Open Access Article
Open Access ArticlePeroxoniobium inhibits leukemia cell growth†
Elene C. Pereira-Maia*a,
Ivina P. Souza‡
a,
Kelen J. R. C. Nunes‡a,
Alexandre A. Castro‡b,
Teodorico C. Ramalho‡ b,
Fernando Stefflera,
Helio A. Duarte
b,
Fernando Stefflera,
Helio A. Duarte a,
Ana Pacheli‡
a,
Ana Pacheli‡ a,
Poliana Chagas‡a and
Luiz C. A. Oliveira*a
a,
Poliana Chagas‡a and
Luiz C. A. Oliveira*a
aUniversidade Federal de Minas Gerais, Department of Chemistry, Av. Antonio Carlos 6627, Belo Horizonte, MG 31270-901, Brazil. E-mail: elene@qui.ufmg.br; lcao.ufmg@gmail.com
bUniversidade Federal de Lavras, Department of Chemistry, Lavras, MG 37200-000, Brazil
First published on 14th March 2018
Abstract
A new class of polyoxoniobate complex has been synthesized and characterized as a novel anticancer agent for photodynamic therapy. The complex inhibits the growth of chronic myelogenous leukemia cells with an IC50 value of 30 μM, in the dark. However, upon exposure to light (365 nm) there is a fivefold increase in the cytotoxic activity. Light radiation activate the complex with the formation of radical species capable of interacting with DNA according to our experimental and theoretical data.
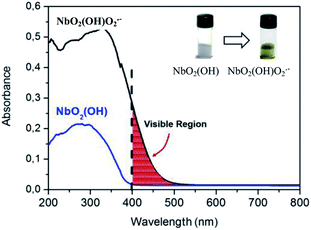
In this work, we prepared a photosensitive peroxoniobium complex presenting a balance with an active radical phase when illuminated with radiation of 365 nm. A versatile niobium species of amorphous structure was obtained by the reaction of niobium ammonium oxalate with ammonium hydroxide up to pH 7. The material obtained, a niobium oxyhydroxide (NbO2(OH)) (white solid),1,2 can be modified with the generation of NbO2(OH)O2˙− peroxo groups (yellow solid).3 The yellow compound is formed by treatment with H2O2. The absorption radiation in the visible region due to the charge transfer transition between the peroxo group and the niobium is shown in Fig. 1.
This complex with the radical as an intermediate is favored in the presence of visible and UV radiation. This property is of interest for photodynamic therapy of cancer (PDT), which involves the exposure of malignant cells containing a photosensitizer molecule to light irradiation, in the presence of oxygen species. The photoactivated drug produces reactive oxygen species that initiate a series of events, resulting in cell death. Selective light activation allows a preferential tumor destruction in comparison to healthy tissues.4 Several metal complexes exhibit photocytotoxicity under UV or visible light,5,6 but data about niobium compounds are very scarce in the literature.7
The polyoxoniobate, generated from niobium oxyhydroxide described here can be very active in the treatment of diseased cells when illuminated with visible or UV radiation due to its light absorption capacity because of the peroxo groups formed. The peroxoniobium complex has some advantages, such as ease synthesis and in mild conditions, high solubility, low activity under light off, and resistance to inactivation by thiol reagents. Moreover, it is nontoxic8 and does not employ noble metals like most of the compounds proposed in the literature. Actually, niobium oxide was tested as a bone implant component and showed absence of inflammatory cells or degeneration of the osteoblasts without any sign of damage to the preexisting bone tissue, showing compatibility with the bone tissue.9–11
The innovative part in the process of obtaining the polyoxoniobate complex presented in this work consists in the leaching of the complex when treating the niobium oxyhydroxide with H2O2. With the treatment, a yellow solid and a leached yellow liquid is obtained, which is the complex containing peroxoniobium in its structure, sensitive to the UV-Vis radiation generating radical species. This species generated with the leaching at neutral pH presents high negative charge and a kinetic volume of 223 nm. The XRD of the lyophilized polyoxoniobate indicated strong amorphous character. However, the polyoxoniobate is known to form well defined polyoxometalates such as Lindqvist ([Nb6O19]8−) and decaniobate ([Nb10O28]6−). Fig. 2 shows a comparison between the experimental and PBE/LANL2DZ/aug-cc-pVDZ DFT IR spectra. It is clear that the pattern of the decaniobate structure is closer to the experimental spectrum. One should keep in mind that DFT frequencies are normally 10% underestimated with respect to the experimental values. The broader absorption below 600 cm−1 can be attributed to the interaction between different decaniobate structures forming the amorphous solid. The calculated peaks at 710, 760 and 860 cm−1 are related to the experimental peaks of 800, 870 and 910 cm−1 indicating that decaniobate is the local arrangement of the polyoxoniobate complex.
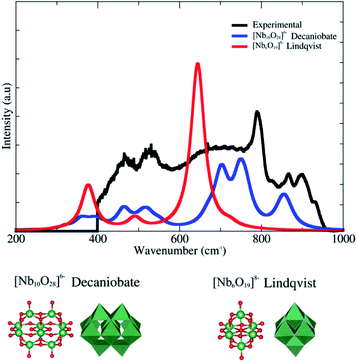 | ||
| Fig. 2 Infrared spectra for experimental procedures, Lindqvist and decaniobate structures (simulated). The line shape chosen was Lorentzian and the half-width is about 20. | ||
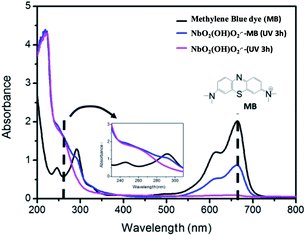
The generation of reactive oxygen under radiation was confirmed by the reaction of the complex with an organic dye, which was monitored by UV-Vis spectroscopy (Fig. 3). The spectrum of the dye solution shows the characteristic peak of the methylene blue (MB) at 663 nm (black trace). It can be clearly seen that in the presence of the peroxoniobium complex and radiation (365 nm) the signal decreased indicating the reaction of the peroxoniobium complex with the dye (blue trace). In the absence of light, there is no decrease in the signal related to the dye, indicating the need of the radiation to activate the oxidation action of the peroxoniobium complex. The equilibrium in which the radical species forms it is not necessary to use a photosensitizer agent, such as porphyrins.4 A further investigation was carried out by 31P NMR (Fig. S1†) using guanosine as model molecule able to react with the peroxoniobium complex. The 31P NMR spectrum shown in Fig. S1-a† corresponds to 5-GMP and revealed that the phosphorus atom in the structure exhibits a chemical shift at δ 5.93. When 5-GMP and the polyoxoniobate are in contact, no significant changes are observed in the 31P spectrum, only a small displacement of the phosphorus signal to δ 5.90 (Fig. S1-b†). However, when the 5-GMP and polyoxoniobate mixture is submitted to radiation (Fig. S1-c†), an interaction between the compounds occurs, giving rise to a new species that presents a different chemical shift in the P spectrum (δ 5.27).
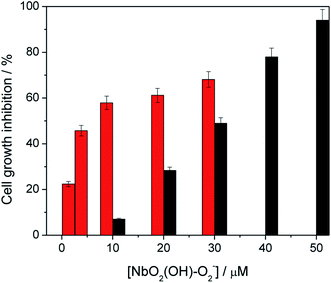
The effect of the peroxoniobium on the growth of K562 cells was evaluated after 4 h of incubation. The compound inhibits K562 cell growth in a concentration-dependent manner, with an IC50 of 30.0 ± 1.5 μmol L−1. Ammonium niobate(V) oxalate was also tested and it has no effect on K562 cells up to 100 μM. The cytotoxic activity of polyoxoniobate increases by 5 times upon 5 min of UV-A light irradiation, with an IC50 value of 6.2 ± 0.4 μmol L−1 (Fig. 4). The higher activity, when exposed to light, associated to the low toxicity of niobium compounds place the peroxoniobium complex as a candidate for photodynamic therapy.
There are few reports in the literature about the cytotoxic activity of niobium compounds. A peroxo niobium complex with ascorbic acid (K3[Nb(Asc)(O2)3]) is moderately active in HL60 human leukemia cells but not in K562 human myelogenous leukemia cells.12 A tetrameric Nb28-containing cluster inhibits the growth of the human breast cancer MCF-7 cells line with an IC50 value of 5.21, after 48 h of incubation.13
Methylene blue (MB) is one of the main photosensitizing agents used in PDT due to its good tissue penetration and low cytotoxicity.14 It is active in several types of tumors upon irradiation with red laser light.15 This fact allied to the ability of the peroxoniobium compound to interact with MB (Fig. 3) prompted us to investigate its effect in the MB photocytotoxicity. We have first checked that exposure to UV-A light did not affect the cytotoxicity of MB in K562 cells (Table 1). Afterwards, the photocytotoxicity of MB in K562 cells was assessed in the presence of the peroxoniobium complex at its IC50 value. Cells were incubated with increasing concentrations of MB and irradiated for 5 min with UV-A light, in the absence and the presence of 6.5 μM of the peroxoniobium complex. Upon light irradiation, 6.5 μM of peroxoniobium complex doubles the effect of MB. Therefore, the peroxoniobium compound prepared is a promising candidate for PDT therapy, alone or in combination with other sensitizing agents, such as methylene blue dye (Table 1).
| Compound | IC50a | IC50 irradiatedb |
|---|---|---|
| a IC50 Methylene blue concentration required to inhibit 50% of cell growth under dark conditions.b IC50 Methylene blue concentration required to inhibit 50% of cell growth after 5 min of UV-A irradiation.c Assays were performed in the presence of 6.5 μM of the peroxoniobium complex. | ||
| MB | 7.3 ± 0.4 | 7.0 ± 0.5 |
| MB + NbO2(OH)–O2−c | 6.3 ± 0.3 | 3.0 ± 0.1 |
The peroxoniobium complex (Fig. S2†) and the DNA/complex systems were thus fully optimized at DFT level,16 with conjugate gradient and quasi-Newton–Raphson algorithms. The final geometries were obtained with the density functional Becke's three-parameter exchange functional and the gradient-corrected functional of Lee, Yang and Paar (B3LYP),17 using LanL2DZ basis set.
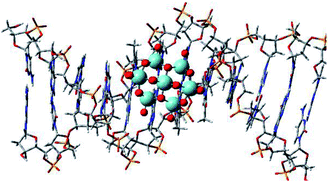
The DICKERSON-DREW B-DNA DODECAMER was obtained from the Protein Data Bank (PDB), with code 4C64 and resolution: 1.32 Å (ref. 18 and 19) and it was chosen as model according to previous works, and has shown suitable for our calculations.20 As previously discussed, the Nb complexes (Fig. S2†) were entirely optimized at the DFT level, to obtain the most stable initial geometries to perform the calculations with the DNA structure. It is important to mention that the more stable complex/DNA system is related to a higher cytotoxicity potential. The following species were considered for this theoretical investigation: Complex a (no radical groups), Complex b (protonated structure) and Complex c (radicals formation). The natural charges of all atoms were elucidated, and according to these data, it is possible to realize more pronounced negative charges referred to the radical species. The protonated substituent (OOH) presented a charge value equals to −0.534 a.u., while the corresponding radical (deprotonated) has shown a charge value significantly lower (−1.170 a.u.). The same is observed for the substituent (OH), with a charge value of −0.379 a.u., and the corresponding radical (−0.739 a.u.). The formation of more negative charges suggests to the highest reactivity of Complex c, in relation to the other complexes. Complex a was put together with DNA in three distinct regions (Fig. S3†), and after performing the optimization, the Nb complex reactivity was analyzed in these zones.
PM6 calculations were performed in order to evaluate the Nb complex (Complex a) affinity in different DNA regions. These results are presented in Table S1.† as relative interaction energy values. The Complex a, when put into different regions of DNA, presented quite relevant changes in relation to the intermolecular interaction energy. Thus, Complex a, when docked into the central region of DNA, showed a more favorable energy (Fig. 5). After the optimization structure, the region with the highest interaction between DNA and Nb complexes was considered for other calculations.
According to our theoretical methodology, we have the energy minimizations for the systems: free DNA (EDNA), free complex (Ecomplex) and DNA-complex system (Esystem). In line with those systems, the affinity energy was calculated using the following equation:
| E = EDNA/complex − Ecomplex − EDNA | (1) |
The results from this methodology are described in Table S2,† also as relative interaction energy values. By using the strategy described at the ESI,† it was possible to analyze the efficiency of Nb complexes towards DNA, evaluating which factors contribute most to this reactivity. Our first results indicate that the Nb complex, in general, presents a significant affinity with DNA, with a pronounced increase in the affinity/reactivity in the presence of radical groups (–OO˙. radical for example). According to our calculations, Complex a showed an intermediate reactivity towards DNA, presenting a significantly higher energy value than that obtained for Complex c. The energy difference between Complex a and c was 2.88 kcal mol−1. On the other hand, the energy difference between Complex b and c was remarkable, about 5.01 kcal mol−1. Complex c presented good interaction potential with DNA, undoubtedly due to the radical groups added to the complex structure, coherent with the experimental observations. According to the computational investigation, we can conclude that all Nb complex species (without and with radical groups) presented reactivity and stability when docked into the DNA crystallographic structure. These results corroborate with the experimental data observed in the reaction of guanosine with the niobium complex shown in the 31P NMR of Fig. S1.† Furthermore, the addition of radical groups substantially increases the affinity of the complex towards DNA, supported by the obtaining of more stable structures for the complex/DNA system (lowest energy values), suggesting higher levels of cytotoxicity.
Conflicts of interest
The authors declare no competing financial interests.Acknowledgements
The authors are grateful to FAPEMIG, CNPq, INCT-MIDAS, INCT-catálise, and UFMG for the financial support.References
- L. C. A. Oliveira, T. C. Ramalho, M. Gonçalves, F. Cereda, K. T. Carvalho, M. S. Nazzarro and K. Sapag, Chem. Phys. Lett., 2007, 446, 133–137 CrossRef CAS.
- A. Esteves, L. C. A. Oliveira, T. C. Ramalho, M. Goncalves, A. S. Anastacio and H. W. P. Carvalho, Catal. Commun., 2008, 10, 330–332 CrossRef CAS.
- L. C. A. Oliveira, M. F. Portilho, A. C. Silva, H. a. Taroco and P. P. Souza, Appl. Catal., B, 2012, 117–118, 29–35 CrossRef CAS.
- B. S. Howerton, D. K. Heidary and E. C. Glazer, J. Am. Chem. Soc., 2012, 134, 8324–8327 CrossRef CAS PubMed.
- T. Bortolotto, P. P. Silva, A. Neves, E. C. Pereira-Maia and H. Terenzi, Inorg. Chem., 2011, 50, 10519–10521 CrossRef CAS PubMed.
- T. Bortolotto, P. P. Silva-Caldeira, C. T. Pich, E. C. Pereira-Maia and H. Terenzi, Chem. Commun., 2016, 7130, 7130–7133 RSC.
- A. Blázquez-Castro, J. C. Stockert, B. López-Arias, A. Juarranz, F. Agulló-López, A. García-Cabañes and M. Carrascosa, Photochem. Photobiol. Sci., 2011, 10, 956–963 Search PubMed.
- N. A. Dsouki, M. P. de Lima, R. Corazzini, T. M. Gáscon, L. A. Azzalis, V. B. C. Junqueira, D. Feder and F. L. A. Fonseca, J. Mater. Sci.: Mater. Med., 2014, 25, 1301–1305 CrossRef CAS PubMed.
- G. V. O. Fernandes, G. G. Alves, a. B. R. Linhares, M. H. Prado da Silva and J. M. Granjeiro, Key Eng. Mater., 2011, 493–494, 37–42 CrossRef.
- M. Tamai, K. Isama, R. Nakaoka and T. Tsuchiya, J. Artif. Organs, 2007, 10, 22–28 CrossRef CAS PubMed.
- M. Kushwaha, X. Pan, J. A. Holloway and I. L. Denry, Dent. Mater., 2012, 28, 252–260 CrossRef CAS PubMed.
- H. Thomadaki, A. Lymberopoulou-Karaliota, A. Maniatakou and A. Scorilas, J. Inorg. Biochem., 2011, 105, 155–163 CrossRef CAS PubMed.
- D. Zhang, Z. Liang, S. Xie, C. Zhang, J. Wang and J. Niu, Inorg. Chem., 2014, 6, 6–11 Search PubMed.
- S. M. L. Gontijo, R. C. Felizali, P. P. G. Guimarães, R. A. S. Santos, R. D. Sinisterra, M. E. Cortés and P. V. Araújo, Photodiagn. Photodyn. Ther., 2017, 18, 252–256 CrossRef CAS PubMed.
- J. P. Tardivo, A. Del Giglio, C. S. De Oliveira, D. S. Gabrielli, H. C. Junqueira, D. B. Tada, D. Severino, R. De Fatima Turchiello and M. S. Baptista, Photodiagn. Photodyn. Ther., 2005, 2, 175–191 CrossRef CAS PubMed.
- C. J. Cramer and D. G. Truhlar, Phys. Chem. Chem. Phys., 2009, 11, 10757–10816 RSC.
- E. F. F. Cunha, E. F. Barbosa, A. A. Oliveira, T. C. Ramalho, A. A. Oliveira and T. C. Ramalho, J. Biomol. Struct. Dyn., 2010, 27, 619–625 Search PubMed.
- L. Lercher, M. A. McDonough, A. H. El-Sagheer, A. Thalhammer, S. Kriaucionis, T. Brown and C. J. Schofield, Chem. Commun., 2014, 50, 1794–1796 RSC.
- U. Issar, T. Kumari and R. Kakkar, J. Comput. Sci., 2015, 10, 166–177 CrossRef.
- P. Haris, V. Mary, M. Haridas and C. Sudarsanakumar, J. Chem. Inf. Model., 2015, 55, 2644–2656 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra00898a |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2018 |