Open Access Article
Open Access ArticleRoles of membrane protein damage and intracellular protein damage in death of bacteria induced by atmospheric-pressure air discharge plasmas
Hao Zhangabc,
Jie Maac,
Jie Shenbc,
Yan Lanbc,
Lili Dinga,
Shulou Qiand,
Weidong Xia*ad,
Cheng Cheng *bc and
Paul K. Chu*e
*bc and
Paul K. Chu*e
aSchool of Life Science, University of Science and Technology of China, Hefei, Anhui Province 230026, People's Republic of China. E-mail: xiawd@ustc.edu.cn
bInstitute of Plasma Physics, Chinese Academy of Sciences, P. O. Box 1126, Hefei, 230031, P. R. China. E-mail: chengcheng@ipp.ac.cn
cCenter of Medical Physics and Technology, Hefei Institutes of Physical Science, Chinese Academy of Sciences, Hefei 230031, People's Republic of China
dDepartment of Thermal Science and Energy Engineering, University of Science and Technology of China, Hefei, Anhui Province 230026, People's Republic of China
eDepartment of Physics and Department of Materials Science and Engineering, City University of Hong Kong, Tat Chee Avenue, Kowloon, Hong Kong, China. E-mail: paul.chu@cityu.edu.hk
First published on 8th June 2018
Abstract
Although plasma sterilization has attracted much attention, the underlying mechanisms and biochemical pathways are still not fully understood. In this work, we investigate the molecular mechanism pertaining to the inactivation of Escherichia coli (E. coli) by air discharge plasmas. The membrane protein YgaP and intracellular protein swc7 are over-expressed in E. coli by genetic recombination and gene inducible expression techniques and plasma exposure is demonstrated to alter the structures of YgaP and swc7 in E. coli. The plasma-induced damage of YgaP and swc7 involves changes in the secondary and tertiary structures instead of the primary structure and the modification effectiveness depends on the storage time after the plasma treatment. Owing to the unique structure of E. coli, YgaP is more susceptible to the plasma treatment than intracellular swc7. Within 1 h after plasma exposure, YgaP is modified but not swc7, but after 1 h or longer, both YgaP and swc7 proteins are indeed modified. By analyzing the plasma-induced antimicrobial efficacy and modification of YgaP and swc7, plasma-induced modification of the membrane proteins is the major cause of bacterial death but there is no identifiable relationship with modification of the intracellular protein. The new results provide insights into the mechanism of multiple plasma-induced damage to bacteria and cells as well as the disinfection mechanism.
1. Introduction
Plasma medicine is an emerging field involving biological and medical applications of non-thermal plasmas,1,2 such as deactivation of microorganisms,3–13 blood coagulation,14 wound healing,15–17 cancer treatment,18,19 and sterilization of medical devices.20 Recent studies have revealed excellent antimicrobial and anticancer effectiveness by deactivation of various types of bacteria and apoptosis of a wide range of cancer cells.21–23 The reactive species (RS) produced by plasmas are considered to play major roles in these biological effects and although there have been efforts to unravel the underlying mechanisms of plasma-induced biological effects in order to fully utilize cold plasmas in biomedical applications, the molecular mechanisms are not well understood.24Toxicity produced by cold plasmas in cancer cells and bacteria may be complex and multiple including physical destruction,6 multiple signaling pathways,25 biosynthetic pathway and metabolic regulation,26 gene expression regulation,23 and so on. Therefore, it is necessary to investigate the underlying mechanisms from different perspectives especially the molecular level. Microorganisms and cells are assemblages of various functional biomolecules, especially proteins which are considered the main vectors in cellular functions and important components in cells.27 Hence, it is necessary to explore the degree of plasma-induced molecular modification or damage in the investigation of plasma sterilization mechanisms. The metabolism and cell response are affected if plasmas modify bio-molecules, leading to metabolic disturbance and false response in sterilization and cell apoptosis. There have been recent studies on the precise molecular mechanism governing the interactions between plasmas with bio-macromolecules and plasmas have been shown to cause loss of biological functions and structural changes in bio-molecules in vitro.27–36 P. Attri et al. studied the structural changes in hemoglobin (Hb) in the presence of RS generated by the dielectric barrier discharge (DBD) plasma in the presence of various gases such as O2, N2, Ar, He, NO (10%) + N2 and air.33 S. Choi et al. used lysozyme as a model enzyme and verified the plasma-induced structural changes by circular dichroism (CD), fluorescence, and X-ray crystallography.36 J. H. Park et al. used Ar, Ar–O2, and Ar–N2 as the working gases in the plasma and investigated the structural changes in proteins (hemoglobin (Hb) and myoglobin (Mb)). Additionally, they also studied the influence of H2O2 on the Hb and Mb structures by molecular dynamic simulation.35 Although these studies unraveled some of the plasma-induced structure changes in some bio-molecules and suggested that plasma generated ROS played major roles in the biological effects, all the model proteins in these studies were prepared in solutions in vitro for the plasma treatment. In fact, the effects of plasmas on functional proteins in bacteria in vivo have been less investigated and are relatively unknown. Furthermore, in the presence of cell membranes and walls, whether plasmas can indeed modify bio-molecules such as proteins, DNA, RNA, and subcellular organelles in E. coli is not clear. There are a variety of proteins, sugar, lipids and other molecules in the cell membrane and cytoplasm and these biomolecules can be classified as membrane molecules and intracellular molecules. Membrane molecules are mainly involved in materials exchange and signal transduction, and intracellular functional molecules regulate complex metabolic pathways. Hence, it is important to study the plasma induced effects on biomolecules in E. coli and determine the roles of different biomolecules changes in plasma triggered bactericidal effects.
In this work, we study the bactericidal effects of air discharge plasma (Fig. 1(a)) as well as plasma-induced biomolecules modification or damage on membrane molecules and intracellular molecules in E. coli. In practice, the bacteria on the skin wound surface are under wet conditions. A typical skin wound environment is very complicated and contains various bacteria, damaged skin cells, inflammatory cells, growth factors, and other organic secretions. A portion of the radicals in the liquid generated by the plasma can be scavenged by growth factors, organic secretions, and cells in the wound fluid except bacteria. In order to study the molecular mechanism related to the inactivation of bacteria in a simple environment, the phosphate buffer solution (PBS) is employed to simulate the wet conditions of the skin wound surface. The E. coli bacteria are dissolved in PBS for plasma treatment while the existence and impact of other factors in the wound fluid except the bacteria are ignored. The transmembrane protein Ygap (sulfurtransferase) and intracellular protein swc7 (transcriptase) are used as model molecules to evaluate the effects of air discharge plasmas on bio-molecules in E. coli, the laboratory model of target protein is shown in Fig. 1(b). By adopting molecular biology procedures, the plasma-treated target molecules are located in E. coli and extracted from cells for structural analysis. The plasma-induced damage on the membrane molecules or membrane and intracellular molecules are studied by analyzing the structure changes of plasma-treated YgaP and swc7 proteins. In addition, by combining plasma-induced sterilization and modification of model molecules, the roles of the membrane protein damage and intracellular protein damage in plasma-induced bacterial death is studied on the molecular level. The complete experimental procedures are described in Fig. 1(c).
2. Methods
2.1. Discharge apparatus and RS measurement
The atmospheric-pressure air plasma jet described in Fig. 1(a) consisted of an electrode with an array with 105 fine copper rods (radius = 1 mm).26 and the plasma discharge was generated between the electrode and surface of the E. coli suspension. The air plasma was powered by 10 kV DC and the discharge current was approximately 5 mA. The average plasma power from the whole array (105 discharges) was about 200 mW.37 During plasma exposure, the 10 ml E. coli suspension was put on a culture dish 60 mm in diameter and the energy density of the air plasma was about 7 mW cm−1.2 The distance between the electrode and liquid surface was 10 mm and the plasma gas temperature was about 30 °C.When the plasma reacted with liquids such as deionized water or phosphate buffer solution (PBS), various RS were produced in the liquids. In this study, the bacteria Escherichia coli (E. coli) were immersed in a liquid and the ROS (reactive oxygen species) and RNS (reactive nitrogen species) generated in the bacterial suspension by the plasma were monitored to investigate their effects on the bacteria. H2O2, O3, NO2−, OH˙, and ONOO− in the liquid were determined by fluorescent probes.38 The Amplex Red Hydrogen Peroxide/Peroxidase Assay kits (Invitrogen) were used in the H2O2 measurement. H2O2 reacted with horseradish peroxidase (HRP) to form peroxy radicals which oxidized Amplex red into fluorescent resorufin. Ozone was assessed using the 00607 test kit in accordance with the manufacturer's instruction (WTW, Germany). The nitrite concentration in the plasma-treated liquid was measured using a Griess reagent kit (Invitrogen). In the Griess assay, sulfanilic acid reacted with nitrite to form a diazonium salt, which was then coupled to a second reagent N-(1-naphthyl) ethylene diamine to form an azo dye. This azo dye could be spectrophotometrically detected by monitoring the absorbance at 548 nm. 3′-p-hydroxyphenyl fluorescein (HPF, Invitrogen), a cell-permeable fluorescent probe, was used to detect OH˙ and ONOO−. Upon reacting with OH˙ and ONOO−, the non-fluorescent HPF was O-dearylated to form fluorescent fluorescein. In this study, fluorescence from the plasma treated probe solution was monitored on a fluorescence spectrophotometer (Cary Eclipse, VARIAN, USA). In addition, 10 ml of PBS containing the probes were used to measure O3, H2O2, and NO2− and 400 μl of PBS were used to measure the amounts of OH˙ and ONOO−.
2.2. Cultivation of bacteria and genetic recombination
The full-length YgaP genes were constructed by site-directed mutagenesis.39 Briefly, full-length genes of YgaP were PCR (polymerase chain reaction) amplified and cloned into a pET-28a-derived vector (modified from pET-28a, Novagen) with an N-terminal hexahistidine tag using the Nde I and Not I restriction sites. The recombinant plasmid was transformed into E. coli. The swc7 gene was amplified from purified genomic DNA extracted from S. cerevisiae S288C. The coding sequence was also cloned into a pET28a-derived expression vector with an N-terminal His 6 tag and over expressed in E. coli strain at 37 °C using the LB culture medium (10 g of NaCl, 10 g of Bacto Tryptone, and 5 g of yeast extract per liter). Both the cloning constructs of YgaP and swc7 were verified by sequencing.402.3. Protein over-expression and plasma treatment
When the YgaP-expressing cells and swc7-expressing cells were grown to an OD600 nm of 0.8, expression of the recombinant proteins was induced with 0.2 mM isopropyl β-D-1-thiogalactopyranoside, and cell growth continued for another 20 h at 16 °C before harvesting. The over-expressed cells were collected by centrifugation at 4000 rpm for 15 min and resuspended in 40 ml of PBS to a concentration 1 × 1010 colony forming units (CFU) per ml for subsequent plasma treatment. In this study, 10 ml of the E. coli suspension on a Petri dish were exposed to the air discharge plasma device for 10 min. The treated suspension of the over-expressed E. coli was stored at 0 °C for 0–6 hours to purify YgaP and swc7. As the model molecule, the induced transmembrane protein Ygap was expressed on the bacterial membrane and the induced intracellular protein swc7 was distributed in the cell matrix.2.4. Protein purification
The over-expressed cells suspension was analyzed to purify the target proteins after the plasma treatment after storing for different time. The plasma-treated cells were resuspended in the lysis buffer (50 mM Tris, 500 mM NaCl, 5% glycerin and pH 8.0) by sonication. After sonication for 20 min and centrifugation at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 30 min, the supernatant containing the soluble target protein was collected and loaded into a nickel-chelating column (GE Healthcare) equilibrated with the binding buffer (100 mM NaCl, 20 mM Tris–Cl, pH 8.0). The target protein was eluted with 300 mM imidazole and further loaded into a HiLoad 16/60 SuperdexTM 200 column (GE Healthcare) pre-equilibrated with 100 mM NaCl, 20 mM Tris–Cl, pH 8.0. Fractions containing the target protein were pooled and concentrated to 10 mg ml−1 for subsequent structural analysis. The protein purity was assessed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and the protein sample was stored at −80 °C. The control was not exposed to the plasma but the other conditions were the same. Fig. 1(c) presents the detailed experimental procedures.
000 × g for 30 min, the supernatant containing the soluble target protein was collected and loaded into a nickel-chelating column (GE Healthcare) equilibrated with the binding buffer (100 mM NaCl, 20 mM Tris–Cl, pH 8.0). The target protein was eluted with 300 mM imidazole and further loaded into a HiLoad 16/60 SuperdexTM 200 column (GE Healthcare) pre-equilibrated with 100 mM NaCl, 20 mM Tris–Cl, pH 8.0. Fractions containing the target protein were pooled and concentrated to 10 mg ml−1 for subsequent structural analysis. The protein purity was assessed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and the protein sample was stored at −80 °C. The control was not exposed to the plasma but the other conditions were the same. Fig. 1(c) presents the detailed experimental procedures.
2.5. Analysis of the primary structure of target proteins
The primary structure of YgaP and swc7 was analyzed by measuring the molecular weight. Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) was conducted to determine the change in the molecular weight of the plasma-treated YgaP and swc7 proteins.2.6. Circular dichroism (CD) spectroscopy
The secondary structure of the controlled and plasma-treated YgaP and swc7 were analyzed by circular dichroism (CD). The CD spectra were acquired in the UV range from 200 to 250 nm on a Jasco-810-CD spectropolarimeter (Japan Spectroscopic Company, Tokyo, Japan) with a 500 μl sample quartz cup at 20 °C. The membrane protein solutions and intracellular protein solutions were diluted from 10 mg ml−1 to 0.15 mg ml−1 with PBS before performing circular dichroism and the secondary structure fractions were calculated from the spectra with the CD software.2.7. Dynamic light scattering and tertiary structure analysis
Before the test, the protein solutions were filtered with inorganic membrane filters (Whatman, Anatop 10 Plus, 0.22 μm), centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 minutes, and degassed while attention was paid to make sure there were no bubbles in the samples. The solutions were put into a sample cell (50 μl) in the Dynapro-MS800 (ATC, England) instrument illuminated with a 25 mW and 750 nm solid-state laser at a fixed scattering angle θ = 90° and equipped with a digital autocorrelator. The measurement was performed at 20 °C and the data were collected after the temperature stabilized for 10 to 15 min. The DynaPro-MS800 was used to estimate the molecular weight and radius according to the model of molecular radius/molecular weight and the particle distribution in the solution was also examined.
000 rpm for 10 minutes, and degassed while attention was paid to make sure there were no bubbles in the samples. The solutions were put into a sample cell (50 μl) in the Dynapro-MS800 (ATC, England) instrument illuminated with a 25 mW and 750 nm solid-state laser at a fixed scattering angle θ = 90° and equipped with a digital autocorrelator. The measurement was performed at 20 °C and the data were collected after the temperature stabilized for 10 to 15 min. The DynaPro-MS800 was used to estimate the molecular weight and radius according to the model of molecular radius/molecular weight and the particle distribution in the solution was also examined.
2.8. Bacteria inactivation assay
Plasma-induced inactivation of E. coli was determined by a colony-forming unit (CFU) assay as shown in the following. After plasma exposure, the E. coli suspensions were serially diluted with deionized water, and 30 μl of each dilution was spread on LB agar plates. The plates were inverted and incubated for 24 h at 37 °C to enable development and subsequent counting of the bacterial colonies. Plating for the CFU assay from the bacterial suspension was processed at different storage time after the plasma treatment.3. Results and discussion
3.1. Plasma-induced RS stress
The bacteria E. coli are immersed in a liquid and so neither charged particles nor RS generated in the plasma (gas) can interact directly with them. However, ROS and RNS in the bacterial suspension produced by the plasma may play important roles in bacterial deactivation.41–43 To evaluate the RS stress, the H2O2, O3, NO2−, OH˙, and ONOO− concentrations in the liquid are measured by fluorescent probes as shown in Fig. 2. The H2O2, O3, and NO2− concentrations increase from 3.16 μM to 327.4 μM, 0.45 μM to 26.5 μM, and 0.35 μM to 54.6 μM, respectively, after plasma exposure for 10 minutes. Meanwhile, the enhanced fluorescence demonstrates that the OH˙ and ONOO− concentrations increase after plasma exposure providing proof that the plasma produces RS in the solution. There are large concentrations of H2O2 (Fig. 2(a)) and O3 (Fig. 2(b)) after storage for 6 h, but the concentration of NO2− (Fig. 2(c)) decreases quickly perhaps because it can be oxidized to NO3− by strong oxidizing RS.27 As long-lived RS, H2O2 and O3 can exist for a relatively long time in the solution.5,27,41 Fluorescence from OH˙ and ONOO− (Fig. 2(d)) return to the original level indicating that the short-lived RS cannot survive be sustained the plasma.44 | ||
| Fig. 2 Reactive species level in the lysis buffer detected by fluorescent probes at 1 h, 3 h, and 6 h after 10 min plasma treatment: (a) H2O2, (b) O3, (c) NO2−, (d) OH˙ and ONOO−. | ||
The increase in RS indicates that the plasma can produce oxidative stress in the E. coli suspension and the oxidative stress can be sustained for a long time (6 h) because of H2O2 and O3. There is direct evidence of ROS generation in PBS or cell culture medium and even in different types of cells.43,45 Generally, RS are the mediator between non-thermal plasmas and cells or bacteria and accumulation of RS triggers sterilization or apoptosis.5,6,23,46 RS are also involved in various important cellular metabolism processes in bacteria and mammalian cells and in fact,47 because of the high reactivity of RS, the plasma-induced effects in cells or bacteria may be multiple and complicated. Plasma-generated RS regulate gene transcription,26 transmembrane signal transduction, and mitochondrial dysfunction in cells or bacteria, and RS-triggered enzyme deactivation in solutions has been studied.27,28 The long-lived RS and short-lived RS may play different roles in the plasma-induced outcome with different effectiveness.27
Here, PBS is employed to simulate the wet conditions at the skin wound surface. To study the plasma efficiency, supplementary experiments on the concentration of H2O2 (serving as a representative of ROS) in PBS added with 5% serum PBS and 10% serum PBS after plasma treatment for 10 minutes are performed. As shown in Fig. 3, after plasma treatment for 10 minutes, PBS has an H2O2 concentration of about 325.2 μM which is the largest among all the plasma-exposed liquids. Meanwhile, the 5% serum PBS contains 247.5 μM of H2O2 while the concentration of the 10% serum PBS is only 220.7 μM. The major cause for the small ROS concentration in PBS in the serum is because a portion of the plasma generated radicals can be scavenged by organic species in the serum. Therefore, if the media are changed, for instance, by adding some serum to the media, the concentration of the plasma generated ROS will change and the plasma efficiency also varies.
 | ||
| Fig. 3 The concentration of H2O2 in different liquid environments (PBS, 5% serum + PBS, 10% serum + PBS) after 10 minutes of plasma treatment. | ||
3.2. Antimicrobial effects of plasma
As shown in Fig. 4, the survival curves of the E. coli suspension after plasma exposure for 10 min and different storage time are obtained by CFU counting. Obvious antimicrobial effects are observed after plasma exposure and after 1 h, the inactivation efficacy corresponds to more than 3-magnitude reduction, meaning that over 99% of the initial population of the bacteria are disinfected. After storage for 3 h and 6 h, the surviving bacteria decline further. Fig. 4 indicates the statistically significant antimicrobial effects on the E. coli compared to the untreated control and the bactericidal effect is mostly observed in the first hour after plasma treatment.As the E. coli bacteria are immersed in the PBS, the plasma-induced antimicrobial effects only stem from ultra-violet (UV) light and dissolved RS. Nevertheless, it has been shown that UV emitted from non-thermal plasmas is below the dose required for bacteria deactivation and hence,48–50 the antimicrobial effects can be attributed to RS stress in the solution in this study. Although RS are considered to be the direct mediator in plasma sterilization, the associated mechanism and why the effects are more prominent in the first hour are not clear. It will be discussed based on RS-induced protein damage later in this paper.
3.3. Protein primary structure
To explore plasma-induced molecular damage in E. coli, the model proteins YgaP and swc7 are over-expressed in E. coli for plasma treatment for 10 min. The YgaP and swc7 are extracted and purified for structural analysis. Both the membrane protein YgaP and intracellular protein swc7 have unique amino acid sequences called the primary structure as the sequence-specific characteristic produces the protein structure and determines the unique functions.51 The key to primary structure changes is whether the plasma can break or destroy the amino-acid chains of the proteins. Here, the amino-acid chains in the plasma-treated YgaP and swc7 aere analyzed by measuring the molecular weight. There will be new small peptide chains or protein fragments in the purified protein solution, if YgaP and swc7 are broken by the plasma treatment. As shown in Fig. 5, the electrophoretic results show that the molecular weights of the membrane protein YgaP and intracellular protein swc7 are not affected after the plasma treatment regardless of storage time. The molecular weights of the treated YgaP and swc7 proteins are consistent with the control suggesting that the plasma cannot break the molecular chains in either the membrane protein or intracellular protein. Plasmas can induce structure changes of protease in vitro and so it is likely that plasmas can modify the structure of YgaP and swc7 proteins in vivo.27,28,30 However, Fig. 5 indicates that the primary structure of YgaP and swc7 proteins is not affected and a possible reason is the protection rendered by the bacteria coating or side chains of the protein.52 | ||
| Fig. 5 Molecular weight changes in the membrane protein YgaP and intracellular protein swc7 after plasma treatment for 10 min and storage from 0 to 6 h. | ||
3.4. Secondary structure
The secondary structure of the treated transmembrane protein YgaP and intracellular protein swc7 is studied by CD at 1 h, 3 h, and 6 h after the plasma treatment. As shown in Fig. 5, the secondary structure of both YgaP and swc7 is modified by the plasma and the degree depends on the storage time. The CD spectra acquired from YgaP indicate that the damage increases gradually with storage time (Fig. 6(a)) and the secondary structure especially the α-helix and the β-sheet which play important roles in the transmembrane region and sulfur transportation of YgaP is altered.39 After storage for 1 h, the α-helix content in YgaP decreases from 34.70 (control) to 28.60%, whereas the β-sheet content increases from 9.50 (control) to 13.20% (Fig. 6(c)). Modification causes structural damage and disfunction of the membrane protein YgaP and the change in the α-helix and β-sheet increases with storage time. The CD spectra obtained from swc7 indicate that the secondary structure is essentially unchanged compared to the control group after storage for 1 h. However, after 3 h and 6 h, structural changes are observed (Fig. 6(b)) supporting Fig. 6(d). Between 1 h and 3 h, the α-helix content decreases from 26.3% to 22.3% and the drops further after 6 h. Each protein has a unique molecular structure which determines the biological function.53 The secondary change can destroy the structural domain of YgaP and swc7 leading to loss of the original molecular functions. On the molecular level, PCD (programmed cell death) of cells can result from long-term accumulation of various types of molecular and cellular damage.54,55 In our study, after plasma treatment for 1 h, the membrane protein YgaP is modified by the plasma, but the intracellular swc7 is not affected. It may be because of the protection offered by the cell walls and membranes of E. coli to intracellular proteins.56 In fact, almost all the E. coli are disinfected in the first hour after plasma treatment and therefore, plasma-induced membrane protein damage may be the major reason for the bacterial death. The communication between bacteria and the surrounding environment relies membrane receptor proteins and membrane channel proteins.57,58 When membrane proteins are damaged by the plasma, bacteria death results in this extreme environment.54In addition, the effects of different plasma exposure time on E. coli (Fig. 7(a) and (b)) are studied. The reason for choosing 10 min plasma exposure is because the effects are moderate. A shorter exposure time produces insufficient modification on the model proteins and so it is difficult to investigate. As shown in Fig. 7, the secondary structure change in the membrane protein YgaP and intracellular protein swc7 is hardly visible after 6 h with 5 min plasma treatment. This may be because the inadequate plasma dosage and ROS generated during exposure for 5 min are not enough to modify the model proteins visibly. Nonetheless, in our future experiments, we will try to investigate the effects of a shorter exposure time on proteins in bacteria by improving the detection techniques.
 | ||
| Fig. 7 Secondary structure percentages of membrane protein YgaP and intracellular protein swc7 after plasma treatment for 5 min and storage for 6 h. | ||
3.5. Tertiary structure
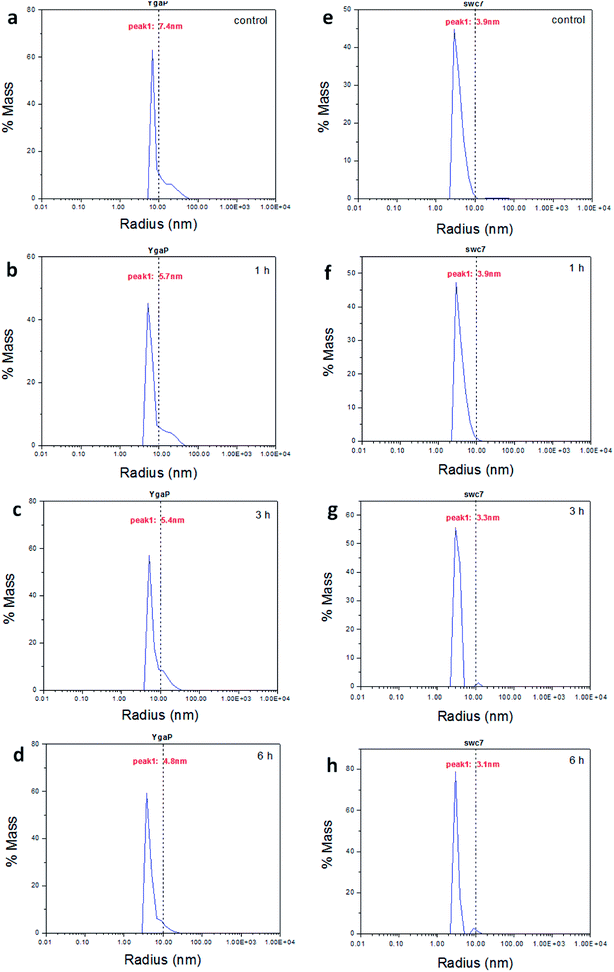
Changes in the secondary structure can alter the tertiary structure and this topic is also explored. The hydrodynamic radius and particle distribution of the controlled and plasma-treated YgaP and swc7 proteins are investigated by DLS as shown in Fig. 8. Fig. 8(a) shows that the pristine membrane protein YgaP has a molecular radius of 7.4 nm. The average hydrodynamic radii of the treated YgaP sulfurtransferase diminish to 5.7 nm, 5.4 nm, and 4.8 nm after storage for 1 h, 3 h, and 6 h, respectively (Fig. 8(b)–(d)). Condensation is typical in the hydrophobic collapse model and the intracellular swc7 of the untreated bacterial suspension has a hydrodynamic radius of 3.9 nm (Fig. 8(e)).59 The hydrodynamic radius of the intracellular swc7 after storage for 1 h is almost the same as that of the control (Fig. 8(f)). Fig. 8(g) shows the plasma treatment effects on intracellular swc7 after 3 h revealing enhanced cohesion of the intracellular swc7 and the hydrodynamic radius of a large part of the swc7 protein decreases to 3.3 nm disclosing that the plasma also modifies the tertiary structure of swc7 producing condensation similar to the membrane protein YgaP. Fig. 8(h) shows further collapse of the swc7 molecule after 6 h. The DLS results confirm that the plasma not only modifies the secondary structure, but also alters the tertiary structure of YgaP and swc7 thus furnishing evidence that the membrane protein is more susceptible to plasma modification than intracellular proteins in the first hour after plasma exposure.In this study, the model proteins are prepared in E. coli. The cell wall and cell membrane of E. coli can protect the cell components from damage to some extent.60 YgaP is modified in the first hour after plasma treatment, whereas modification of swc7 takes a longer storage time (3 h and 6 h), suggesting that the cell wall and cell membrane can delay the plasma-induced modification of intracellular molecules. In addition, the non-thermal dielectric barrier discharge (DBD) plasmas can modify the structure of lactate dehydrogenase (LDH) enzyme via direct interaction with bio-macromolecule. It has been shown that the α-helix content of LDH decreases from 33.20 to 15.40%, whereas the β-sheet content increases from 12.30 to 26.60% after treatment for 5 min and storage for 6 h.27 However, the degree of plasma modification on YgaP and swc7 in E. coli is weaker than the change of the DBD treated LDH solution because the cell wall and membrane attenuates the plasma potency.
Fig. 2 indicates that the plasma can produce various RS which can damage cell molecules. Excess RS cause oxidative stress which results in irreversible injury on both the molecular and cellular levels.61 Although the plasma-generated RS cannot modify the primary structure of the model proteins, they are still regulators for the secondary structure and tertiary structural changes in YgaP and swc7, but owing to the protection by the cell wall and cell membrane, only long-lasting RS such as H2O2, O3, and NO2− may play a key role.27 These long-standing RS cross cell membranes through aquaporins or by diffusion and work on swc7.62 Although the oxidizing properties of short-lived OH˙ and ONOO− are very strong, the decay is quick. Hence, there is not enough time for OH˙ and ONOO− to penetrate the cell wall and membrane to modify the intracellular swc7 and so the impact on swc7 is limited. However, for the membrane protein YgaP, both long-standing and short-lived RS can damage its structure. In addition, the microstructure of the cell membrane is damaged and the membrane permeability increases resulting in leakage.63 The RS can penetrate the membrane more easily to trigger protein damage. For example, H2O2 oxidize proteins effectively causing denaturing and many amino acids can be modified by ozone.64,65 Therefore, the plasma-induced protein damage may result from the combined action of the various RS.66 It is noted that RS also can damage DNA, RNA, and other important functional molecules giving rise to metabolic abnormalities.62,67
In this study, plasma induced ROS generation in the liquid phase and inactivation of E. coli are investigated and modification of the membrane protein YgaP and intracellular protein swc7 are also studied. ROS produced by plasmas are considered to play major roles in inactivation of bacteria. O. Lunov et al. suggested that in air plasmas, ozone played an important role in ROS induction and subsequent inactivation of bacteria.3 In addition, S. Maheux et al. indicated that formation of NH4+ species in the saline solution treated with a cold atmospheric plasma was mainly responsible for fast bacterial inactivation.9 As for the sterilization mechanism, O. Lunov et al. found that plasma induced bacteria death might arise from membrane damage6 and R. A. Chinga et al. surmised that plasma might contribute to bacteria death by causing DNA and biomolecule damage.13 Here, our results reveal damage of the membrane and intracellular proteins in E. coli. Based on the comparison between the death time of E. coli after the plasma treatment and storage time, the possible sterilization mechanism can be postulated. As shown in Fig. 9, a short storage time (1 h) after plasma exposure is enough to damage the membrane proteins but not intracellular protein. The damaged membrane triggers death of most bacteria and both the long-lived and short-lived reactive oxygen species are involved in the membrane alteration. Plasma triggered cellular membrane damage is found to be the most likely and pivotal step in this disinfection mechanism. During this period, because of the protection rendered by the cell walls and membranes of E. coli to intracellular proteins, there is not enough time for ROS to modify the swc7. For the longer storage time (3 h and 6 h), both the membrane and intracellular proteins are modified, including other biomolecules such as DNA and mRNA. The plasma-induced toxicity on functional molecules also leads to destruction and death of the bacteria. In this time frame, only the remaining long-lived reactive species modify the intracellular biomolecules.
4. Conclusion
An air discharge plasma jet is employed to modify membrane proteins YgaP and intracellular proteins swc7 as the model biomolecules in E. coli suspensions. The plasma treatment modifies the secondary structure and tertiary structure of YgaP and swc7 in E. coli, but does not affect the primary structure. The damage on YgaP and swc7 depends on the storage time. The plasma treatment produces structural change in YgaP in 1 h after plasma exposure, but 3 h to 6 h for swc7. Protein damage involves dissolved RS and onset of oxidative stress in the E. coli suspension. The RS are reactive and toxic producing oxidative destruction of the biomolecules and eventually cell death.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China under Grant No.11675177, No.11475174 and No. 51777206, Natural Science Foundation of Anhui Province Grant No. 1708085MA13, as well as Hong Kong Research Grants Council (RGC) General Research Funds (GRF) No. CityU 11301215 and 11205617.References
- M. G. Kong, G. Kroesen, G. Morfill, T. Nosenko, T. Shimizu, J. Van Dijk and J. L. Zimmermann, New J. Phys., 2009, 11, 115012 CrossRef.
- G. Fridman, G. Friedman, A. Gutsol, A. B. Shekhter, V. N. Vasilets and A. Fridman, Plasma Processes Polym., 2008, 5, 503–533 CrossRef.
- O. Lunov, V. Zablotskii, O. Churpita, A. Jäger, L. Polívka, E. Syková, N. Terebova, A. Kulikov, Š. Kubinová and A. Dejneka, RSC Adv., 2016, 6, 25286 RSC.
- Z. Xu, J. Shen, Z. Zhang, J. Ma, R. Ma, Y. Zhao, S. Qiang, S. Qian, Z. Hao, L. Ding, C. Cheng and P. K. Chu, Plasma Processes Polym., 2015, 12, 827–835 CrossRef.
- J. Shen, Q. Sun, Z. Zhang, C. Cheng, Y. Lan, H. Zhang, Z. Xu, Y. Zhao, W. Xia and P. K. Chu, Plasma Processes Polym., 2015, 12, 252–259 CrossRef.
- O. Lunov, V. Zablotskii, O. Churpita, A. Jäger, L. Polívka, E. Syková, L. Dejneka and S. Kubinov, Biomaterials, 2016, 82, 71–83 CrossRef PubMed.
- X. Lu, G. V. Naidis, M. Laroussi and K. Ostrikov, Phys. Rep., 2014, 540, 123–166 CrossRef.
- C. Cheng, S. Jie, X. De-Zhi, X. Hong-Bing, L. Yan, F. Shi-Dong, M. Yue-Dong and P. K. Chu, Chin. Phys. B, 2014, 23, 075204 CrossRef.
- S. Maheux, D. Duday, T. Belmonte, C. Penny, H. M. Cauchie, F. Clement and P. Choquet, RSC Adv., 2015, 5, 42135 RSC.
- K. Bazaka, M. V. Jacob, W. Chrzanowski and K. Ostrikov, RSC Adv., 2015, 5, 48739 RSC.
- N. Puac, M. Miletic, M. Mojovic, A. Popovic-Bijelic, D. Vukovic, B. Milicic, D. Maletic, S. Lazovic, G. Malovic and Z. L. Petrovic, Open Chem., 2015, 13, 332–338 Search PubMed.
- T. Itarashiki, N. Hayashi and A. Yonesu, Vacuum, 2014, 110, 213–216 CrossRef.
- R. A. Chinga, J. Lin and S. Roy, IEEE Trans. Plasma Sci., 2014, 42, 1861–1869 CrossRef.
- S. P. Kuo, O. Tarasenko, J. Chang, S. Popovic, C. Y. Chen, H. W. Fan, A. Scott, M. Lahiani, J. D. Drake, P. Alusta and M. Nikolic, New J. Phys., 2009, 11, 115016 CrossRef.
- D. H. Lee, J. O. Lee, W. Jeon, I. G. Choi, J. S. Kim, J. H. Jeong, T. Kang and C. H. Seo, Appl. Phys. Lett., 2011, 99, 203701 CrossRef.
- V. Vosmanská, K. Kolářová, S. Rimpelová, Z. Kolská and V. Švorčík, RSC Adv., 2015, 5, 17690 RSC.
- D. Dobrynin, G. Fridman, G. Friedman and A. Fridman, New J. Phys., 2009, 11, 115020 CrossRef.
- N. K. Kaushik, N. Kaushik, K. C. Yoo, N. Uddin, J. S. Kim, S. J. Lee and E. H. Choi, Biomaterials, 2016, 87, 118–130 CrossRef PubMed.
- N. Kumar, P. Attri, E. H. Choi and H. S. Uhm, RSC Adv., 2015, 5, 14670 RSC.
- J. Ehlbeck, U. Schnabel, M. Polak, J. Winter, T. Woedtke, R. Brandenburg, T. Von, D. Hagen and K. D. Weltmann, J. Phys. D: Appl. Phys., 2011, 44, 013002 CrossRef.
- K. Duske, L. Jablonowski, I. Koban, R. Matthes, B. Holtfreter, A. Sckell, J. B. Nebe, T. V. Woedtke, K. D. Weltmann and T. Kocher, Biomaterials, 2015, 52, 327–334 CrossRef PubMed.
- O. Lunov, O. Churpita, V. Zablotskii, I. G. Deyneka, I. K. Meshkovskii, A. Jäger, E. Sykova, S. Kubinov and A. Dejneka, Appl. Phys. Lett., 2015, 106, 053703 CrossRef.
- S. U. Kang, J. H. Cho, J. W. Chang, Y. S. Shin, K. I. Kim, J. K. Park, S. S. Yang, J. S. Lee, E. Moon, K. Lee and C. H. Kim, Cell Death Dis., 2014, 5, e1056 CrossRef PubMed.
- J. Shen, C. Cheng, S. Fang, H. Xie, Y. Lan, G. Ni, Y. Meng, J. Luo and X. Wang, Appl. Phys. Express, 2012, 5, 036201 CrossRef.
- J. W. Chang, S. U. Kang, Y. S. Shin, K. I. Kim, S. J. Seo, S. S. Yang, J. S. Lee, E. Moon, S. J. Baek, K. Lee and C. Kim, Arch. Biochem. Biophys., 2014, 545, 133–140 CrossRef PubMed.
- Z. Xu, J. Wei, J. Shen, Y. Liu, R. Ma, Z. Zhang, S. Qian, J. Ma, Y. Lan, H. Zhang, W. Xia, Q. Sun, C. Cheng and P. K. Chu, Appl. Phys. Lett., 2015, 106, 034005 Search PubMed.
- H. Zhang, Z. Xu, J. Shen, X. Li, L. Ding, J. Ma, Y. Lan, W. Xia, C. Cheng, Q. Sun, Z. Zhang and P. K. Chu, Sci. Rep., 2015, 5, 10031 CrossRef PubMed.
- Z. Ke and Q. Huang, Plasma Processes Polym., 2013, 10, 731–739 CrossRef.
- H. P. Li, L. Y. Wang, G. Li, L. H. Jin, P. S. Le, H. X. Zhao, X. H. Xing and C. Y. Bao, Plasma Processes Polym., 2011, 8, 224–229 CrossRef.
- E. Takai, K. Kitano, J. Kuwabara and K. Shiraki, Plasma Processes Polym., 2012, 9, 77–82 CrossRef.
- B. Surowsky, A. Fischer, O. Schlueter and D. Knorr, Innovative Food Sci. Emerging Technol., 2013, 19, 146–152 CrossRef.
- H. Tolouie, M. A. Mohammadifar, H. Ghomi and M. Hashemi, Crit. Rev. Food Sci. Nutr., 2017, 1–15 CrossRef PubMed.
- P. Attri, T. Sarinont, M. Kim, T. Amano, K. Koga, A. E. Cho, H. C. Eun and M. Shiratani, Sci. Rep., 2015, 5, 17781 CrossRef PubMed.
- R. Zhou, R. Zhou, J. Zhuang, Z. Zong, X. Zhang, D. Liu, K. Bazaka and K. Ostrikov, PLoS One, 2016, 11, e0155584 Search PubMed.
- J. H. Park, M. Kim, M. Shiratani, A. E. Cho, E. H. Choi and P. Attri, Sci. Rep., 2016, 6, 35883 CrossRef PubMed.
- S. Choi, P. Attri, I. Lee, J. Oh, J. H. Yun, J. H. Park, H. C. Eun and W. Lee, Sci. Rep., 2017, 7, 1027 CrossRef PubMed.
- Z. M. Xu, C. Cheng, J. Shen, Y. Lan, S. H. Hu, W. Han and P. K. Chu, Bioelectrochemistry, 2018, 121, 125–134 CrossRef PubMed.
- P. A. Krishna and M. C. Alisa, Plasma Processes Polym., 2011, 8, 1154–1164 CrossRef.
- S. L. Ling, W. Wang, L. Yu, J. H. Peng, X. Y. Cai, Y. Xiong, Z. Hayati, L. Zhang, Z. Zhang, L. Song and C. Tian, Sci. Rep., 2016, 6, 20025 CrossRef PubMed.
- X. H. Bai, H. J. Chen, Y. L. Jiang, Z. Wen, Y. Huang, W. Cheng, Q. Li, L. Qi, J. Zhang, Y. Chen and C. Zhou, J. Biol. Chem., 2014, 289, 23403–23416 CrossRef PubMed.
- J. Ma, H. Zhang, C. Cheng, J. Shen, L. Bao and W. Han, Plasma Processes Polym., 2017, 14, 7 Search PubMed.
- G. J. Kim, W. Kim, K. T. Kim and J. K. Lee, Appl. Phys. Lett., 2010, 96, 021502 CrossRef.
- H. M. Joh, S. J. Kim, T. H. Chung and S. H. Leem, Appl. Phys. Lett., 2012, 101, 053703 CrossRef.
- Z. C. Liu, D. X. Liu, C. Chen, D. Li, A. J. Yang, M. Z. Rong, H. L. Chen and M. G. Kong, J. Phys. D: Appl. Phys., 2015, 48, 495201 CrossRef.
- N. Mastanaiah, J. A. Johnson and S. Roy, PLoS One, 2013, 8, e70840 Search PubMed.
- F. X. Liu, P. Sun, N. Bai, Y. Tian, H. X. Zhou, S. C. Wei, Y. Zhou, J. Zhang, W. Zhu, K. Becker and J. Fang, Plasma Processes Polym., 2010, 7, 231–236 CrossRef.
- R. Mittler, S. Vanderauwera, N. Suzuki, G. Miller, V. B. Tognetti, K. Vandepoele, M. Gollery, V. Shulaev and F. V. Breusegem, Trends Plant Sci., 2011, 16, 300 CrossRef PubMed.
- M. Laroussi, Plasma Processes Polym., 2005, 2, 391–400 CrossRef.
- Z. Machala, L. Chladekova and M. Pelach, J. Phys. D: Appl. Phys., 2010, 43, 222001 CrossRef.
- M. Laroussi and F. Leipold, Int. J. Mass Spectrom., 2004, 233, 81–86 CrossRef.
- F. Jacob and J. Monod, J. Mol. Biol., 1961, 3, 318–356 CrossRef PubMed.
- B. S. Berlet and E. R. Stadtman, J. Biol. Chem., 1997, 272, 20313–20316 CrossRef.
- R. H. Holm, P. Kennepohl and E. I. Solomon, Chem. Rev., 1996, 96, 2239–2314 CrossRef PubMed.
- K. Kannan and S. K. Jain, Pathophysiology, 2000, 7, 153–163 CrossRef PubMed.
- K. Lewis, Microbiol. Mol. Biol. Rev., 2000, 64, 503–514 CrossRef PubMed.
- G. D. Shockman, L. Daneo-Moore and M. L. Higgins, Ann. N. Y. Acad. Sci., 1974, 235, 161–197 CrossRef PubMed.
- G. Schatz and B. Dobberstein, Science, 1996, 271, 1519 Search PubMed.
- N. M. Kumar and N. B. Gilula, Cell, 1996, 84, 381–388 CrossRef PubMed.
- R. Zhou, X. Huang, C. J. Margulis and B. J. Berne, Science, 2004, 305, 1605–1609 CrossRef PubMed.
- M. E. Goetz and A. Luch, Cancer Lett., 2008, 266, 73–83 CrossRef PubMed.
- E. Cabiscol, J. Tamarit and J. Ros, Int. Microbiol., 2010, 3, 3–8 Search PubMed.
- F. Antunes and E. Cadenas, FEBS Lett., 2000, 475, 121–126 CrossRef PubMed.
- H. Liu, Y. Du, X. Wang and L. Sun, Int. J. Food Microbiol., 2004, 95, 147–155 CrossRef PubMed.
- J. Nordberg and E. S. Arner, Free Radicals Biol. Med., 2001, 31, 1287–1312 CrossRef PubMed.
- F. Cataldo, Polym. Degrad. Stab., 2005, 89, 274–281 CrossRef.
- K. A. Dill, Biochemistry, 1990, 29, 7133–7155 CrossRef PubMed.
- H. Wiseman and B. Halliwell, Biochem. J., 1996, 313, 17 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2018 |