Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Highly efficient synthesis of arbutin esters catalyzed by whole cells of Candida parapsilosis†
Xiaofeng Li ab,
Haixia Xuab,
Guanglei Zhao
ab,
Haixia Xuab,
Guanglei Zhao *a,
Hui Wu
*a,
Hui Wu b,
Yigang Yub,
Furao Laib and
Xinglong Xiao*b
b,
Yigang Yub,
Furao Laib and
Xinglong Xiao*b
aState Key Laboratory of Pulp and Paper Engineering, South China University of Technology, Wushan Road 381, Guangzhou 510640, China. E-mail: glzhao@scut.edu.cn; Tel: +86-20-87111770
bSchool of Food Science and Engineering, South China University of Technology, Guangzhou 510640, China. E-mail: fexxl@scut.edu.cn
First published on 13th March 2018
Abstract
Acylation modification of phenol glycosides is currently of great interest due to the improved bioavailability and multiple functions. In this work, mono- or diesters of arbutin, an important phenol glycoside derivative, can be controllably synthesized by using whole-cell biocatalytic systems. Among fourteen microbial strains selected, Candida parapsilosis cells showed the best catalytic activity and high organic solvent tolerance. Compared with the best pure solvent tetrahydrofuran, the use of a binary solvent pyridine-isooctane gave a slightly lower conversion (98.3% vs. 97.2%) and selectivity (85.3% vs. 80.5%) and much higher substrate solubility (37.1 vs. 214.0 mg mL−1), in a 24 h bioconversion of arbutin with a VP-arbutin molar ratio of 15 and whole cell dosage of 30 mg mL−1. The production of various arbutin esters with different fatty acid chain lengths can be realized by using this whole-cell strategy, with the substrate conversion and 6′-regioselectivity of 54.1–98.3% and 83.2–99.0%, respectively.
1. Introduction
Arbutin (hydroquinone-o-β-D-glucopyranosid, a natural phenol glycoside, is ubiquitous in edible plants such as strawberry plants,1,2 Bergenia crassifolia3 and pear trees.4 Over the last decades, it has attracted significant attention due to its versatile biological activities which include anti-inflammatory treatment,5 scavenging free radicals6 and lowering blood sugar.7 Furthermore, it is well-known for its high tyrosinase-inhibiting activity through direct combination with tyrosinase, inhibiting the formation of melanin and accelerating its decomposition. This activity has found considerable applications in the cosmetic industry as a powerful vegetable-based whitening agent.8,9 However, arbutin suffers from low bioavailability due to its poor cell membrane penetration. Recently, many studies have indicated that acylation modification of arbutin could increase its antimelanogenesis and antioxidant activities. Nagai et al.10 investigated the antioxidative effects of various acylated arbutins and found that the oxidation of methyl linoleate was inhibited by the arbutin esters. Tokiwa et al.11 reported a much higher inhibitory effect of arbutin esters on mushroom tyrosinase than unmodified arbutin and indicated that the esters mainly inhibited the latter part of the tyrosinase catalyzed reaction.Generally, ester derivatives of phenol glycoside are prepared chemically or enzymatically.12 Chemical acylation methods require complex procedures with multiple protection/deprotection steps, consuming a large amount of reagents, resulting in adverse environmental effects. Enzymatic acylation is superior to commonly used chemical approaches considering its high regioselectivity, mild reaction conditions, and environmental friendliness.13,14 However, these enzymatic methods suffer from the high cost, low stability, and the lack of enzyme recyclability. Thus, great effort has been devoted to screening for low-cost enzymes, development of enzyme immobilization strategies, and searching for new enzymes with high stability.11,15–17
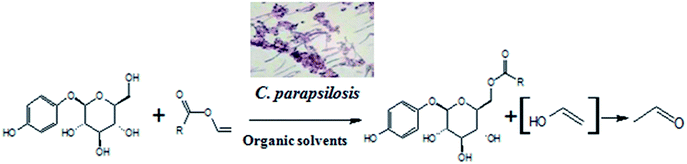
Whole cells are another kind of biocatalyst, different from pure enzymes, and exert catalytic functions based on the cell-bound/endoenzymes. This kind of biocatalyst has the advantages inherent in enzymatic processes, and has its own advantages,18–21 including higher stability compared to pure enzymes,22 coenzyme regeneration for multi-step biotransformation reaction,23 etc. To the best of our knowledge, no studies have been reported on whole-cell-catalyzed acylation of arbutin. Herein, we report, for the first time, a facile and highly efficient biocatalytic method for the acylation of arbutin with vinyl propionate (VP) using microbial whole-cells (Scheme 1). The origin of the cells and solvent effects on the reaction were investigated. In addition, the established biocatalytic process was used for the synthesis of arbutin esters with different fatty acid chain lengths.
2. Methods and materials
2.1 Biological and chemical materials
Arbutin was purchased from Aladdin. Rhizomucor miehei GIM 3.510, Rhizopus chinensis GIM 3.144, Mucor circinelloides GIM 3.521, Rhizopus oryzae GIM 3.509, Aspergillus niger GIM 3.25, Rhizopus oligosporus GIM 3.515, Candida parapsilosis GIM 2.190, Pseudomonas putida GIM 1.193, Bacillus subtilis GIM 1.135, Pseudomonas fluorescent GIM 1.209, Pseudomonas cepacia GIM 1.139, Pseudomonas stutzeri GIM 1.273, Aspergillus oryzae GIM 3.5232, Pseudomonas aeruginosa GIM 1.46, Geotrichum candidum GIM 2.12 were supplied by the Guangdong Culture Collect Centre. The acyl donors all were purchased from TCI (Japan). All other chemicals were from commercial sources and were of the highest purity available.2.2 Preparation of whole-cell biocatalysts
The bacterial and yeast strains were activated using a nutrient broth medium at 37 °C and 180 rpm for 24 h. Fungal strains were activated with a potato-dextrose-agar culture medium at 28 °C for 60 h. After activation, 5% (v/v) seed cultures were inoculated into the culture medium containing 5.0 g L−1 yeast extract, 5.0 g L−1 (NH4)2SO4, 1.0 g L−1 K2HPO4, 0.2 g L−1 MgSO4·7H2O, and 5.0 g L−1 soybean oil. The cultivation was carried out on a rotary shaker at 37 °C and 180 rpm for 48 h. Then the bacterial and yeast cells were collected by centrifugation (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, 3 min) and the supernatant fractions were discarded. For the fungal stains, the cells were collected by vacuum filtration. Then the cells achieved were washed twice with distilled water to remove the residual medium, freeze-dried at −45 °C for 24 h and stored at 4 °C.
000 rpm, 3 min) and the supernatant fractions were discarded. For the fungal stains, the cells were collected by vacuum filtration. Then the cells achieved were washed twice with distilled water to remove the residual medium, freeze-dried at −45 °C for 24 h and stored at 4 °C.
2.3 General procedure for whole cell mediated acylation of arbutin
In a typical experiment, arbutin (20 mM L−1), VP (900 mM L−1) and freeze-dried cells (40 mg mL−1) were added to 2 mL of anhydrous organic solvents and the mixture incubated at 40 °C in an orbital air-bath shaker (200 rpm). Aliquots were took at specified time intervals from the reaction mixture, the whole-cell catalyst was separated by centrifugation (12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, 5 min), then taking 40 μL of supernatant diluted with chromatographic grade methanol 25-fold for HPLC detection with a 20 μL sample injection volume. The initial rate was calculated according to the reduction of substrate in the initial stage of 1 hour. Regioselectivity was defined as the molar ratio of the desired product to the total amount of ester products formed. All data are averages of at least two separate experiments.
000 rpm, 5 min), then taking 40 μL of supernatant diluted with chromatographic grade methanol 25-fold for HPLC detection with a 20 μL sample injection volume. The initial rate was calculated according to the reduction of substrate in the initial stage of 1 hour. Regioselectivity was defined as the molar ratio of the desired product to the total amount of ester products formed. All data are averages of at least two separate experiments.
2.4 Operational stability
A total of 2 mL of anhydrous THF containing arbutin (20 mM L−1), VP (300 mML−1) and the whole-cell catalyst (30 mg mL−1) incubated at 40 °C in an orbital air-bath shaker (200 rpm). After 24 h, the whole-cell catalyst was separated by centrifugation (12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, 5 min), then taking 40 μL of supernatant diluted with chromatographic grade methanol 25-fold for HPLC detection with a 20 μL sample injection volume. After 24 h of reaction, the rest of the whole-cell catalyst was washed with anhydrous THF three times, and dried for 2 h at 40 °C. This regenerated the whole-cell catalyst for use in the next catalytic reaction batch, and this process was repeated 6 times.
000 rpm, 5 min), then taking 40 μL of supernatant diluted with chromatographic grade methanol 25-fold for HPLC detection with a 20 μL sample injection volume. After 24 h of reaction, the rest of the whole-cell catalyst was washed with anhydrous THF three times, and dried for 2 h at 40 °C. This regenerated the whole-cell catalyst for use in the next catalytic reaction batch, and this process was repeated 6 times.
2.5 HPLC analysis
The reaction mixture was analyzed by RP-HPLC on a 4.6 mm × 250 mm (5 μm) Zorbax SB-C18 column (Agilent Technologies Industries Co., Ltd., USA) using a Waters 600 Four element pump and a UV detector at 283 nm. The mobile phase is a mixture of methanol and water at 0.9 mL min−1. The volumetric ratio of methanol and water and the retention times for arbutin and its 6′-O-monoester were 40/60, 3.063 and 3.279 min (acetylation), 55/45, 3.080 and 3.740 min (propionylation), 70/30, 3.075 and 7.548 min (octanoylation), 80/20, 3.066 and 10.751 min (lauroylation), 3.066 and 6.334 min (10-undecenoylation), 85/15, 3.055 and 7.302 min (myristoylation), 90/10, 3.044 and 6.334 min (palmitoylation), 95/5, 3.054 and 10.809 min (stearylation), respectively.2.6 Separation and structural characterization of the products
After the reaction, the whole-cell catalyst was separated by centrifugation, and the supernatant was evaporated under vacuum to remove solvent. The residue was isolated to different component with thin-layer chromatography (TLC) method using chloroform/methanol as mobile phase. After crystallization under vacuum drying, the powder of products were obtained. The acylation position of the products were determined by 13C NMR (Bruker AVANCE Digital 400 MHz Nuclear Magnetic Resonance Spectrometer, Bruker Co., Germany) at 100 MHz. DMSO-d6 was used to dissolve substrate and product, the ppm shifts indicate chemical shifts. The mass spectra of products were obtained on Bruker maXis impact high-resolution mass spectrometry.2.7 Determination of the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) p values of the ester derivatives of arbutin
p values of the ester derivatives of arbutin
The 1-octanol-water partition coefficients (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) p) of the acylated derivatives were measured according to a previously published method.24 Each derivative was dissolved in 1-octanol (100 μM, 2 mL) and then mixed vigorously with 2 mL of water. After equilibration for 10 min, the derivatives in each layer were measured and their amounts were assessed by using the HPLC analytical process. The log
p) of the acylated derivatives were measured according to a previously published method.24 Each derivative was dissolved in 1-octanol (100 μM, 2 mL) and then mixed vigorously with 2 mL of water. After equilibration for 10 min, the derivatives in each layer were measured and their amounts were assessed by using the HPLC analytical process. The log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) p was calculated by taking the logarithm of the ratio of the HPLC peak area of each acylated derivative in octanol to the corresponding peak area in water.
p was calculated by taking the logarithm of the ratio of the HPLC peak area of each acylated derivative in octanol to the corresponding peak area in water.
2.8 Determination of arbutin solubility
To determine its solubility, 20 mg of arbutin standard was dissolved in 1 mL of methanol to prepare a 20 mg mL−1 standard stock solution. A 50 μL standard storage solution was diluted to 1000 μg mL−1, and 0.1, 0.2, 0.4, 0.6, 0.8, and 1 mL of the standard solution (1000 μg mL−1) were diluted to 100, 200, 400, 600, 1000 μg mL−1, respectively, and a sample volume of 20 μL was used for HPLC detection.Arbutin saturated solution configuration: to 1 mL of organic solvent, an excess of arbutin was added. After mixing at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm in a centrifuge for 3 min, 20 μL of the supernatant was used for HPLC detection, and its solubility was calculated.
000 rpm in a centrifuge for 3 min, 20 μL of the supernatant was used for HPLC detection, and its solubility was calculated.
3 Results and discussions
3.1 Catalytic behaviors of different strains for arbutin acylation
Table 1 shows the catalytic behaviors of seven bacterial strains, six fungal strains and one yeast strain possessing lipase activity. All the fungal strains tested (entries 1–7) showed the catalytic ability towards catalysis of arbutin propionylation. Whole-cell of Rhizomucor miehei showed the highest activity with a substrate conversion up to 27.2%. Among the six bacterial strains (entries 8–13), only Pseudomonas putida and Bacillus subtilis were able to catalyze the reaction with the conversion rate being of 39.8% and 8.0%, respectively. Among the fourteen strains tested, Candida parapsilosis (entry 14) showed the best catalytic activity, and the conversion rate reached 98.3% after 24 h.| Entry | Strains | V0 (mM h−1 L−1) | Conversiona (%) | 6′-Regioselectivity (%) |
|---|---|---|---|---|
| a The reactions were carried out in THF (2 mL) with arbutin (20 mM L−1), VP (900 mM L−1) and whole-cell catalysts (40 mg mL−1) at 40 °C and 200 rpm. | ||||
| 1 | Rhizomucor miehei GIM 3.510 | 3.9 ± 0.2 | 27.2 ± 0.1 | >99.0 |
| 2 | Rhizopus chinensis GIM 3.144 | 2.5 ± 0.0 | 22.2 ± 0.5 | >99.0 |
| 3 | Mucor circinelloides GIM 3.521 | NA | 15.5 ± 0.5 | >99.0 |
| 4 | Rhizopus oryzae GIM 3.509 | 1.5 ± 0.3 | 13.6 ± 0.6 | >99.0 |
| 5 | Aspergillus niger GIM 3.25 | NA | 6.5 ± 0.3 | >99.0 |
| 6 | Rhizopus oligosporus GIM 3.515 | NA | 5.9 ± 0.7 | >99.0 |
| 7 | Geotrichum candidum GIM 2.12 | NA | NA | NA |
| 8 | Pseudomonas putida GIM 1.193 | 1.8 ± 0.1 | 39.8 ± 0.4 | >99.0 |
| 9 | Bacillus subtilis GIM 1.135 | NA | 8.0 ± 0.3 | >99.0 |
| 10 | Pseudomonas fluorescent GIM 1.209 | NA | NA | NA |
| 11 | Pseudomonas cepacia GIM 1.139 | NA | NA | NA |
| 12 | Pseudomonas stutzeri GIM 1.273 | NA | NA | NA |
| 13 | Pseudomonas aeruginosa GIM 1.46 | NA | NA | NA |
| 14 | Candida parapsilosis GIM 2.190 | 23.7 ± 0.3 | 98.3 ± 0.1 | 85.3 ± 0.9 |
The products were separated with thin-layer chromatography (TLC) method and structural analyzed by HRMS and 13C NMR spectrums. HRMS analysis of the separated products showed that only two kinds of esters produced in the acylation process. The acylation positions of acylated products were further confirmed based on the significant chemical shift differences between arbutin and the ester products. Results showed that a mono-acylation occurred on the C6′ of sugar moiety of arbutin, which led to a downfield shift of the C6′ signal of 2.60 ppm and an upfield shift of the directly neighboring carbon atom C5′ signal of 2.84 ppm. In addition, the signal peak of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O appeared at 173.91 ppm or 170.66 ppm. And the carbon atoms of the fatty acid chain also appeared at 27.35 ppm (–CH–) and 9.44 ppm (CH3). The 13C NMR spectrum of another kind of ester shows downfield shifts of two carbon atoms C3′ and C6′ (3.27 ppm and 2.98 ppm, respectively) and upfield chemical shifts of their neighboring carbon atoms C4′ and C5′ (1.32 ppm and 2.60 ppm, respectively). Besides, two signal peaks of C
O appeared at 173.91 ppm or 170.66 ppm. And the carbon atoms of the fatty acid chain also appeared at 27.35 ppm (–CH–) and 9.44 ppm (CH3). The 13C NMR spectrum of another kind of ester shows downfield shifts of two carbon atoms C3′ and C6′ (3.27 ppm and 2.98 ppm, respectively) and upfield chemical shifts of their neighboring carbon atoms C4′ and C5′ (1.32 ppm and 2.60 ppm, respectively). Besides, two signal peaks of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O appeared at 173.91 ppm, confirming that 3′,6′-diester of arbutin was formed.
O appeared at 173.91 ppm, confirming that 3′,6′-diester of arbutin was formed.
Interestingly, whole cells of C. parapsilosis catalyzed the reaction producing both mono- and di-esters, while the other molds and bacteria tested showed high 6′-regioselectivity (>99.0%) producing only monoesters. The results indicated that the cell-bound enzymes from various microbial sources have different substrate recognition characteristics.25–27
3.2 Effect of the reaction media
Generally speaking, organic solvents have a great impact on non-aqueous catalysis,28 affecting the stability and activity of the biocatalysts, and substrate solubility.Table 2 (entries 1–5) shows the influence of different pure solvents on the whole-cell mediated acylation of arbutin. As a polar compound, arbutin is highly soluble in three polar pure solvents (DMSO, DMF, and pyridine). However, no cellular catalytic activity was tested in DMF and DMSO, which is likely because highly polar solvents deactivate the cell-bound enzymes by stripping off the essential water layer, causing protein aggregation and precipitation as well as “interfacial inactivation”.29–31 Although high initial production rates and conversions were observed in solvents with lower polarity, the solubility of arbutin was lower than 37 mg mL−1. To increase the solubility of arbutin in hydrophobic organic solvents and improve the reaction efficiency, a series of binary solvent mixtures containing the polar solvent pyridine and various hydrophobic solvents (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) were tested as reaction media for the whole-cell Candida parapsilosis catalyzed transesterification of arbutin with VP. As shown in Table 2 (entries 6–10), compared with the pure solvents system of THF and t-pentanol, the solubility of arbutin in their corresponding binary solvents was greatly improved, 4–5 times higher than in the pure hydrophobic solvents. In pyridine/acetone, both the initial rate and conversion of the reaction were much higher than pure pyridine. This may be due to the addition of hydrophobic organic solvents reducing the polarity of the reaction system, thereby reducing the toxic effects on the cells and the cell-bound lipases.32 Among the five binary solvents tested, pyridine-isooctane gave the best initial rate (37.2 mM h−1 L−1) and substrate conversion (97.2%), similar to those achieved in pure THF. The results indicate that a suitable binary solvent (pyridine-isooctane) can be used instead of the pure solvents to achieve a highly efficient reaction with a greatly increased substrate concentration. Interestingly, 6′-regioselectivity was negatively correlated with conversion yield in pure solvents, consistent with the report of Li et al.33 Herein, Candida parapsilosis cells exhibited relatively high organic solvent tolerance, which can be used as a promising catalysts for non-aqueous reaction.
1, v/v) were tested as reaction media for the whole-cell Candida parapsilosis catalyzed transesterification of arbutin with VP. As shown in Table 2 (entries 6–10), compared with the pure solvents system of THF and t-pentanol, the solubility of arbutin in their corresponding binary solvents was greatly improved, 4–5 times higher than in the pure hydrophobic solvents. In pyridine/acetone, both the initial rate and conversion of the reaction were much higher than pure pyridine. This may be due to the addition of hydrophobic organic solvents reducing the polarity of the reaction system, thereby reducing the toxic effects on the cells and the cell-bound lipases.32 Among the five binary solvents tested, pyridine-isooctane gave the best initial rate (37.2 mM h−1 L−1) and substrate conversion (97.2%), similar to those achieved in pure THF. The results indicate that a suitable binary solvent (pyridine-isooctane) can be used instead of the pure solvents to achieve a highly efficient reaction with a greatly increased substrate concentration. Interestingly, 6′-regioselectivity was negatively correlated with conversion yield in pure solvents, consistent with the report of Li et al.33 Herein, Candida parapsilosis cells exhibited relatively high organic solvent tolerance, which can be used as a promising catalysts for non-aqueous reaction.
| Entry | Solventsa | Solubility (mg mL−1) | V0 (mM h−1 L−1) | Conversion (%) | 6′-Regioselectivity (%) |
|---|---|---|---|---|---|
a The reaction conditions for pure solvents: arbutin 20 mM L−1, VP 900 mM L−1 and whole-cells 40 mg mL−1, 40 °C and 200 rpm; the reactions for binary solvents: solvent ratio 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/v), arbutin 20 mM L−1, VP 150 mM L−1, whole-cells 30 mg mL−1, 40 °C and 180 rpm.b No activity. 1 (v/v), arbutin 20 mM L−1, VP 150 mM L−1, whole-cells 30 mg mL−1, 40 °C and 180 rpm.b No activity. |
|||||
| 1 | DMF | 275.9 ± 0.1 | NAb | NA | NA |
| 2 | DMSO | 227.0 ± 0.3 | NA | NA | NA |
| 3 | Pyridine | 192.6 ± 0.5 | 6.5 ± 0.2 | 42.9 ± 0.9 | 96.1 ± 0.8 |
| 4 | THF | 37.1 ± 0.5 | 23.7 ± 0.3 | 98.3 ± 0.1 | 85.3 ± 0.9 |
| 5 | t-Pentanol | 35.2 ± 0.2 | 15.1 ± 0.3 | 89.3 ± 0.2 | 87.7 ± 0.9 |
| 6 | Pyridine/t-butanol | 134.0 ± 0.7 | 16.6 ± 0.1 | 86.5 ± 0.3 | 92.9 ± 0.7 |
| 7 | Pyridine/t-pentanol | 160.3 ± 0.4 | 12.6 ± 0.0 | 88.8 ± 0.4 | 89.1 ± 1.0 |
| 8 | Pyridine/acetone | 163.0 ± 0.5 | 5.6 ± 0.2 | 89.1 ± 0.4 | 86.2 ± 0.3 |
| 9 | Pyridine/THF | 191.8 ± 0.6 | 19.7 ± 0.2 | 85.6 ± 0.5 | 90.0 ± 0.3 |
| 10 | Pyridine/isooctane | 214.0 ± 0.3 | 37.2 ± 0.0 | 97.2 ± 0.5 | 80.5 ± 0.4 |
3.3 Effects of reaction conditions on the whole-cell catalyzed acylation of arbutin
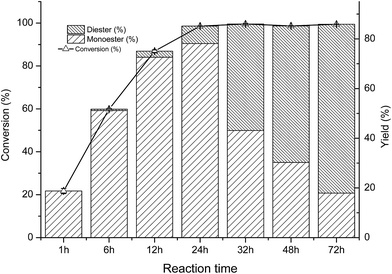
As shown in Fig. 1a, the initial rate of the reaction increased along with the increased of the VP/arbutin molar ratio. When the molar ratio of VP/arbutin increased from 0 to 5 mM mM−1, the substrate conversion increased sharply and then slowly until it reached above 90.0%. These results are consistent with those of Li et al.33 who found that increased molar ratio of substrates resulted in higher initial reaction rates and conversion yields of the enzymatic acylation of dihydromyricetin. The rate increased sharply, and further increases in the substrate molar ratio only resulted in small gains in reaction rate and conversion. The acylation of arbutin is a reversible reaction in non-aqueous media, accompanied by the hydrolysis of the enol ester (a side reaction that evidently consumes the acyl donor). Increasing the VP ratio may provide a sufficient amount of acyl donor and favor of the acylation of arbutin.34 However, the 6′-regioselectivity decreased with increasing of the VP/arbutin molar ratio, which may be due to the high acyl donor concentration results in an increase in the formation of di-esters. Higher acyl donor concentration promoted the formation of the acyl-enzyme intermediate and increased the rate of the nucleophilic attack of different alcohol ends (arbutin) on the carbonyl carbon of the intermediate.35 The result is similar to those reported by Chebil et al.25 who found that with an increasing molar ratio of vinyl acetate to quercetin, the proportion of the monoester decreased. In contrast, Li et al.33 reported that the molar ratio of acyl donor to dihydromyricetin showed only a minor effect on the proportion of dihydromyricetin-3-acetate in the total acylation products.As shown in Fig. 1b, with increasing whole cell dosage, the initial rate gradually increased. The conversion rate increased more quickly when the whole cell dosage was increased from 20 to 30 mg mL−1 and then increased slowly until it reached a relatively constant value of 90% at whole-cell dosages above 50 mg mL−1. This is mainly because of the stronger mass-transfer limitations induced by the higher cell dosages. When the enzymes are saturated, increasing the amount of enzyme present has little influence on substrate conversion. Thus, to conserve the whole-cell catalyst, choosing a suitable whole cell dosage is necessary. Interestingly, the results show a trend towards decreasing the 6′-regioselectivity of the reaction with increasing whole cell dosage. According to classical enzymatic theory, this can be ascribed to the formation of arbutin monoester-enzyme intermediate promoted by higher whole-cell dosages, which results in increased production of the double ester.
3.4 Time course of the whole-cell catalyzed acylation of arbutin
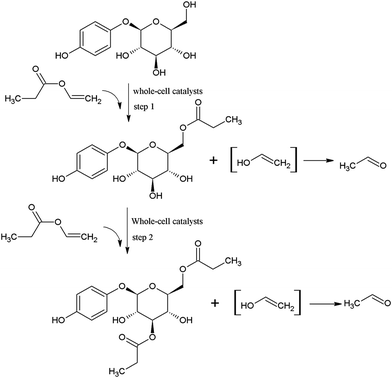
As shown in Fig. 2, at the initial stage of the reaction, the conversion of arbutin was significantly increased, and the acylation reaction reached equilibrium after approximately 24 h. It is interesting to note that only the monoester was formed at the early stage of the reaction (0–1 h). After that, the yield of monoester increased while a small amount of the diester was produced and continuously increased. When the reaction time was prolonged greater than 24 h, the monoester content was reduced and the di-ester yield increased significantly. Similar results were obtained by Chebil et al.25 during the synthesis of isoquercitrin esters with a lipase PSL-C and Sin et al.36 achieved similar results during the synthesis of fructose esters with Pseudomonas sp.Therefore, it is likely that the whole-cell catalyzed acylation of arbutin contained two sequential steps (Fig. 3). First, the primary hydroxyl group of arbutin is proposed to attack the acyl-enzyme intermediate to form the 6′-monoester of arbutin. Then, the 3′-hydroxyl group of the sugar moiety of the 6′-monoester attacks the acyl-enzyme intermediates, forming a 3′,6′-diester. This also explains the decrease in the 6′-regioselectivity, as shown in Fig. 1a and b, which increases the rate of step 2.
3.5 Regioselective acylation of arbutin with various acyl donors
In addition to the propionate acylation of arbutin, whole-cell biocatalysis was applied to the acylation of arbutin with acyl donors of differing fatty acid chain lengths. Several studies have shown that the long chain fatty acid esters of phenol glycosides have higher liposolubility and stronger bioactivities.27 Table 3 shows that when the length of carbon chain was increased from 2 to 18, both the initial rate and substrate conversion significantly decreased. The increase in carbon chain length significantly enhanced the steric hindrance of the acyl donor molecules, preventing the formation of the acyl-enzyme intermediate and reducing the efficiency of the enzymatic reaction. For fatty acid vinyl esters with higher carbon-chain lengths (C3–C18), the conversion yield dropped constantly, and the whole-cell catalysis was most efficient for C3, with a conversion of 98.3% after 24 h. However, the 6′-regioselectivity of the reaction increased, which can be attributed to the steric hindrance of the reaction. With increasing fatty acid chain length of the acyl donors, the steric hindrance of the monoester increases, and preventing monoester attack of the acyl donor-enzyme intermediates to form the diester.| Acyl donor | Structural formula of acyl donors | V0 (mM h−1 L−1) | Conversion (%) | 6′-Regioselectivity (%) | log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) p of the monoester p of the monoester |
|---|---|---|---|---|---|
| a The reactions were carried out in THF (1 mL) with arbutin (20 mM L−1), VP (900 mM L−1) and whole-cell catalysts (40 mg mL−1) at 40 °C and 200 rpm. | |||||
| Vinyl acetate | 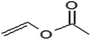 |
45.8 ± 0.3 | 97.6 ± 0.6 | 83.2 ± 0.3 | −0.8 ± 0.1 |
| Vinyl propionate | 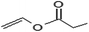 |
23.7 ± 0.3 | 98.3 ± 0.1 | 85.3 ± 0.9 | −0.3 ± 0.3 |
| Vinyl caprylate | 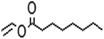 |
6.8 ± 0.9 | 92.4 ± 1.0 | 94.7 ± 0.7 | 2.1 ± 0.3 |
| Vinyl 10-undecenoate | 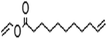 |
4.7 ± 0.2 | 90.3 ± 0.3 | >99.0 | 2.2 ± 0.5 |
| Vinyl laurate | 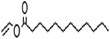 |
3.0 ± 0.5 | 71.1 ± 0.5 | >99.0 | 2.4 ± 0.7 |
| Vinyl myristate | 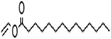 |
3.8 ± 0.1 | 69.7 ± 0.3 | >99.0 | 4.0 ± 0.9 |
| Vinyl palmitate | 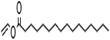 |
2.5 ± 0.7 | 55.9 ± 0.1 | >99.0 | 5.0 ± 0.4 |
| Vinyl stearate |  |
7.7 ± 0.9 | 54.1 ± 0.1 | >99.0 | 5.3 ± 0.3 |
The log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) p value is a common parameter reflecting the liposolubility of a compound determined by its distribution in oil and water phases.37 Table 3 shows that when the fatty acid chain length of the acyl donor increased, the log
p value is a common parameter reflecting the liposolubility of a compound determined by its distribution in oil and water phases.37 Table 3 shows that when the fatty acid chain length of the acyl donor increased, the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) p values of the acylated derivatives increased gradually from −0.8 to 5.3. This suggests that the enzymatic acylation could effectively improve the lipophilicity of the hydrophilic arbutin. It also indicates that the acylation of phenol glycoside is an effective method of increasing liposolubility and several studies have reported that acylated derivatives have higher activity.38–40
p values of the acylated derivatives increased gradually from −0.8 to 5.3. This suggests that the enzymatic acylation could effectively improve the lipophilicity of the hydrophilic arbutin. It also indicates that the acylation of phenol glycoside is an effective method of increasing liposolubility and several studies have reported that acylated derivatives have higher activity.38–40
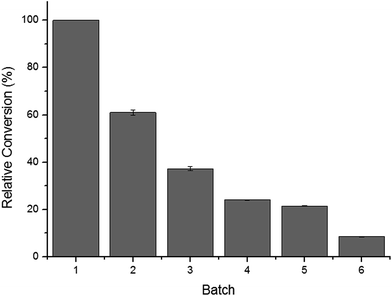
3.6 Operational stability of the whole cell biocatalyst
Fig. 4 shows the operational stability of the whole-cell biocatalysts. It was observed that the cells retained high catalytic activity in their second use. After repeated used (three cycles), the activity of the whole cells was reduced to nearly 60%. In the fourth cycle, the conversion rate decreased rapidly below 40%, likely due to the toxic effects of the organic solvents to the cell structures and cell-bound enzymes. One of the major shortcomings of free enzymes is that they can only be used once. Thus, great effort has been made on the development of immobilization materials and strategies for enzymes. The results presented herein demonstrated that C. parapsilosis whole-cell catalysts are advantageous over free enzymes in their operational stability. The cell structure may serve as a natural carrier for loading and protecting enzymes, avoiding further immobilization and lowering the total costs of processing.4. Conclusions
This study details a facile and efficient biocatalytic method for the synthesis of arbutin esters using C. parapsilosis whole-cells as a catalyst. With this approach, high yield and high regioselectivity of 6′-monoesters of arbutin were obtained within 24 h, while high yields of diesters can be obtained by prolonging the reaction to greater than 72 h. The catalytic performance and operational stability of the whole-cells for the acylation of arbutin may provide new opportunities to develop green industrial processes for novel phenol glycoside derivative production. Investigations on the bioactivities of the ester products and a comparative study of the phenol glycoside compounds with similiar structures to arbutin on the reactions are in progress.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work is supported by the National Natural Science Foundation of China (No. 21676105), Self Determined Research Fund of SCUT from the College Basic Research and Operation of MOE (No. 2015ZZ111, 2015ZZ123), Self-determined Project of State Key Laboratory of Pulp and Paper Engineering (2015C11) and the Science and Technology Program Foundation of Guangdong Province (No. 2015A030401025).References
- K. Jurica, I. B. Karačonji, S. Šegan, D. M. Opsenica and D. Kremer, Arh. Hig. Rada Toksikol., 2015, 66, 197–202 CrossRef CAS PubMed.
- Y. Tian, J. Liimatainen, A. Alanne, A. Lindstedt, P. Liu, J. Sinkkonen, H. Kallio and B. Yang, Food Chem., 2017, 220, 266–281 CrossRef CAS PubMed.
- B. Żbikowska, R. Franiczek, A. Sowa, G. Połukord, B. Krzyżanowska and Z. Sroka, Microb. Drug Resist., 2017, 23, 771–780 Search PubMed.
- C. Sasaki, Y. Yoshida, C. Asada and Y. Nakamura, J. Mater. Cycles Waste Manage., 2016, 18, 385–392 CrossRef CAS.
- H. Lee and K. Kim, Inflammation Res., 2012, 61, 817–825 CrossRef CAS PubMed.
- F. Khadir, M. Pouramir, S. G. Joorsaraee, F. Feizi, H. Sorkhi and F. Yousefi, Caspian J. Intern. Med., 2015, 6, 196–200 Search PubMed.
- H. Takii, K. Matsumoto, T. Kometani, S. Okada and T. Fushiki, Biosci., Biotechnol., Biochem., 1997, 61, 1531–1535 CrossRef CAS PubMed.
- J. Takebayashi, R. Ishii, J. Chen, T. Matsumoto, Y. Ishimi and A. Tai, Free Radical Res., 2010, 44, 473–478 CrossRef CAS PubMed.
- Y. H. Jin, S. J. Lee, M. H. Chung, J. H. Park, Y. I. Park, T. H. Cho and S. K. Lee, Arch. Pharmacal Res., 1999, 22, 232–236 CrossRef CAS.
- M. Nagai, Y. Watanabe and M. Nomura, Biosci., Biotechnol., Biochem., 2009, 73, 2501–2505 CrossRef CAS PubMed.
- Y. Tokiwa, M. Kitagawa, T. Raku, S. Yanagitani and K. Yoshino, Bioorg. Med. Chem. Lett., 2007, 17, 3105–3108 CrossRef CAS PubMed.
- E. Hermankova-Vavrikova, A. Krenkova, L. Petraskova, C. S. Chambers, J. Zapal, M. Kuzma, K. Valentova and V. Kren, Int. J. Mol. Sci., 2017, 18, 1074 CrossRef PubMed.
- P. M. L. Gonçalves, S. M. Roberts and P. W. H. Wan, Tetrahedron, 2004, 60, 927–932 CrossRef.
- M. Ferrero and V. Gotor, Chem. Rev., 2000, 100, 4319–4348 CrossRef CAS PubMed.
- Y. Watanabe, M. Nagai, K. Yamanaka, K. Jose and M. Nomura, Biochem. Eng. J., 2009, 43, 261–265 CrossRef CAS.
- R. Yang, N. Li, R. Li, T. J. Smith and M. Zong, Bioresour. Technol., 2010, 101, 1–5 CrossRef CAS PubMed.
- R. Yang, N. Li, M. Ye and M. Zong, J. Mol. Catal. B: Enzym., 2010, 67, 41–44 CrossRef CAS.
- A. S. Staebler, A. Cruz, W. van der Goot, H. M. Pinheiro, J. Cabral and P. Fernandes, J. Mol. Catal. B: Enzym., 2004, 29, 19–23 CrossRef CAS.
- N. C. Mhetras, K. B. Bastawde and D. V. Gokhale, Bioresour. Technol., 2009, 100, 1486–1490 CrossRef CAS PubMed.
- P. Kafarski and B. Lejczak, J. Mol. Catal. B: Enzym., 2004, 29, 99–104 CrossRef CAS.
- C. Razak, R. Musani, M. Basri and A. B. Salleh, J. Am. Oil Chem. Soc., 1999, 76, 171–174 CrossRef CAS.
- X. Li, Z. Zhu, G. Zhao, Y. Yu, F. Lai and H. Wu, Appl. Microbiol. Biotechnol., 2012, 93, 143–150 CrossRef PubMed.
- W. A. Duetz, J. B. van Beilen and B. Witholt, Curr. Opin. Biotechnol., 2001, 12, 419–425 CrossRef CAS PubMed.
- M. Ardhaoui, A. Falcimaigne, S. Ognier, J. M. Engasser, P. Moussou, G. Pauly and M. Ghoul, J. Biotechnol., 2004, 110, 265–271 CrossRef CAS PubMed.
- L. Chebil, J. Anthoni, C. Humeau, C. Gerardin, J. Engasser and M. Ghoul, J. Agric. Food Chem., 2007, 55, 9496–9502 CrossRef CAS PubMed.
- L. Chebil, C. Humeau, A. Falcimaigne, J. Engasser and M. Ghoul, Process Biochem., 2006, 41, 2237–2251 CrossRef CAS.
- Z. Wang, Y. Bi, X. Li and M. Zong, Process Biochem., 2013, 48, 1208–1211 CrossRef CAS.
- M. Yang, H. Wu, Z. Lu, X. Li, F. Lai and G. Zhao, Bioorg. Med. Chem. Lett., 2014, 24, 3377–3380 CrossRef CAS PubMed.
- N. Doukyu and H. Ogino, Biochem. Eng. J., 2010, 48, 270–282 CrossRef CAS.
- R. Sinha and S. K. Khare, Extremophiles, 2014, 18, 1057–1066 CrossRef CAS PubMed.
- K. P. Dhake, K. M. Deshmukh, Y. S. Wagh, R. S. Singhal and B. M. Bhanage, J. Mol. Catal. B: Enzym., 2012, 77, 15–23 CrossRef CAS.
- D. Jain and S. Mishra, J. Mol. Catal. B: Enzym., 2015, 117, 21–30 CrossRef CAS.
- W. Li, H. Wu, B. Liu, X. Hou, D. Wan, W. Lou and J. Zhao, J. Biotechnol., 2015, 199, 31–37 CrossRef CAS PubMed.
- M. H. Katsoura, A. C. Polydera, L. Tsironis, A. D. Tselepis and H. Stamatis, J. Biotechnol., 2006, 123, 491–503 CrossRef CAS PubMed.
- A. C. Mathpati, K. C. Badgujar and B. M. Bhanage, Enzyme Microb. Technol., 2016, 84, 1–10 CrossRef CAS PubMed.
- Y. M. Sin, K. W. Cho and T. H. Lee, Biotechnol. Lett., 1998, 20, 91–94 CrossRef CAS.
- M. Zhang, X. Xin, F. Lai, X. Zhang, X. Li and H. Wu, J. Agric. Food Chem., 2017, 65, 7424–7432 CrossRef CAS PubMed.
- V. Kumar, F. Jahan, R. V. Mahajan and R. K. Saxena, Bioresour. Technol., 2016, 218, 1246–1248 CrossRef CAS PubMed.
- M. Stompor, M. Switalska, R. Podgorski, L. Uram, D. Aebisher and J. Wietrzyk, Acta Biochim. Pol., 2017, 64, 577–583 CrossRef CAS PubMed.
- X. Xing, X. Li, X. Xiao, Y. Tang and G. Zhao, ACS Sustainable Chem. Eng., 2017, 5, 10662–10672 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra00595h |
| This journal is © The Royal Society of Chemistry 2018 |