Open Access Article
Open Access ArticleMetabolic profiling putatively identifies plasma biomarkers of male infertility using UPLC-ESI-IT-TOFMS†
F. J. Zenga,
H. C. Jia,
Z. M. Zhanga,
J. K. Luob,
H. M. Lu *a and
Y. Wang*b
*a and
Y. Wang*b
aCollege of Chemistry and Chemical Engineering, Central South University, Changsha, China. E-mail: hongmeilu@csu.edu.cn
bDepartment of Integrated Traditional Chinese and Western Medicine, Male Department of Integrated Traditional Chinese and Western Medicine, Xiangya Hospital, Central South University, Changsha, China. E-mail: wangyang_xy87@csu.edu.cn
First published on 19th July 2018
Abstract
Male infertility has become a global health problem. Currently, the diagnosis of male infertility depends on the results of semen quality or requires invasive surgical intervention. The process is complex and time-consuming. Metabolomics is an emerging platform with unique advantages in disease diagnosis and pathological mechanism research. In this study, ultra-performance liquid chromatography-electrospray ionization-ion trap-time of flight mass spectrometry (UPLC-ESI-IT-TOFMS) combined with chemometrics methods was used to discover potential biomarkers of male infertility based on non-targeted plasma metabolomics. Plasma samples from healthy controls (HC, n = 43) and various types of infertile patients, i.e., patients having oligozoospermia (OS, n = 36), asthenospermia (AS, n = 56) and erectile dysfunction (ED, n = 45) were analyzed by UPLC-ESI-IT-TOFMS. Principal component analysis (PCA) and orthogonal partial least squares discriminant analysis (OPLS-DA) were performed. The results of OPLS-DA showed that HCs could be discriminated from infertile patients including OS (R2 = 0.903, Q2 = 0.617, AUC = 0.992), AS (R2 = 0.985, Q2 = 0.658, AUC = 0.999) or ED (R2 = 0.942, Q2 = 0.500, AUC = 0.998). Some potential biomarkers were successfully discovered by variable selection methods and variable important in the projection (VIP) in combination with the T-test. Statistical significance was set at p < 0.05; the Benjamini–Hochberg false discovery rate was used to reduce type 1 errors resulting from multiple comparisons. The identified biomarkers were associated with energy consumption, hormone regulation and antioxidant defenses in spermatogenesis. To elucidate the pathophysiology of male infertility, relative metabolic pathways were studied. It was found that male infertility is closely related to disturbed phospholipid metabolism, amino acid metabolism, steroid hormone biosynthesis metabolism, metabolism of fatty acids and products of carnitine acylation, and purine and pyrimidine metabolisms. Plasma metabolomics provides a novel strategy for the diagnosis of male infertility and offers a new insight to study pathogenesis mechanism.
Introduction
Infertility is defined as the failure of a couple to conceive after one year of regular sexual activity without a contraceptive method.1 Infertility has become a common and important global health issue, affecting 13–15% of couples; male infertility (MI) is directly or indirectly responsible for 60% of these cases.2 According to the presence or absence of sexual function, MI patients are classified into two phenotypes: sexually abnormal MI patients such as those with erectile dysfunction (ED) and sexually normal MI patients such as those with azoospermia, cryptozoospermia, oligozoospermia (OS), teratospermia, asthenospermia (AS) and necrospermia.3MI is caused by many factors including genetic defects, environmental factors, cryptorchidism, endocrine dyscrasia, lifestyle, and testis pathologies.4–8 Due to complexities of the causes, a comprehensive inspection of the concerned male should be performed.9 Routine evaluations for MI include physical examination, karyotype analysis, semen analysis, endocrine detection, medical history questionnaire, Y-chromosome microdeletion analysis, and additional tests such as sperm DNA fragmentation and genetic screening.10–12 Unfortunately, for many people, these routine screenings give normal results, leading to a diagnosis of idiopathic infertility and inconclusive etiology.3,13 A definitive diagnosis must then be pursued with surgical intervention in the form of a testicular biopsy, which carries inherent complications.14 These diagnostic approaches are time consuming, costly, uncomfortable and sometimes unacceptable for patients. Furthermore, the molecular mechanisms underlying MI remain obscure. Therefore, minimally invasive methods to diagnose specific etiologies of MI are essential, and it is necessary to find robust biomarkers in molecular mechanism studies of MI.
Metabolomics is an emerging field that quantitatively measures altered metabolites resulting from pathophysiological changes; it is rapidly becoming a discovery method for new diagnostic and prognostic biomarkers of human diseases.15 Metabolomics is usually divided into targeted metabolomics and non-targeted metabolomics. Non-targeted metabolomics is a comprehensive analysis of all measurable small molecule metabolites in biological samples. Thus, non-targeted metabolomics is appropriate to systematically analyze MI. Seminal fluid and urine are usually used in non-targeted metabolomics studies of MI. However, the production of seminal fluid can be an embarrassing, difficult and stressful experience; thus, it is usually not accepted by the patients. Urine is easily influenced by diet and environment. Instead, plasma may be a better potential biologic matrix as it is more stable and convenient to acquire.16 Furthermore, changes in plasma metabolites can indicate global functional changes. Spermatogenesis is a process of high energy consumption. As an important human body fluid, plasma is closely related to the body's energy metabolism. Therefore, abnormal metabolism of plasma directly affects the quality of sperm. Previous studies have shown that plasma has a strong association with many complex diseases including a variety of cancers,17–19 diabetes,20 metabolic syndrome21 and sepsis-induced acute lung injury.22 Therefore, plasma is suitable for the systematic analysis of MI.
Several analytical techniques are available for non-targeted metabolomics studies. Nuclear magnetic resonance (NMR) and mass spectrometry coupled with either gas chromatography or liquid chromatography are the most frequently used techniques. The main disadvantages of NMR are its low sensitivity and limited dynamic range, due to which only the most abundant components can be observed.23 Liquid chromatography-mass spectrometry (LC-MS) and gas chromatography-mass spectrometry (GC-MS) are analytical techniques widely used to resolve complex biological mixtures with high separation efficiency, sensitive detection and good reproducibility; these two techniques are highly complementary because each detects metabolites with different physicochemical properties and generally, only a limited number of compounds are simultaneously detected by both platforms.24,25 Compared with GC-MS, LC-MS allows the detection and quantification of thousands of putative metabolites with high precision and accuracy.26 LC-MS can analyze hundreds to thousands of metabolites at once, allowing identification of potential disease metabolite markers.27 Moreover, LC-MS analyses, for the most part, do not require derivatization.28 Our previous study has reported through GC-MS studies that plasma metabolites are associated with MI.29 However, to the best of our knowledge, few MI plasma metabolic profiling studies based on LC-MS have been reported.
To study the metabolic status of MI in depth, we performed a detailed subclass analysis of patients with sperm abnormalities. A medium-sized case control study was designed to discover potential biomarkers of the most common MI patterns (OS, AS and ED). Uncommon sperm abnormalities such as teratospermia, necrospermia, cryptozoospermia, and azoospermia were not analyzed because the number of collected samples failed to reach the metabolomics study significance. An ultra-performance liquid chromatography-electrospray ionization-ion trap-time of flight mass spectrometry (UPLC-ESI-IT-TOFMS) platform was used to acquire metabolic profiles. Possible biological significances of these potential metabolic biomarkers were further explained.
Experimental
Reagents and chemicals
HPLC grade methanol and acetonitrile were purchased from Merck (Darmstadt, Germany). LC-MS grade formic acid was purchased from Sigma-Aldrich (St. Louis, MO, USA). Distilled water (18.25 MΩ) was prepared using the Milli-Q water purification system (Millipore, Billerica, MA). All standards were purchased from Sigma-Aldrich (St. Louis, MO, USA).Biological samples
The samples were obtained from Xiangya Hospital of Central South University, Hunan Province, China. The clinical study was conducted according to the principles of the Helsinki Declaration. All experiments were approved by the Medical Ethics Committee of Xiangya Hospital; all participants were Han Chinese, and they provided written informed consent. Referring to the fifth edition of the World Health Organization (WHO) Laboratory Manual for the Examination and Processing of Human Semen, oligozoospermia is defined as total sperm number <40 million per ejaculate; asthenozoospermia is defined with the following ranges: rapid progressive motility (a) <25% or rapid progressive motility (a) + slow progressive motility (b) <50%. According to the International Index of Erectile Function (IIEF), erectile dysfunction is defined as IIEF < 21. Two hundred and three individuals selected from June 2015 to December 2015 were divided into 2 groups: one group containing 160 men who attended Xiangya Hospital because of conception failure for at least 12 months was defined as MI. The MI patients included 45 ED patients, 56 AS patients, 36 OS patients, 9 teratospermia patients, 4 necrospermia patients, 3 cryptozoospermia patients, and 7 azoospermia patients. The other group containing 43 age-matched fertile men was defined as healthy controls (HC). Blood was collected into heparinized tubes and centrifuged at 4000g and 4 °C for 20 min. The plasma was separated and frozen at −80 °C until analysis. Before participants were included in the analysis, it was confirmed that they did not have metabolic diseases. Clinical data of the participants are presented in the supplemental material.Sample preparation
A volume of 400 μL of methanol was added to 100 μL of plasma. After vortexing, the mixture was centrifuged at 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 15 min at 4 °C for deproteinization. The supernatant (450 μL) was transferred into a 5 mL glass centrifugation tube and evaporated to dryness under N2 gas. The residue was reconstituted in 100 μL of 5% acetonitrile aqueous solution. Next, the mixture was vortex-mixed for 60 s and centrifuged for 15 min (18
000g for 15 min at 4 °C for deproteinization. The supernatant (450 μL) was transferred into a 5 mL glass centrifugation tube and evaporated to dryness under N2 gas. The residue was reconstituted in 100 μL of 5% acetonitrile aqueous solution. Next, the mixture was vortex-mixed for 60 s and centrifuged for 15 min (18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g, 4 °C). The final supernatant was taken for UPLC-ESI-IT-TOFMS analysis. All samples were analyzed using UPLC-ESI-IT-TOFMS in random order after being preprocessed to ensure that no systematic biases were present.
000g, 4 °C). The final supernatant was taken for UPLC-ESI-IT-TOFMS analysis. All samples were analyzed using UPLC-ESI-IT-TOFMS in random order after being preprocessed to ensure that no systematic biases were present.
A quality control (QC) sample was prepared by pooling 100 μL aliquots from each plasma sample and vortex mixing. Sample preparation for the QC sample was performed as described above.
UPLC-ESI-IT-TOFMS analysis
All sample analyses were carried out on a LC/IT-TOFMS system (Shimadzu, Japan). Chromatographic separation was achieved on a Waters ACQUITY UPLC HSS C18 column (100 mm × 2.1 mm, i.d. 1.8 μm) at 40 °C. A binary gradient elution system composed of solvent A (0.1% formic acid in water, v/v) and solvent B (0.1% formic acid in acetonitrile, v/v) was used under the following gradient program: 5% B increased to 40% B in 6 min and to 85% B in 14 min. The value ramped to 100% B in 3 min and was maintained for 7 min; then, it decreased to 5% in 1 min and finally maintained at 5% B for 5 min. Five microliter aliquot of each sample was injected into UPLC-ESI-IT-TOFMS for analysis. The flow rate was 0.4 mL min−1. An LC eluent was introduced into the MS system and individually detected in ESI positive (ESI+) and ESI negative (ESI−) ion modes. IT-TOF mass conditions were as follows: CDL temperature, 200 °C; heater block temperature, 200 °C; nebulizing gas (N2) flow rate, 1.5 L min−1; dry gas (N2) pressure, 100 kPa; ion trap pressure, 1.8 × 10−5 kPa; and ion accumulated time, 56 ms. The detector voltage was set at −3.5 kV in the negative ion electrospray mode and 4.5 kV in the positive ion electrospray mode.Data processing and analysis
All LC-MS data were processed using open-source XCMS30 to yield a data matrix containing retention times, accurate masses and peak intensities. To reduce variations from sample preparation and instrument factors, analytical noise was removed with the blank sample. To remove analytes that were not reproducibly detected, analytes for which more than half of the values were missing in QC samples or for which QC samples had a relative standard deviation of intensity larger than 30% were removed from the data set. The matrix was then imported into the SIMCA-P software (Version 14.1, Umetrics, Umea, Sweden) for multivariate statistical analysis. After dataset pre-treatment, mean-centered and pareto scaled mathematical methods, principal component analysis (PCA) and orthogonal partial least squares discriminant analysis (OPLS-DA)31 were performed. The parameters R2 and Q2 were used to evaluate goodness of fit and predictive ability of the constructed model, respectively. Permutation testing was also used to evaluate the constructed OPLS-DA model. The order of the Y-variable randomly permuted the specified number of times, and separate models were fitted to all the permuted Y-variables, extracting as many components as was done with the original Y matrix. The permutation plot then displayed the correlation coefficient between the original Y-variable and the permuted Y-variable on the X-axis versus cumulative R2 and Q2 on the Y-axis, and it also displayed the regression line. The intercept is a measure of the overfit. The criteria for validity are as follows: all permuted R2 and Q2 values to the left are lower than the original point to the right, and the regression line of the Q2 points has a negative intercept. The area under the receiver operating characteristic curve (ROC) (AUC) was used as a metric to assess prediction ability. A model is good when AUC ranges from 0.9 to 1, moderate if AUC ranges from 0.7 to 0.9, and poor if AUC ranges from 0.5 to 0.7. We used MetaboAnalyst 3.0 (ref. 32) for pathway analysis. The significance of the mean difference between two groups was obtained using T-test. To reduce false discovery rate (FDR) after the T-test, P-values were adjusted using the Benjamini–Hochberg (BH) procedure.33 The BH procedure is defined as Pm ≤ m × q/M, where M = total number of tested metabolites, q = FDR (set at 5% in the present paper), Pm = individual P-value's rank, and m = individual rank of the tested metabolite.Biomarker identification
Briefly, QC samples were analyzed to acquire accurate mass information and MS/MS spectra for potential biomarkers. Possible formulas of potential biomarkers were generated using the formula predict software of Shimadzu LCMS solution. Element number restriction, Lewis check, isotopic pattern, and hydrogen/carbon element ratio check were used to reduce the number of candidate formulas. The structures of potential biomarkers were researched with Human Metabolome Database,34 Metlin35 and MS FINDER.36 Twenty-three metabolites were considered to be potential biomarkers, of which 10 metabolites were identified by their corresponding chemical standard substances. Detailed identifying information is available in the ESI.†Results and discussion
LC-MS profiles of plasma samples
The separation conditions of plasma on column were optimized. Using the LC-MS conditions described above, all samples were subjected to both positive and negative ion modes for metabolic profiling analysis. Typical total ion chromatography (TIC) results of plasma samples with positive ion modes are shown in Fig. 1. As can be seen in Fig. 1, the plasma metabolites in all groups were similar, whereas the levels of some metabolites were different. The results illustrated that these LC-MS profiles could represent the differences among 4 groups.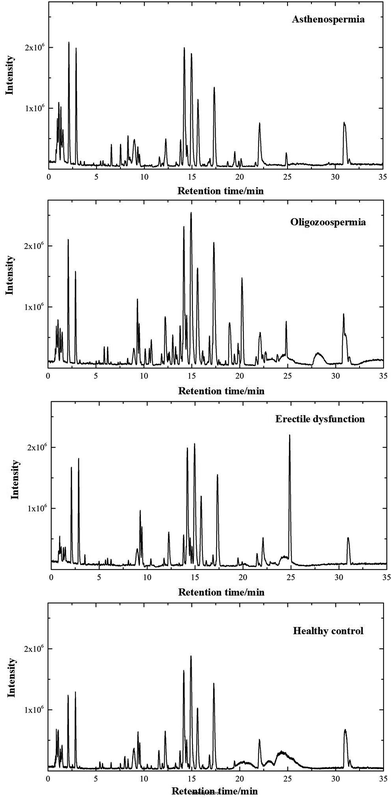 | ||
| Fig. 1 Positive ion mode typical total ion chromatograms of asthenospermia (AS), oligozoospermia (OS), erectile dysfunction (ED) and healthy control (HC) plasma samples. | ||
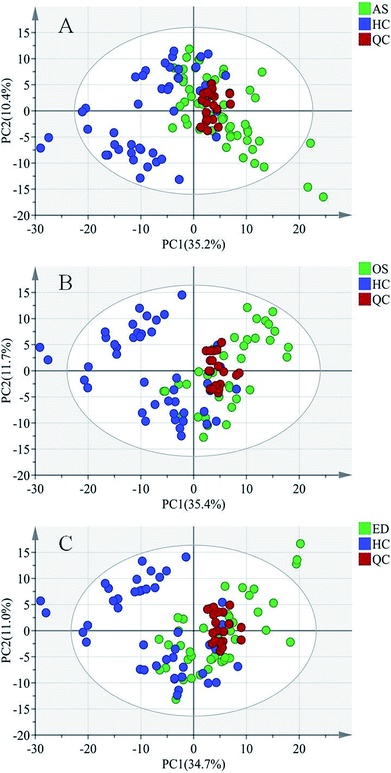
For metabolomics analysis, reliability of the analytical method is very important to obtain valid data. QC samples were employed to ensure that the acquired data were suitable for the following data analysis. As QC samples used here were pooled from study samples, any technical variation introduced during sample preparation, data acquisition and data preprocessing was reflected in the data acquired for QC samples. PCA, an unsupervised statistical method, was used to detect the robustness of the analytical method and the inherent trends within the data. As shown in Fig. 2, QC samples were tightly clustered, which suggested that the LC-MS workflow is robust with good precision, stability, and repeatability. As shown in Fig. 1s of the ESI,† 3D graphics provided more information about the clustering of QC samples. Therefore, the dataset quality was considered to be good for further analysis. As shown in Fig. 2, a clear separation trend could be observed between MI patient groups and HC groups, indicating inherent metabolic changes in MI patients compared to those in controls.
Pattern recognition of HC, ED, AS and OS
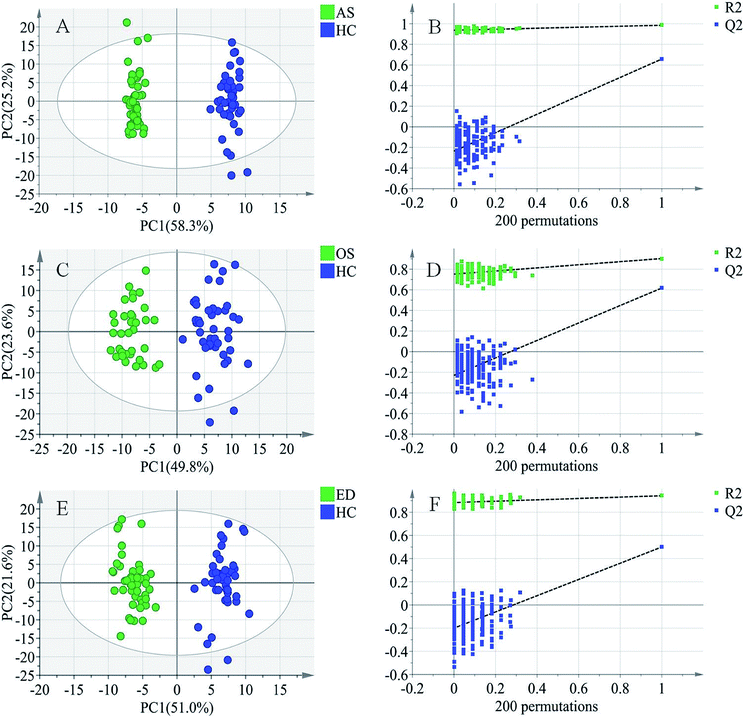
To further study the differences between the MI groups and HC and to find potential biomarkers, the supervised multivariate statistical method OPLS-DA was subsequently used. The classification results are shown in Fig. 3. The samples from HC and AS patient groups (R2 = 0.985, Q2 = 0.657, Fig. 3A), HC and OS patient groups (R2 = 0.902, Q2 = 0.617, Fig. 3C), and HC and ED patient groups (R2 = 0.942, Q2 = 0.500, Fig. 3E) were unambiguously distinguished based on global metabolic profiles.The values of R2 and Q2 indicate that OPLS-DA can provide good classification and prediction results for MI.
To guard against model overfitting, permutation tests (200 random permutations) were performed. These permutation tests were used to contrast the goodness of fit of the original model with the goodness of fit of randomly permuted models. As shown in Fig. 3B, D and F, the validation plots strongly indicated that the original combined models were valid. No overfitting was observed.
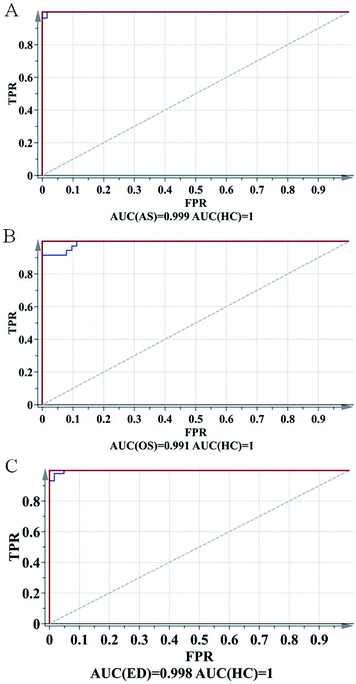
To further evaluate the predictive ability of the OPLS-DA model, ROC analysis was performed with the SIMCA-P software. The ROC plot is a tool for visualizing and summarizing the performance of classification and discrimination models. The ROC plot displays the true positive classification rate (TPR) of a classifier model plotted against the corresponding false positive classification rate (FPR) at various threshold settings of the criterion parameter. As a quantitative measure of classification success, the area under the (ROC) curve (AUC) is computed and visualized in the plot. As shown in Fig. 4, AUCs of AR, OS and ED were 0.999, 0.991 and 0.998, respectively, indicating good predictive ability.
Candidate biomarker discovery and identification
Biomarker discovery is the critical step for metabolomics research. To discover potential biomarkers among hundreds of variables, parameter variable importance in the projection (VIP) was employed to reflect the importance of variables. Variables were selected based on the criterion of VIP value greater than one. Potential biomarkers were validated using the T-test, where p values of <0.05 were considered to be statistically significant and then, they were subjected to qualitative analysis. To reduce false discovery rate (FDR) after the T-test, P-values were adjusted using the Benjamini–Hochberg procedure. Fold change was calculated from the mean values of each; it reflects the change in the trend of MI patient metabolites compared with that in HC. A fold change value greater than one indicates relatively higher concentration present in MI patients, whereas less than one means a relatively lower concentration compared to that in HC. The qualitative and quantitative analysis results of potential biomarkers are summarized in Table 1, and they include HMDB ID, exact mass, molecular formula, metabolite, VIP value, pm value, m × q/M and fold change. Detailed identify information is provided in the supplemental material.| HMDB ID | Exact mass | Formula | Metabolite | VIP | Pm value | m × q/M | Fold change | |
|---|---|---|---|---|---|---|---|---|
| a Identified by standard substances. M = total number of analyzed metabolites; q = FDR; m = individual rank of tested metabolite; Pm = individual P-value. BHAfter Benjamini–Hochberg adjustment, the p-values of metabolites with Pm < m × q/M remained significantly different. | ||||||||
| AS vs. HC | HMDB10379 | 467.3117 | C22H46NO7P | LysoPC(14:0)a | 1.88 | 0.03348 | 0.02313 | 1.69 |
| HMDB10384 | 523.3724 | C26H54NO7P | LysoPC(18:0)a,BH | 2.78 | 0.00743 | 0.00933 | 1.56 | |
| HMDB11523 | 529.2065 | C27H48NO7P | LysoPE(22:4)BH | 2.22 | 0.01886 | 0.01903 | 3.15 | |
| HMDB10406 | 605.8496 | C32H64NO7P | LysoPC(24:1) | 1.61 | 0.03348 | 0.02313 | 1.81 | |
| HMDB11520 | 537.7196 | C27H56NO7P | LysoPE(22:0)a | 2.13 | 0.04192 | 0.02910 | 1.87 | |
| HMDB10402 | 569.3612 | C30H52NO7P | LysoPC(22:5) | 1.95 | 0.04192 | 0.02910 | 1.34 | |
| HMDB29335 | 251.2387 | C12H13NO5 | N-Phenylacetylaspartic acida | 1.78 | 0.03161 | 0.02239 | 0.81 | |
| HMDB0607 | 425.1131 | C13H22N4O8S2 | S-Glutathionyl-L-cysteine | 1.99 | 0.03549 | 0.02425 | 2.65 | |
| HMDB05084 | 481.6480 | C25H39NO6S | N-Acetyl-LTE4 | 2.11 | 0.03549 | 0.02425 | 2.16 | |
| HMDB00121 | 441.0869 | C19H19N7O6 | Folic acida | 1.62 | 0.04157 | 0.02873 | 0.66 | |
| HMDB00705 | 260.1854 | C13H25NO4 | Hexanoylcarnitinea | 1.92 | 0.04491 | 0.03097 | 0.73 | |
| HMDB00723 | 344.2805 | C19H38NO4 | Carnitine (12:0)a | 1.82 | 0.03348 | 0.02313 | 0.95 | |
| OS vs. HC | HMDB11523 | 529.2065 | C27H48NO7P | LysoPE(22:4)BH | 2.37 | 0.00013 | 0.00037 | 2.93 |
| HMDB11520 | 537.7196 | C27H56NO7P | LysoPE(22:0)a,BH | 2.27 | 0.00034 | 0.00112 | 2.04 | |
| HMDB10381 | 481.3149 | C23H48NO7P | LysoPC(15:0)BH | 1.62 | 0.01754 | 0.01940 | 1.76 | |
| HMDB11516 | 503.3391 | C25H46NO7P | LysoPE(20:3)BH | 1.56 | 0.01752 | 0.01903 | 2.22 | |
| HMDB11513 | 505.3572 | C25H48NO7P | LysoPE(20:2) | 1.44 | 0.04686 | 0.03433 | 1.90 | |
| HMDB10384 | 523.3724 | C26H54NO7P | LysoPC(18:0)a | 1.81 | 0.04468 | 0.03209 | 2.87 | |
| HMDB29335 | 251.2380 | C12H13NO5 | N-Phenylacetylaspartic acida | 1.57 | 0.04036 | 0.03134 | 0.78 | |
| HMDB0607 | 425.1131 | C13H22N4O8S2 | S-Glutathionyl-L-cysteineBH | 1.83 | 0.01502 | 0.01716 | 3.25 | |
| HMDB05084 | 481.6480 | C25H39NO6S | N-Acetyl-LTE4BH | 1.92 | 0.00614 | 0.00932 | 1.96 | |
| HMDB02833 | 368.4874 | C19H28O5S | Testosterone sulfateBH | 2.36 | 0.00025 | 0.00074 | 3.90 | |
| HMDB06278 | 370.5071 | C19H30O5S | 5α-Dihydrotestosterone sulfateBH | 2.07 | 0.00108 | 0.00261 | 1.74 | |
| HMDB00014 | 227.0788 | C9H13N3O4 | Deoxycytidinea,BH | 1.84 | 0.02791 | 0.02836 | 1.64 | |
| HMDB00089 | 243.2190 | C9H13N3O5 | Cytidinea,BH | 2.50 | 0.00013 | 0.00037 | 0.74 | |
| HMDB00071 | 251.0210 | C10H12N4O4 | DeoxyinosineBH | 1.93 | 0.00877 | 0.01194 | 2.27 | |
| HMDB00296 | 244.2338 | C9H12N2O6 | Uridinea,BH | 2.05 | 0.02455 | 0.02537 | 0.91 | |
| HMDB00722 | 483.3078 | C26H45NO5S | Lithocholytaurine | 1.50 | 0.04607 | 0.03395 | 2.78 | |
| HMDB01413 | 488.1828 | C14H26N4O11P2 | CiticolineBH | 1.98 | 0.02196 | 0.02201 | 2.87 | |
| ED vs. HC | HMDB10381 | 481.3149 | C23H48NO7P | LysoPC(15:0) | 2.16 | 0.04106 | 0.02313 | 2.16 |
| HMDB11523 | 530.2065 | C27H48NO7P | LysoPE(22:4)BH | 1.98 | 0.01780 | 0.01978 | 2.66 | |
| HMDB11520 | 537.7196 | C27H56NO7P | LysoPE(22:0)a,BH | 2.22 | 0.00573 | 0.00634 | 1.90 | |
| HMDB02833 | 368.4874 | C19H28O5S | Testosterone sulfateBH | 2.49 | 0.00138 | 0.00298 | 3.42 | |
| HMDB06278 | 370.5071 | C19H30O5S | 5α-Dihydrotestosterone sulfateBH | 1.93 | 0.00027 | 0.00037 | 1.70 | |
Association between identified biomarkers and MI
There is few published literature regarding the application of plasma metabolomics in MI. In this study, we observe that a plasma metabolome pattern can discriminate MI patients from HC groups. A series of potential biomarkers were identified with functionally important roles in energy production and antioxidant and hormone regulation in spermatogenesis. From a metabolic viewpoint, these biomarkers are involved in amino acid metabolism, phospholipid metabolism, metabolism of the products of carnitine acylation, steroid hormone biosynthesis metabolism and purine and pyrimidine metabolisms.The levels of a series of LysoPCs and LysoPEs in plasma of infertile male (including AS, ED or OS) patients significantly increased (Table 1); they play important roles in the phospholipid metabolism pathway, and their increase indicates abnormal phospholipid metabolism in MI. Phospholipids are a very important class of lipids for the construction of cell membranes. Metabolites of phospholipid decomposition also participate in maintaining normal physiological function. Phospholipid metabolism has been demonstrated to affect the regulation of the signaling step, leading to neutrophil activation,37 which is the source of proteolytic enzymes and reactive oxygen species (ROS). LysoPCs and LysoPEs are considered important intercellular signaling molecules. In the metabolic pathway of the organism, LysoPCs and LysoPEs are hydrolyzed to fatty acids under the action of lysophospholipase A; then, fatty acids decompose in the mitochondria to provide energy for the human body. In addition, LysoPCs can be converted into lysophospholipid acids (LPA) under the action of lysophospholipase D. LPA can induce endogenous ROS, forming oxidative stress. Oxidative stress can generate DNA damage, leading to sperm damage, sperm deformity and eventually male infertility.38–40 Furthermore, many lipids may be involved in cell proliferation.41 Thus, significant increase in LysoPC and LysoPE contents in MI patients suggests the occurrence of disorders including intercellular signal transduction, sperm cell proliferation, and reactive oxygen species and energy metabolism of MI patients, which disturbs normal spermatogenesis.
The levels of testosterone sulfate and 5α-dihydrotestosterone sulfate in plasma of ED or OS patients were significantly decreased (Table 1). These hormones play important roles in the steroid hormone biosynthesis metabolic pathway, and they are closely related to other sex hormone syntheses.42 The main sources of steroidogenesis in humans are the adrenal glands, gonads (ovaries and testes), and placenta. Steroid hormones are lipophilic low-molecular weight compounds. In human blood, steroid hormones are primarily present in their sulfonated form, and they do not have any physiological activity.43 Sulfonated forms can release free testosterone under catalysis of the sulfotransferase enzyme.44 Free testosterone, 5α-testosterone, and other steroids transported via the blood system affect target tissues by binding to cell nuclei or cytosolic receptors;45 they can control fundamental physiological functions such as body growth, metabolism, sexual development, inflammation, ion homeostasis and reproduction.46 Steroid hormone metabolism often activates mechanisms affecting cellular proliferation, survival, migration and invasiveness. In the present study, there is significant decrease in the levels of sulfate conjugates of sex hormones including testosterone sulfate and 5α-testosterone sulfate. This trend may reflect the disturbance of the normal steroid hormone biosynthesis metabolic pathway. Sex hormone contents of OS and ED patients do not reach the normal standard for spermatogenesis.
The levels of folic acid, hexanoylcarnitine and carnitine C12:0 in the plasma of AS patients significantly decreased (Table 1). T-test analyses showed that folic acid, hexanoylcarnitine and carnitine C12:0 were correlated with AS; after p-value correction using the Benjamini–Hochberg procedure (Pm > m × q/M), no significant associations were observed; despite these results, their relationship with male infertility has been reported in many studies. Folic acid is reported to be strongly associated with male infertility.47 Carnitines play important roles in fatty acid β-oxidation in mammals. Carnitine provides a shuttle system for free fatty acids and derivatives of acyl-CoA through membranes within mitochondria to generate adenosine triphosphate (ATP). Meanwhile, acyl groups are temporarily transferred to carnitine to produce acylcarnitine. Carnitine and acylcarnitine provide ready energy for spermatozoa during spermatogenesis. In addition, carnitine and acetylcarnitine exert substantial antioxidant actions in spermatogenesis,48 providing protective effects against lipid peroxidation in phospholipid membranes. Carnitines act as the primary defence barriers to remove excessive intracellular toxic acetyl-CoA, protecting spermatozoa from oxidative damage. An earlier study reported that the acetylcarnitine level in the semen of oligozoospermic infertile men was significantly lower than that in fertile control subjects.16 In the present study, lower levels of carnitines are also observed in AS patients compared with those in HC. This suggests that the energy requirements and the antioxidant system of normal spermatogenesis in patients with AS are interrupted.
The levels of cytidine and uridine in plasma of OS patients significantly decreased (Table 1), and the levels of deoxyinosine and deoxycytidine in plasma of OS patients significantly increased (Table 1); these four chemicals are the key metabolites of the purine and pyrimidine metabolism pathway. Purines and pyrimidines are the major biomolecules for energy storage in the form of ATP and GTP, and they are required for the transfer of genetic information; they are involved in cell signaling as cAMP and cGMP, and they also act as cofactors (NADH, NADPH and coenzyme A) for many enzymes.49 The disorders of purine and pyrimidine metabolism have been reported and mentioned in general literature. Among them, uridine is identified as a candidate biomarker for autism and polycystic ovary syndrome.50,51 Uridine has also been identified as an oxidative stress biomarker of male infertility in urinary and seminal plasma;52,53 this is consistent with the result of plasma metabolomics in our study. Cytidine is partly produced from cytosine and is converted to uridine by cytidine deaminase. Cytidine may be an important biomarker for esophageal adenocarcinoma, type 2 diabetes and gastric cancer.54–56 Deoxycytidine kinase has been reported to have connection with pancreatic cancer.57 In the present study, several metabolite levels in purine and pyrimidine metabolic pathways changed in the plasma of males with OS infertility; they may reflect increased oxidative stress in spermatogenesis, eventually leading to infertility. However, specific molecular mechanisms still require further study.
Conclusions
In summary, metabolic alterations in AS, ED and OS patients were characterized using a LC-MS-based metabolomics approach. We established a classification diagnostic model for MI. The established model achieved reasonably high accuracy and provided theoretical support for the development of new methods for the clinical diagnosis of male infertility. We used T-test combined with Benjamini–Hochberg procedure and multivariate statistical method OPLS-DA to screen potential biomarkers. These potential biomarkers were identified, and their related metabolic pathways were tentatively analyzed. The potential biomarkers revealed the complexities of metabolic alterations in AS, ED and OS patients, and these may reveal the major molecular mechanisms defining male infertility outcomes. These significantly altered metabolites are associated with phospholipid metabolism, steroid hormone biosynthesis metabolism, carnitine metabolism and purine and pyrimidine metabolisms. The identified biomarkers may reflect functional response to reduced fertility, and they are strongly associated with energy consumption, hormone regulation and antioxidant defense in spermatogenesis. This study also confirmed the feasibility of using an LC-MS-based plasma metabolomics platform for characterization of MI. Compared with previously reported biomarkers, the potential biomarkers found in this study were not identical, but the metabolic pathways they affect were consistent. These potential biomarkers reflect deregulation of plasma metabolism in MI individuals, which might be beneficial to the diagnosis and further pathogenesis research of MI. Although these metabolic insights are in accordance with the results of previous studies, the results are preliminary and need further testing. The involved pathways need to be demonstrated using an in vivo model to provide a theoretical guidance for the etiology and medication of MI.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors gratefully thank the National Natural Science Foundation of China for support of the projects (Grant No. 21375151, 21305163 and 21675174).References
- L. C. Callister, Global Health Nursing, 2010, 35, 174 Search PubMed.
- S. C. Esteves, R. Miyaoka and A. Agarwal, Clinics, 2011, 66, 691–700 CrossRef PubMed.
- M. Abu-halima, M. Hammadeh and J. Schmitt, Fertil. Steril., 2012, 99, 1249–1255 CrossRef PubMed.
- S. Wang, G. Huang and Q. Hu, Biochim. Biophys. Acta, 2016, 1860, 2716–2724 CrossRef PubMed.
- K. L. O. O'Brien, A. C. Varghese and A. Agarwal, Fertil. Steril., 2010, 93, 1–12 CrossRef PubMed.
- N. E. Skakkebæk, M. E. Rajpertde and K. M. Main, Hum. Reprod., 2001, 16, 972–978 CrossRef.
- K. K. Kesari, S. Kumar and J. Nirala, Cell Biochem. Biophys., 2013, 65, 85–96 CrossRef PubMed.
- D. Santi and S. Vezzani, Environ. Res., 2016, 150, 144–153 CrossRef PubMed.
- J. M. Bieniek, A. P. Drabovich and K. C. Lo, Asian J. Androl., 2016, 18, 426–433 CrossRef PubMed.
- C. Y. Chen, T. C. Chiang and C. M. Lin, Analyst, 2013, 138, 4967 RSC.
- R. Nosrati, M. M. Gong and M. C. San Gabriel, Anal. Methods, 2016, 8, 6260–6264 RSC.
- Z. Zhang, Y. Zhang and C. Liu, Int. J. Mol. Sci., 2017, 18, 1–14 Search PubMed.
- J. R. Kovac, A. W. Pastuszak and D. J. Lamb, Fertil. Steril., 2013, 99, 998–1007 CrossRef PubMed.
- F. Deepinder, H. T. Chowdary and A. Agarwal, Expert Rev. Mol. Diagn., 2007, 7, 351–358 CrossRef PubMed.
- G. Xie, L. Lu and Y. Qiu, J. Proteome Res., 2015, 14, 1195–1202 CrossRef PubMed.
- J. Zhang, Z. Huang and M. Chen, Fertil. Steril., 2014, 102, 44–53.e12 CrossRef PubMed.
- O. Turkoglu, A. Zeb and S. Graham, Metabolomics, 2016, 12, 1–16 CrossRef PubMed.
- S. Miyamoto, S. L. Taylor and D. K. Barupal, Metabolites, 2015, 5, 192–210 CrossRef PubMed.
- Q. Liang, C. Wang and B. Li, Biotechnol. Appl. Biochem., 2015, 176, 2170–2184 CrossRef PubMed.
- L. Liu, Y. Li and C. Guan, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2010, 878, 2817–2825 CrossRef PubMed.
- Z. Lin, C. M. Vicente Gonçalves and L. Dai, Anal. Chim. Acta, 2014, 827, 22–27 CrossRef PubMed.
- K. A. Stringer, N. J. Serkova and A. Karnovsky, Am. J. Physiol., 2011, 300, L4–L11 Search PubMed.
- E. J. Want, G. O'Maille, C. A. Smith, T. R. Brandon, W. Uritboonthai, C. Qin, S. A. Trauger and G. Siuzdak, Anal. Chem., 2006, 78, 743–752 CrossRef PubMed.
- M. Wang, R. H. Han and X. Han, Anal. Chem., 2013, 85, 9312–9320 CrossRef PubMed.
- D. Saigusa, Y. Okamura, I. N. Motoike, Y. Katoh and M. Yamamoto, PLoS One, 2016, 11, 1–18 CrossRef PubMed.
- K. Skov, N. Hadrup, J. Smedsgaard and H. Frandsen, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2015, 978, 83–88 CrossRef PubMed.
- T. Basak, S. Varshney, Z. Hamid, S. Ghosh, S. Seth and S. Sengupta, J. Proteomics, 2015, 127, 169–177 CrossRef PubMed.
- D. Vuckovic, Anal. Bioanal. Chem., 2012, 403, 1523–1548 CrossRef PubMed.
- X. Zhou, Y. Wang, Y. Yun, Z. Xia, H. Lu and J. Luo, Talanta, 2016, 147, 82–89 CrossRef PubMed.
- C. A. Smith, E. J. Want and G. O'Maille, Anal. Chem., 2006, 78, 779–787 CrossRef PubMed.
- M. Bylesjo, M. Rantalainen and O. Cloarec, J. Chemom., 2006, 20, 341–351 CrossRef.
- J. Xia, I. V. Sinelnikov and B. Han, Nucleic Acids Res., 2015, 43, W251–W257 CrossRef PubMed.
- B. Yoav and H. Yosef, J. R. Statist. Soc. B, 1995, 57, 289–300 Search PubMed.
- D. S. Wishart, T. Jewison and A. C. Guo, Nucleic Acids Res., 2013, 41, D801–D807 CrossRef PubMed.
- C. A. Smith, G. O'Maille and E. J. Want, Ther. Drug Monit., 2005, 27, 747–751 CrossRef PubMed.
- T. Cajka, W. Tanaka and D. Yukihira, Anal. Chem., 2016, 88, 7946–7958 CrossRef PubMed.
- K. Mayer, C. Fegbeutel, K. Hattar, U. Sibelius, H. J. Krämer, K. Heuer, B. Temmesfeld-wollbrück, S. Gokorsch, F. Grimminger and W. Seeger, Intensive Care Med., 2003, 29, 1472–1481 CrossRef PubMed.
- Z. Zhao, Y. Xiao and P. Elson, J. Clin. Oncol., 2007, 25, 2696–2701 CrossRef PubMed.
- K. Tremellen, Hum. Reprod. Update, 2008, 14, 243–258 CrossRef PubMed.
- M. B. Shamsi, S. Venkatesh and M. Tanwar, Mutat. Res., 2009, 665, 29–36 CrossRef PubMed.
- W. H. Moolenaar and A. Perrakis, Nat. Rev. Mol. Cell Biol., 2011, 12, 674–679 CrossRef PubMed.
- F. Badoud, E. Grata and J. Boccard, Anal. Bioanal. Chem., 2011, 400, 503–516 CrossRef PubMed.
- C. E. Galuska, M. F. Hartmann and A. Sánchez-Guijo, Analyst, 2013, 138, 3792 RSC.
- A. Sánchez-Guijo, V. Oji and M. F. Hartmann, J. Lipid Res., 2015, 56, 1843–1851 CrossRef PubMed.
- E. Kalogera, C. Pistos, X. Provatopoulou, S. Athanaselis, C. Spiliopoulou and A. Gounaris, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2013, 940, 24–34 CrossRef PubMed.
- N. Mannowetz, M. R. Miller and P. V. Lishko, PNAS, 2017, 114, 5743–5748 CrossRef PubMed.
- J. Zhang, X. Mu and Y. X. Xia, J. Proteome Res., 2014, 13, 3088–3099 CrossRef PubMed.
- C. M. Ng, M. R. Blackman and C. Wang, Ann. N. Y. Acad. Sci., 2004, 1033, 177–188 CrossRef PubMed.
- I. Chitrakar, D. M. Kim-holzapfel, W. Zhou and J. B. French, J. Struct. Biol., 2017, 197, 354–364 CrossRef PubMed.
- X. Zhao, F. Xu, B. Qi, S. Hao, Y. Li, Y. Li, L. Zou, C. Lu, G. Xu and L. Hou, J. Proteome Res., 2014, 13, 1101–1111 CrossRef PubMed.
- J. B. Adams, T. Audhya, S. Mcdonough-means, R. A. Rubin, D. Quig, E. Geis, E. Gehn, M. Loresto, J. Mitchell, S. Atwood, S. Barnhouse and W. Lee, Nutr. Metab., 2011, 8, 1–34 CrossRef PubMed.
- A. D. Maher, P. Patki and J. C. Lindon, Clin. Chem., 2008, 54, 2063–2066 CrossRef PubMed.
- H. Shen, W. Xu and J. Zhang, Environ. Sci. Technol., 2013, 47, 8843–8851 Search PubMed.
- W. Y. Lo, L. Bin Jeng and C. C. Lai, Clin. Chim. Acta, 2014, 428, 57–62 CrossRef PubMed.
- J. Xia, Z. Wang and Q. Liang, Clin. Chim. Acta, 2011, 412, 940–945 CrossRef PubMed.
- D. Djukovic, H. R. Baniasadi and R. Kc, Rapid Commun. Mass Spectrom., 2010, 24, 3057–3062 CrossRef PubMed.
- K. Ohmine, K. Kawaguchi, S. Ohtsuki, F. Motoi, H. Ohtsuka and J. Kamiie, Mol. Pharm., 2015, 13, 3282–3291 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra01897a |
| This journal is © The Royal Society of Chemistry 2018 |