that miR-449a could inhibit tumor growth and metastasis at clinical, cellular and molecular levels, as well as in experimental animal models. This study provided detail experimental data that miR-449a involved in GC by inhibiting SGPL1 expression. Studies have shown that miR-449a plays a tumor suppressive role in liver cancer, prostate cancer, lung cancer, neuroblastoma and endometrial cancer.15,21–24 We found that the expression of miR-449a in GC tissues and GC cell lines was downregulated. Overexpression of miR-449a in MKN-28 GC cells significantly reduced the proliferation and invasion of cells in vivo and in vitro, which suggested the potential therapeutic effect of miR-449a on GC. Our study showed that the opposite results were obtained when anti-miR-449a inhibited the expression of miR-449a. The above results suggest that miR-449a may be a tumor suppressor in human GC. MiRNA results in the fragmentation of target mRNA or suppression of mRNA translation via combining with complementary seed sequence of target mRNA perfectly or imperfectly. We found that SGPL1 was the target gene of miR-449a. The combination of miR-449a and SGPL1 mRNA has incomplete complementarity, which leads to the translation inhibition of SGPL1 mRNA and but had no effect on the mRNA overall stability. Previous study has shown that SGPL1 gene is associated with drug resistance in GC cells.25 In this study, we found that miR-449a is the potential upstream regulator of SGPL1. Overexpression of miR-449a reduced the activity of luciferase reporter gene containing SGPL1 3′UTR region, while the variation of seed area in SGPL1 3′UTR could reverse the regulation role of miR-449a. When we transfected with the anti-miR-449a, the opposite results were observed. Compared with non-tumor tissue, the expression of miR-449a in human GC tissue was significantly decreased, while the expression of SGPL1 protein was significantly increased. Overexpression of miR-449a can reduce the expression of SGPL1 at the protein level, and downregulation of miR-449a could increase the expression of SGPL1 protein. Overexpression of miR-449a is closely related to the proliferation and metastasis of GC, indicating that the function of miR-449a overlaps with SGPL1.The results of this study showed that miR-449a can regulate the generation, proliferation, metastasis and local invasion of GC. Our findings also suggested that miR-449a may be used as a potential target for the treatment of GC.
Ethics approval and consent to participate
All experiments were performed in accordance with the Guidelines of the Care and Use of Laboratory Animals Monitoring Committee of Fudan University, and Experiments were approved by the ethics committee at Fudan University. Informed consents were obtained from human participants of this study.Conflicts of interest
The authors declare that they have no competing interests.References
- X. Ma, M. Huang, Z. Wang, B. Liu, Z. Zhu and C. Li, ZHX1 Inhibits Gastric Cancer Cell Growth through Inducing Cell-Cycle Arrest and Apoptosis, J. Cancer, 2016, 7, 60–68 CrossRef PubMed.
- S. Zhang, P. Chen, Z. Huang, X. Hu, M. Chen, S. Hu, Y. Hu and T. Cai, Sirt7 promotes gastric cancer growth and inhibits apoptosis by epigenetically inhibiting miR-34a, Sci. Rep., 2015, 5, 9787 CrossRef PubMed.
- K. Lee, H. Hwang and K. T. Nam, Immune response and the tumor microenvironment: how they communicate to regulate gastric cancer, Gut Liver, 2014, 8, 131–139 CrossRef PubMed.
- E. Rippa, G. La Monica, R. Allocca, M. F. Romano, M. De Palma and P. Arcari, Overexpression of gastrokine 1 in gastric cancer cells induces Fas-mediated apoptosis, J. Cell. Physiol., 2011, 226, 2571–2578 CrossRef PubMed.
- D. Santini, B. Vincenzi, M. E. Fratto, G. Perrone, R. Lai, V. Catalano, C. Cass, P. A. Ruffini, C. Spoto, P. Muretto, S. Rizzo, A. O. Muda, J. R. Mackey, A. Russo, G. Tonini and F. Graziano, Prognostic role of human equilibrative transporter 1 (hENT1) in patients with resected gastric cancer, J. Cell. Physiol., 2010, 223, 384–388 Search PubMed.
- A. Cimini, L. Brandolini, R. Gentile, L. Cristiano, P. Menghini, A. Fidoamore, A. Antonosante, E. Benedetti, A. Giordano and M. Allegretti, Gastroprotective effects of L-lysine salification of ketoprofen in ethanol-injured gastric mucosa, J. Cell. Physiol., 2015, 230, 813–820 CrossRef PubMed.
- X. Feng and S. Huang, Effect and mechanism of lncRNA HOTAIR on occurrence and development of gastric cancer, J. Cell. Biochem., 2017, 13, 560–567 Search PubMed.
- H. Pan, T. Li, Y. Jiang, C. Pan, Y. Ding, Z. Huang, H. Yu and D. Kong, Overexpression of Circular RNA ciRS-7 Abrogates the Tumor Suppressive Effect of miR-7 on Gastric Cancer via PTEN/PI3K/AKT Signaling Pathway, J. Cell. Biochem., 2018, 119, 440–446 CrossRef PubMed.
- X. Zhang, B. Shen and Y. Cui, Signature microRNAs of nuclear Sm complex associated with breast cancer tumorigenesis, J. Cell. Biochem., 2018, 08, 342–349 Search PubMed.
- F. Rehfeld, D. Maticzka, S. Grosser, P. Knauff, M. Eravci, I. Vida, R. Backofen and F. G. Wulczyn, The RNA-binding protein ARPP21 controls dendritic branching by functionally opposing the miRNA it hosts, Nat. Commun., 2018, 9, 1235 CrossRef PubMed.
- R. Chauhan, D. Datzkiw, S. Varma Shrivastav and A. Shrivastav, In silico identification of microRNAs predicted to regulate N-myristoyltransferase and Methionine Aminopeptidase 2 functions in cancer and infectious diseases, PLoS One, 2018, 13, e0194612 CrossRef PubMed.
- J. Sheu-Gruttadauria and I. J. MacRae, Phase Transitions in the Assembly and Function of Human miRISC, Cell, 2018, 04, 224–231 Search PubMed.
- R. Chhabra, R. Dubey and N. Saini, Cooperative and individualistic functions of the microRNAs in the miR-23a∼27a∼24-2 cluster and its implication in human diseases, Mol. Cancer, 2010, 9, 232 CrossRef PubMed.
- S. P. Chen, B. X. Liu, J. Xu, X. F. Pei, Y. J. Liao, F. Yuan and F. Zheng, MiR-449a suppresses the epithelial-mesenchymal transition and metastasis of hepatocellular carcinoma by multiple targets, BMC Cancer, 2015, 15, 706 CrossRef PubMed.
- J. You, Y. Zhang, Y. Li, N. Fang, B. Liu, L. Zu and Q. Zhou, MiR-449a suppresses cell invasion by inhibiting MAP2K1 in non-small cell lung cancer, Am. J. Cancer Res., 2015, 5, 2730–2744 Search PubMed.
- A. Kimura, K. Ogata, B. Altan, T. Yokobori, M. Ide, E. Mochiki, Y. Toyomasu, N. Kogure, T. Yanoma, M. Suzuki, T. Bai, T. Oyama and H. Kuwano, Nuclear heat shock protein 110 expression is associated with poor prognosis and chemotherapy resistance in gastric cancer, Oncotarget, 2016, 7, 18415–18423 CrossRef PubMed.
- Y. Yao, J. Ma, Y. Xue, P. Wang, Z. Li, Z. Li, Y. Hu, X. Shang and Y. Liu, MiR-449a exerts tumor-suppressive functions in human glioblastoma by targeting Myc-associated zinc-finger protein, Mol. Oncol., 2015, 9, 640–656 CrossRef PubMed.
- S. M. Berger, B. Pesold, S. Reber, K. Schonig, A. J. Berger, I. Weidenfeld, J. Miao, M. R. Berger, O. J. Gruss and D. Bartsch, Quantitative analysis of conditional gene inactivation using rationally designed, tetracycline-controlled miRNAs, Nucleic Acids Res., 2010, 38, e168 CrossRef PubMed.
- M. Guled, L. Lahti, P. M. Lindholm, K. Salmenkivi, I. Bagwan, A. G. Nicholson and S. Knuutila, CDKN2A, NF2, and JUN are dysregulated among other genes by miRNAs in malignant mesothelioma -A miRNA microarray analysis, Genes, Chromosomes Cancer, 2009, 48, 615–623 CrossRef PubMed.
- L. Di Lisio, G. Gomez-Lopez, M. Sanchez-Beato, C. Gomez-Abad, M. E. Rodriguez, R. Villuendas, B. I. Ferreira, A. Carro, D. Rico, M. Mollejo, M. A. Martinez, J. Menarguez, A. Diaz-Alderete, J. Gil, J. C. Cigudosa, D. G. Pisano, M. A. Piris and N. Martinez, Mantle cell lymphoma: transcriptional regulation by microRNAs, Leukemia, 2010, 24, 1335–1342 CrossRef PubMed.
- A. Mao, Y. Liu, Y. Wang, Q. Zhao, X. Zhou, C. Sun, C. Di, J. Si, L. Gan and H. Zhang, miR-449a enhances radiosensitivity through modulating pRb/E2F1 in prostate cancer cells. Tumour biology, Int. Soc. Oncodev. Biol. Med., 2016, 37, 4831–4840 CrossRef PubMed.
- Y. Liu, Y. Wang, X. Sun, C. Mei, L. Wang, Z. Li and X. Zha, miR-449a promotes liver cancer cell apoptosis by downregulation of Calpain 6 and POU2F1, Oncotarget, 2016, 7, 13491–13501 Search PubMed.
- Z. Zhao, X. Ma, D. Sung, M. Li, A. Kosti, G. Lin, Y. Chen, A. Pertsemlidis, T. H. Hsiao and L. Du, microRNA-449a functions as a tumor suppressor in neuroblastoma through inducing cell differentiation and cell cycle arrest, RNA Biol., 2015, 12, 538–554 CrossRef PubMed.
- W. Ye, J. Xue, Q. Zhang, F. Li, W. Zhang, H. Chen, Y. Huang and F. Zheng, MiR-449a functions as a tumor suppressor in endometrial cancer by targeting CDC25A, Oncol. Rep., 2014, 32, 1193–1199 CrossRef PubMed.
- K. Matula, E. Collie-Duguid, G. Murray, K. Parikh, H. Grabsch, P. Tan, S. Lalwani, R. Garau, Y. Ong, G. Bain, A. D. Smith, G. Urquhart, J. Bielawski, M. Finnegan and R. Petty, Regulation of cellular sphingosine-1-phosphate by sphingosine kinase 1 and sphingosine-1-phosphate lyase determines chemotherapy resistance in gastroesophageal cancer, BMC Cancer, 2015, 15, 762 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2018 |