Open Access Article
Open Access ArticleAntimicrobial activity of nano-sized silver colloids stabilized by nitrogen-containing polymers: the key influence of the polymer capping†
Carin C. S. Batistaa,
Lindomar J. C. Albuquerquea,
Iris de Araujoa,
Brunno L. Albuquerqueb,
Fernanda D. da Silvaa and
Fernando C. Giacomelli *a
*a
aCentro de Ciências Naturais e Humanas, Universidade Federal do ABC, Santo André, Brazil. E-mail: fernando.giacomelli@ufabc.edu.br
bDepartamento de Química, Universidade Federal de Santa Catarina, Florianópolis, 88040-900, Brazil
First published on 19th March 2018
Abstract
Synthesis of stable silver colloids was achieved using nitrogen-containing polymers acting simultaneously as a reducing and stabilizer agent. The polymers polyethyleneimine (PEI), polyvinylpyrrolidone (PVP) and poly(2-vinyl pyridine)-b-poly(ethylene oxide) (PEO-b-P2VP) were used in the procedures. The influence of the surface chemistry and chemical nature of the stabilizer on the cytotoxicity and antimicrobial properties have been evaluated. The produced nanomaterials were found to be non-toxic up to the highest evaluated concentration (1.00 ppm). Nevertheless, at this very low concentration, the AgNPs stabilized by PVP and PEO-b-P2VP were found to be remarkable biocides against bacteria and fungus. On the other hand, we have surprisingly evidenced negligible antimicrobial activity of AgNPs stabilized by positively charged PEI although both (AgNPs and PEI) materials separately are known for their antimicrobial activity as also evidenced in the current investigation. The evidence is claimed to be related to the blocking of Ag+ kinetic release. Accordingly, the antimicrobial effect of nano-sized silver colloids largely depends on the chemical nature of the polymer coating. Possibly, the outstanding colloid stabilization provided by polyethyleneimine slows down Ag+ release thereby hampering its biological activity whereas the poorer stabilization and good ionic transport property of PVP and PEO-b-P2VP allows much faster ion release and cell damage.
Introduction
Noble metal nanoparticles have proven to be very useful in medical applications due to their well-known antibacterial, antifungal and antiviral properties.1–6 Particularly considering silver nanoparticles (AgNPs), they are nowadays popular as coatings for surgical clothes, dressings and other medical devices.7,8 Additionally, AgNPs are also being added to a variety of everyday products including food packaging, textiles and detergents.9,10 Indeed, the biocide effect of silver salts has been known since the 19th century whereas such a property for AgNPs was discovered recently.11 There are up-to-date investigations reporting the effect of size,12 shape13 and surface charge14 on the resulting toxicity of silver colloids. Nevertheless, the actual mechanism of action governing their toxicity remains heavily debated.15,16 Possibly, the antimicrobial property arises from the contribution of silver ions (Ag+) and bulk silver existing as nanocolloids (the nanoparticles per se).17 The interaction of Ag+ with thiol groups in enzymes and proteins is accepted to be the main responsible for the antimicrobial action, although their lethal action can, at least to some extent, be also linked to inhibition of cell division and binding to bases of the DNA chains.18 Simultaneously, the adhesion of AgNPs onto the cell membrane of microorganisms and further cell internalization can lead firstly to changes in membrane permeability and later to damages in intracellular structures and biomolecules, ultimately causing cell death.19 Hence, the antimicrobial property of AgNPs can be influenced by the features of the nano-sized silver colloids and by the presence of dissolved silver ions. Taking into account the above mentioned issues, clearly, the biological activity of AgNPs is at least indirectly sensitive to size, shape, surface charge and surface chemistry (chemical nature of the stabilizer). These physicochemical parameters are driven by the strategies to produce the colloidal suspensions. Presently, the most versatile methods towards the manufacturing of metallic nanoparticles involve the reduction of a metallic salt in the presence of a reducing agent and a stabilizer required to provide colloidal stability. In such way, the use of polymers and copolymers is an interesting route for the fabrication of polymer-stabilized metallic nanoparticles. The polymeric materials are used almost exclusively as stabilizers in the presence of different reducing agents. Polysaccharides,20 biodegradable21 and temperature-responsive22 polymers, polycations,23 polyamino acids24 as well as other non-ionic water-soluble polymers25 were successfully employed for such a purpose with resulting biological activity of the hybrid materials evaluated. Alternatively, when nitrogen-containing polymers are incorporated into the process, they can simultaneously act as reducing and stabilizer agent while imparting unique properties to the manufactured colloidal systems.26 Indeed, the use of nitrogen-containing polymers acting simultaneously as reducing and stabilizer agent is an insightful approach and it can be also employed for instance in one-pot synthesis of metallic colloids directly in the aqueous media without the aid of any other external agent expect the ionic precursor. Such method was successfully applied by us in the synthesis of nano-sized silver colloids produced and stabilized using a variety of nitrogen-containing polymers.27 The reactions were carried out in ethylene glycol and in such a case, the reduction of Ag+ was promoted by the nitrogen atoms of the polymer chains and also by the oxygen atoms of the solvent. The polymer chains subsequently chemisorb onto the newly generated particles promoting colloidal stability. The procedures led to nearly spherical structures stabilized by lengthy hydrophilic shells where the dimension is primarily governed by the molecular weight of the polymers. Likewise, the surface charge of the resulting hybrid materials depends on the polymer chemical structure. Herein, we relate the produced assemblies to the cytotoxicity and antimicrobial properties. The biological assays were investigated regarding AgNPs stabilized by PEI (polyethylenimine), PVP (polyvinylpyrrolidone) and PEO-b-P2VP (poly(2-vinyl pyridine)-b-poly(ethylene oxide)). The antibacterial and antifungal properties were evaluated in three different bacteria strains (Gram-negative or Gram-positive) and against Candida albicans which is a type of yeast commonly found at the human gut flora. Additionally, the cytotoxicity was evaluated into two different mammalian cell lines. The nanoparticle stabilization provided by the capping agent significantly impacts the biocide properties of the produced polymer-stabilized silver colloids.Experimental
Materials
Silver nitrate (AgNO3) and ethylene glycol (EG) were of analytical grade purchased from Sigma-Aldrich and used as received. The polymers PEI (polyethyleneimine) and PVP (polyvinylpyrrolidone) were also purchased from Sigma-Aldrich whereas PEO-b-P2VP block copolymer (poly(ethylene oxide)-b-poly(2-vinyl pyridine)) was purchased from Polymer Source Inc. The water was pretreated with Milli-Q® Plus System (Millipore Corporation). The Fig. 1 portrays the molecular structure of the reducing/stabilizer polymers.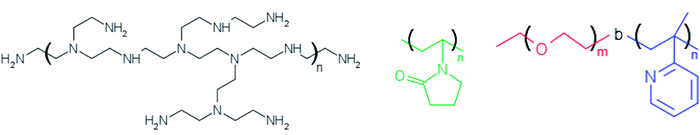 | ||
| Fig. 1 Molecular structure of branched polyethyleneimine (left), polyvinylpyrrolidone (middle) and poly(ethylene oxide)-b-poly(2-vinyl pyridine) (right). | ||
The reducing/stabilizer polymers are herein referred to as BLPEI (Mn = 1200 g mol−1, Mw/Mn = 1.1), BHPEI (Mn = 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g mol−1, Mw/Mn = 2.5), PVP (Mn = 40
000 g mol−1, Mw/Mn = 2.5), PVP (Mn = 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g mol−1, Mw/Mn = 2.0) and PEO-b-P2VP (Mn = 21
000 g mol−1, Mw/Mn = 2.0) and PEO-b-P2VP (Mn = 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000-b-13
000-b-13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 g mol−1, Mw/Mn = 1.1). The labels B, L and H stand respectively for branched, lower molecular weight and higher molecular weight.
500 g mol−1, Mw/Mn = 1.1). The labels B, L and H stand respectively for branched, lower molecular weight and higher molecular weight.
Nanoparticle synthesis
The synthesis of the AgNPs was carried out following the procedure described previously.27 The silver colloids were produced in EG environment containing molecularly dissolved PEI, PVP or PEO-b-P2VP. The silver nitrate (11.0 mg, 1.0 mL) was slowly added to a solution (3.0 mL) containing EG and the reducing/stabilizer polymers (1.6 mg). The mixture was then homogenized, heated up to 50 °C and the reactions were carried out for 2 hours. The produced AgNPs were further precipitated in acetone followed by centrifugation (4 °C, 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, 30 min) and resuspension in water.
000 rpm, 30 min) and resuspension in water.
Nanoparticle characterization
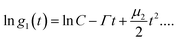 | (1) |
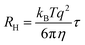 | (2) |
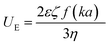 | (3) |
Nanoparticle quantification
The mass of silver nanoparticles at each formulation was determined based on the methodology previously reported by Paramelle et al. and Kalishwaralal et al.29,30 Firstly, the molar concentration of AgNPs (CAgNPs) was determined taking into account the measured absorbance:
 | (4) |
| NL = CAgNPsNA | (5) |
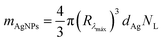 | (6) |
Biological assays
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 U mL−1 and streptomycin 10
000 U mL−1 and streptomycin 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 μg mL−1) at 37 °C in 5% CO2 atmosphere.
000 μg mL−1) at 37 °C in 5% CO2 atmosphere.The cytotoxicity of the silver nanoparticles and pure polymers were evaluated in both cell lines. The cells were cultured into 96-well plates at a density of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well for 24 h and afterwards treated with 90 μL of fresh DMEM plus 10 μL of different concentrations of silver nanoparticles. The cells were incubated for 24 h at 37 °C, washed with PBS and subjected to MTT assay. Briefly, 100 μL of MTT (3-(4,5-dimethylthiazol-2yl)-2,5-diphenyl-tetrazolium bromide) reagent solution (0.5 mg mL−1) was added to each well for 4 h. The medium was aspirated and violet crystals of formazan generated by living cells at each well were dissolved in 100 μL of dimethyl sulfoxide (DMSO). The absorbance at 570 nm was measured using a Synergy microplate reader. The relative cell viability (%) was determined by comparing the absorbance at 570 nm with control wells containing untreated cells (100%).
000 cells per well for 24 h and afterwards treated with 90 μL of fresh DMEM plus 10 μL of different concentrations of silver nanoparticles. The cells were incubated for 24 h at 37 °C, washed with PBS and subjected to MTT assay. Briefly, 100 μL of MTT (3-(4,5-dimethylthiazol-2yl)-2,5-diphenyl-tetrazolium bromide) reagent solution (0.5 mg mL−1) was added to each well for 4 h. The medium was aspirated and violet crystals of formazan generated by living cells at each well were dissolved in 100 μL of dimethyl sulfoxide (DMSO). The absorbance at 570 nm was measured using a Synergy microplate reader. The relative cell viability (%) was determined by comparing the absorbance at 570 nm with control wells containing untreated cells (100%).
 | (7) |
The use of human blood sample in this study was approved by the Universidade Federal do ABC under protocol 66527417.2.0000.5594.
 | (8) |
Results
Structural features of the produced AgNPs
We recently demonstrated the production of nano-sized silver colloids simultaneously reduced and stabilized by nitrogen-containing polymers.27 The most relevant parameters of the manufactured assemblies are summarized in Table 1.| Stabilizer | RH (nm) | PDI | RTEM (nm) | Wshell (nm) | ζ-potential (mV) |
|---|---|---|---|---|---|
| a Wshell = RH − RTEM. | |||||
| BLPEI | 30.6 | 0.24 | 19.0 | 11.6 | +14.2 |
| BHPEI | 37.4 | 0.15 | 19.0 | 18.6 | +42.6 |
| PVP | 40.3 | 0.16 | 11.0 | 29.3 | −9.5 |
| PEO-b-P2VP | 56.1 | 0.16 | 14.0 | 42.1 | +1.6 |
The dynamic light scattering data revealed radius ranging from 30.6 to 56.1 nm. The smallest AgNPs are those stabilized by polymers of lower molecular weight (BLPEI and BHPEI) and respectively, larger sizes were monitored for AgNPs stabilized by PVP and PEO-b-P2VP. The radii reflect essentially the thickness of the polymeric stabilizing shells (Wshell) calculated as Wshell = RH − RTEM and given in Table 1. Besides being influence by the molecular weight of the polymers, the parameter Wshell also reflects the hydrophilicity of the polymer chains. Since PEI is a less hydrophilic polymer compared to PEO and PVP, the polycation is supposed to be less stretched towards the solvent (water) due to unfavorable interactions whereas for the case of PEO and PVP, the energetically favorable interactions between solvent and polymer segments lead to the expansion of the stabilizing chains. These differences presumably influence the silver ion release from the produced AgNPs and consequently the biological activity of the entities (discussed hereafter).
The scattering data have been complemented by TEM analysis. The images are portrayed in Fig. 2 and they evidence that the AgNPs are reasonable polydisperse. Indeed, in agreement with the TEM images, the values of PDI reported in Table 1 also reveal that all the samples are polydisperse (PDI ≥ 0.15).
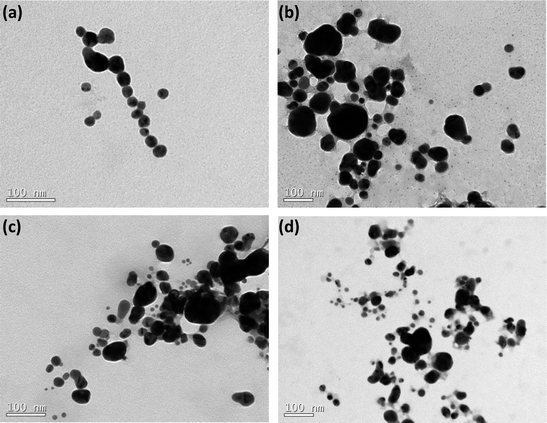 | ||
| Fig. 2 TEM images of AgNPs coated by nitrogen-containing polymers: (a) BLPEI, (b) BHPEI, (c) PVP and (d) PEO-b-P2VP. | ||
Moreover, the monitored sizes are smaller compared to those reported by DLS. This also confirms that the silver cores are stabilized by lengthy polymeric shells regardless the polymer features. Indeed, the polymeric shell is hardly seen by TEM and that is the main reason of discrepancies comparing the measured dimensions. The values of ζ-potential point out that the nanoparticles stabilized by BPEI hold a positive surface charge whereas the surface is respectively nearly neutral and negatively charged when the metallic nanoparticles are stabilized by PVP and PEO-b-P2VP. The negative ζ-potential of PVP-stabilized AgNPs is presumably the consequence of charge partitioning on carbonyl groups present at the PVP polymer backbone. The PEO-b-P2VP provides a nearly neutral ζ-potential to the assemblies since the outer PEO shell does not exhibit ionizable groups and there is no charge partitioning that could possibly lead to a negative or positive surface. The positively charged PEI-stabilized AgNPs is straightforward and due to the ionization of tertiary amines of the PEI chains. The higher positive value for BHPEI compared to BLPEI is most probably linked to its higher molecular weight and therefore a higher number of ionized amines making part of the shell. Furthermore, the PEI-stabilized AgNPs were produced significantly faster and the slowest reaction rate was monitored in PEO-b-P2VP environment. This behavior is supposed to be connected to stronger Ag-PEI interaction compared to the strengths of Ag-PVP and Ag-PEO-b-P2VP pairs.
Biological assays
The previous step before performing the biological assays was the quantification of the mass of silver nanoparticles at each formulation. This was done based on the UV-vis spectra reported in Fig. 3 and the use of eqn (4)–(6). The Table 2 reports the amount of AgNPs at the starting stock solutions along to the values of extinction coefficients used. The concentrations are in the range 4–40 ppm. The whole set of samples were normalized by dilution to an equal starting concentration of 1.00 ppm. This was the highest concentration of AgNPs evaluated in biological assays.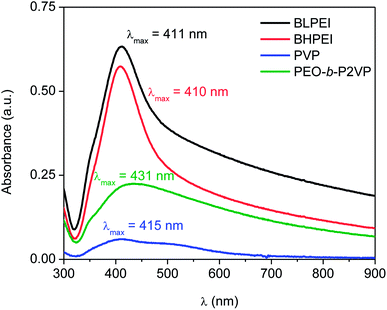 | ||
| Fig. 3 UV-spectra of AgNPs suspended in water and stabilized by using different nitrogen-containing polymers according to the legend. | ||
| Stabilizer | Absorbance | λmáx (nm) | ε (108 M−1 cm−1) | CUV-vis (ppm) | CICP-OES (ppm) |
|---|---|---|---|---|---|
| BLPEI | 0.633 | 411 | 295 | 38.9 | 41.3 |
| BHPEI | 0.574 | 410 | 295 | 35.3 | 39.3 |
| PVP | 0.060 | 415 | 416 | 4.1 | 5.2 |
| PEO-b-P2VP | 0.225 | 431 | 739 | 21.8 | 26.0 |
The total silver concentration in the hybrid systems was also accessed by ICP-OES. The monitored values are about 15–20% higher than those determined by UV-vis most probably due the presence of silver ions (Ag+) not taken into account in the determinations based on UV-vis data.
Subsequently, the cytotoxicity of the silver colloids and the antimicrobial performance were evaluated.
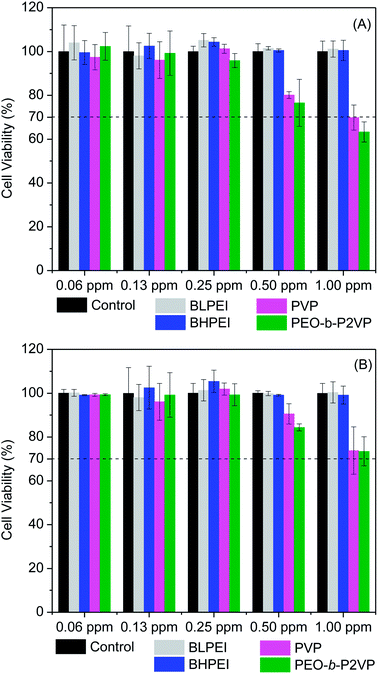
The cell viability was probed by using the straightforward MTT protocol where polymer-stabilized AgNPs as well as the pure polymers were evaluated in contact with HeLa and Telo-RF cells and the results are given in Fig. 4 (AgNPs) and Fig. S1† (polymers). The data clearly demonstrate that the polymer-stabilized AgNPs are not cytotoxic at least up to the highest concentration evaluated (1.00 ppm). The percentage of viable cells is always higher than 70% as requested by the ISO 10993-5 (2009) to characterize the material as non-toxic. Interestingly, even the AgNPs stabilized by BPEI did not demonstrated substantial levels of cytotoxicity although the pure polymers are known to hold high levels of cytotoxicity depending on concentration as can be evidenced in Fig. S1.† The intrinsic cytotoxicity of catiomers is usually attributed to electrostatic interactions with a variety of negatively charged lipids present in several cell organelle31,32 thereby inducing, usually, cell apoptosis.33 The current results evidence that only in contact with dilute BPEI solutions (∼0.3 mg mL−1) the percentage of viable cells is higher than 70%. On the other hand, the polymers PEO-b-P2VP and PVP are non-toxic even at reasonable high concentrations (∼2 mg mL−1) as well as the AgNPs stabilized by such polymers. Regarding the absence of cytotoxicity of the BPEI-stabilized AgNPs, the behavior could be related to the low concentration of the polycation at the final suspension. The starting polymer concentration at EG environment was 0.4 mg mL−1 and certainly this value is smaller after the following steps of precipitation, centrifugation and resuspension in water as described in the experimental section. Additionally, strong electrostatic, hydrophobic and van der Waals interactions between BPEI and silver could to some extent weaken the ability of PEI chains in promoting cell damage.
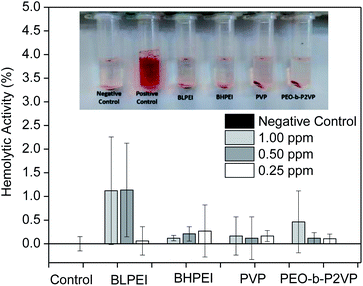
Complementary to MTT assays, the influence of the structural features of the produced AgNPs on the hemolytic behavior was investigated by placing them in contact with red blood cells (RBCs) for 30 min and further measuring the absorbance of released hemoglobin at λ = 545 nm. The dose-dependent hemolytic behavior is reported in Fig. 5. The ISO 10993-5 (2009), that describes the biological evaluation of medical devices, considers values of hemolytic activity below 5% as satisfactory. The current results evidence that the differences in optical density of experimental samples as compared to the control was never higher than ∼2% regardless the concentration and whatever the system thus demonstrating biocompatibility of the produced AgNPs also with RBCs at the investigated concentration range. Accordingly, as similar evidenced by cell viability assays, the produced AgNPs did not show significant hemolytic effect even at the highest concentration tested (1.00 ppm) demonstrating biocompatibility also when in contact with the bloodstream (hemolytic effect < 2%).
The analogous results as evaluating the pure polymers are given in the ESI File (Fig. S2†) where a slightly higher hemolytic effect was monitored when RBCs were in contact with BHPEI.
Antimicrobial activity
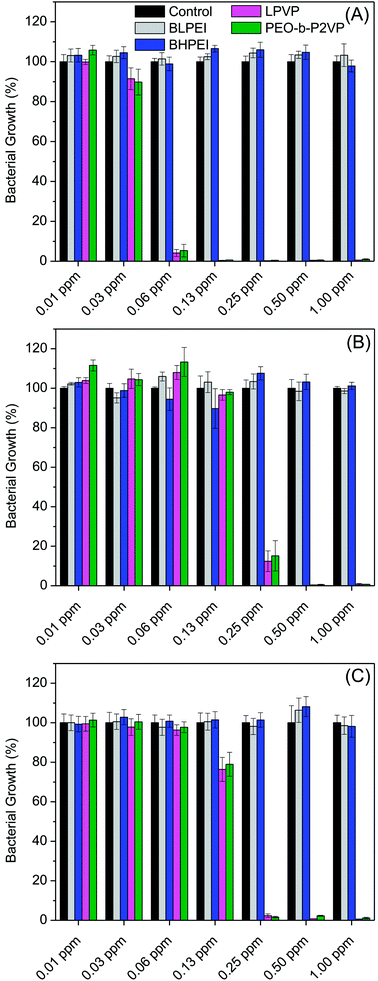
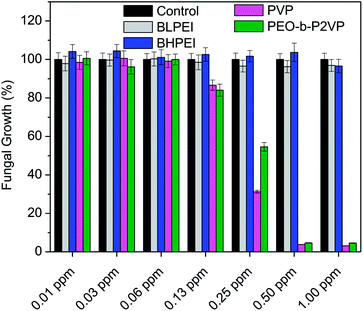
The antibacterial performance of the produced polymer-stabilized AgNPs was evaluated by liquid growth inhibition assay against two Gram-negative (Escherichia coli and Enterobacter cloacaes and one Gram-positive bacteria strains (Bacillus megaterium). The results are portrayed in Fig. 6A–C. Gram-negative and Gram-positive bacteria present membrane composition differences which may have particular influence on the mechanism of action of the proposed antimicrobial agents. Essentially, the Gram-negative species hold higher contents of the zwitterionic lipid phosphatidylethanolamine in the membranes whereas the Gram-positive bacteria membranes are constituted by high amount of the anionic phosphatidylglycerol and cardiolipin lipids.34,35 Besides the evaluation against bacteria strains, the AgNPs were also tested against the fungus Candida albicans and the results are given in Fig. 7.Firstly, the results portrayed in Fig. 6 evidence a remarkable antibacterial activity of the PVP and PEO-b-P2VP stabilized AgNPs when the concentrations are above 0.06 ppm and 0.25 ppm, respectively against Enterobacter cloacae and Escherichia coli. These data regards to the activity against Gram-negative bacteria. On the other way around, and very surprisingly, the BPEI-stabilized AgNPs are not effective as antibacterial agent. The results are to some extent similar when evaluating the performance against the Gram-positive bacterium Bacillus megaterium. In this case, it is observed antibacterial effect of PVP and PEO-b-P2VP-stabilized AgNPs in concentrations higher than 0.13 ppm whereas the BPEI-stabilized AgNPs are not effective in the whole range of concentration evaluated. The results are also similar in the evaluation of the performance against the fungus Candida albicans, although a slightly higher resistance of the microorganism is evidenced (Fig. 7). The minimum growth at the evaluated concentration range was ∼3–4% when the concentration of silver colloids was 0.50 ppm or higher. The BPEI-stabilized AgNPs also does not have any effect against such microorganism. The quantitative values of antimicrobial activity are given in Table 3. The values of MIC90 and MIC99 are defined as the lowest concentrations of the antimicrobial agent at which respectively 90% and 99% of the strains are inhibited.
| Microorganism | BLPEI | BHPEI | PVP | PEO-b-P2VP |
|---|---|---|---|---|
| MIC90 | ||||
| Enterobacter cloacae | — | — | 0.06 ppm | 0.06 ppm |
| Escherichia coli | — | — | 0.50 ppm | 0.50 ppm |
| Bacillus megaterium | — | — | 0.25 ppm | 0.25 ppm |
| Candida albicans | — | — | 0.50 ppm | 0.50 ppm |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| MIC99 | ||||
| Enterobacter cloacae | — | — | 0.13 ppm | 0.13 ppm |
| Escherichia coli | — | — | 0.50 ppm | 0.50 ppm |
| Bacillus megaterium | — | — | 0.50 ppm | 1.00 ppm |
| Candida albicans | — | — | — | — |
The values are within the range reported in the literature for AgNPs manufactured by using different approaches.2,36–38 The same evaluations were performed using the pure polymers where BPEI was found to be a very powerful antimicrobial agent whereas PVP and PEO-b-P2VP are not the responsible for the antimicrobial effect at least in the concentration range 1–0.06 mg mL−1. It is accordingly obvious that the antimicrobial effect of PVP and PEO-b-P2VP stabilized AgNPs comes from the presence of the metallic silver cores (or silver ions) since the polymers themselves do not hold such property (Fig. S3–S4†).
The data reported in Table 3 evidences that the PVP or PEO-b-P2VP stabilized AgNPs possesses essentially equivalent antimicrobial activity for a given microorganism whereas both BPEI-stabilized AgNPs demonstrated to be ineffective against all tested strains in the range of concentration evaluated. Comparing the antimicrobial activity against Gram-negative and Gram-positive strains, one observes that the lowest MIC99 values are reported for Enterobacter cloacae which is a Gram-negative bacterium and the highest MIC99 values are reported for Bacillus megaterium. The understanding of such behavior may linked to the phospholipid compositions of bacteria membranes. The Gram-negative bacteria contain both anionic and zwitterionic phospholipids whereas Gram-positive bacteria contain predominantly zwitterionic phospholipids. For instance, the Enterobacter cloacae membranes consist of 80% zwitterionic phosphatidylethanolamine (PE), 15% anionic phosphatidylglycerol (PG) and 5% anionic cardiolipin.39 The lipid composition of Escherichia coli is similar.40 On the other hand, the Bacillus megaterium membranes consist of roughly 53% zwitterionic phosphatidylethanolamine (PE), 43% anionic phosphatidylglycerol (PG) and 3% anionic cardiolipin.41 Accordingly, electrostatic repulsions between the more negatively charged bacterium membrane and the negatively charge surface of the assemblies can at least partially explain the higher resistance of Bacillus megaterium. Therefore, the surface charge of the hybrid material might have a marginal influence in the observed behavior, although the BPEI-stabilized AgNPs are non-toxic against the tested microorganisms. Amongst the several factors influencing the antimicrobial activity of AgNPs, the stability provide by the capping agent is supposed to, directly or indirectly, severely affect the efficacy. Despite the fact that the antimicrobial activity of pure hyperbranched PEI and of AgNPs is known, the hybrid BPEI-stabilized AgNPs are herein shown to be inert. Presumably, this is due to the highly stable assemblies produced which protects the silver core and consequently does not allow reasonable amounts of Ag+ ions to be released to promote cell damage. The great inhibition of Ag+ release promoted by PEI surface coating was recently confirmed.42 Thus, the PEI shielding restrict metal oxidation resulting in less ionic Ag+ release compared to the anionic and nearly neutral coatings. The presence of ring at the molecular structure of PVP and PEO-b-P2VP may also contribute to a poorer stabilization of the manufactured AgNPs due to stearic effects enabling faster Ag+ release. Additionally, faster Ag+ release can also be explained by the ionic transport property of PVP and PEO-b-P2VP. On the other hand, electrostatic repulsion between Ag+ and the positively charged PEI chains may also eventually slow down Ag+ release. Accordingly, the prime effect of silver ions (Ag+) in such property seems to overall much better describe the reported behaviors as suggested by the literature42–45 whereas the extent to which nanoparticle's features directly affect the toxicity of AgNPs still remains as an open question. This has been also addressed by Sotiriou and Pratsinis46 who concluded that bactericidal activity is dominated by Ag+ rather than by AgNPs, particularly for particles smaller than 10 nm. The paramount importance of silver ions on the toxicity of silver nanoparticles seems to be presently well accepted.15,17,47,48
At this time, it is of due relevance to highlight that there are some reports in the literature highlighting the antimicrobial effect of PEI-stabilized AgNPs.14,23,49 Nevertheless, in those investigations, the PEI chains were used as stabilizer whereas silver reduction was promote by ascorbic acid or sodium borohydride. However, the current report makes us believe that such systems can maybe be considered biocide only at much higher concentrations, if so. The herein employed methodology ruled out the initial presence of unreduced Ag+ and free BPEI chains which could themselves damage the microorganisms. This is true because the colloids were produced in EG environment and further transferred to water. Taking into account that Ag+ and branched PEI chains are highly toxic, the presence of unreduced Ag+ and freely diffusing polymer chains can certainly conduct to misinterpretation of the actual antimicrobial effect of hybrid PEI-stabilized AgNPs. Accordingly, in order to evaluate the effect of PEI-coated AgNPs, one has to unquestionably get rid of all remaining reactants. At this condition, such as in the current investigation, we evidence that the AgNPs stabilized by polyethyleneimine are not effective as an antimicrobial agent at least in the concentration range evaluated. In summary, the antimicrobial effect of AgNPs seems to be governed by the availability of Ag+ and, the stability promoted by the capping agent possibly allows faster Ag+ release from PVP and PEO-b-P2VP-coated AgNPs due to the weaker stabilization provided by such polymers and due to their ionic transport property particularly for silver ions. On the other hand, the PEI coatings seem to greatly inhibit the Ag+ release thereby making the PEI-stabilized systems ineffective in the time scale assessed.
Bacterial inactivation mechanism
The time–kill curves were explored towards a better understanding of the bacterial inactivation mechanism. Accordingly, the suspension of Escherichia coli was subject to treatment and the absorbance was measured as a function of time. The results are portrayed in Fig. 8.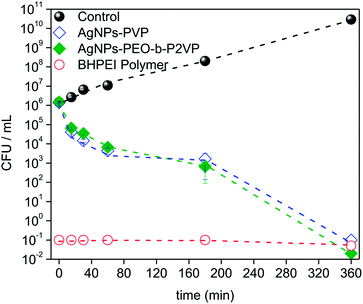 | ||
| Fig. 8 Time–kill plots (CFU mL−1 versus time) for Escherichia coli strain exposed to MIC99 concentrations of distinct antimicrobial agents according to the legend (CFU: colony-forming units). | ||
The results evidence that the strain is progressively inactivated as a function of time when treated by using the AgNPs regardless the stabilizer (PVP or PEO-b-P2VP). One sees that after 3 h treatment the value of log CFU mL−1 drops about three times and it reaches a value seven times smaller when the incubation time doubles (6 h). The profiles reported in Fig. 8 can be used to distinguish between a bacteriostatic and a bactericidal agent. The pure BHPEI polymer is a strong bactericidal agent since more than 99% reduction in CFU mL−1 is instantaneously observed and it defines an antibiotic as bactericidal. On the other hand, the observed effect for the polymer-stabilized AgNPs is time-dependent. Indeed, one of the most important characteristics of an antibiotic is the distinction between the bactericidal or bacteriostatic effect. Essentially, antibiotics that kill bacteria are bactericidal whereas those that inhibit their growth are called bacteriostatic. Although one observes time-dependent killing in Fig. 8, the AgNPs not only inhibit bacterial growth but also kill the large majority of them within the time scale of 6 h and thus the systems can be also considered as bactericidal since the extent of reduction in bacterial numbers is remarkably. Presumably, the required time to promote the biocide effect is linked to the time-dependent release of Ag+.
Conclusions
The use of nitrogen-containing polymers acting simultaneously as reducing and stabilizer agent in the course of silver colloids synthesis is demonstrated. The method regards to an interesting approach and it can be also used for instance in one-pot synthesis of metallic colloids directly in aqueous media without the aid of any other external agent expect the ionic precursor. The entire set of manufactured polymer-stabilized AgNPs were found to be non-toxic to mammalian and red blood cells whereas those stabilized by PVP and PEO-b-P2VP were found to be remarkably biocide against bacteria and fungus strains even at very low concentrations (c ≤ 1.00 ppm). On the other hand, the antimicrobial activity of AgNPs stabilized by positively charged BHPEI and BLPEI was evidenced to be negligible. These findings highlight that the known biocide effect of AgNPs can be suppressed depending on the chemical nature of the capping agent used to provide colloidal stabilization. Furthermore, although this issue will certainly remain heavily debated, the results suggest that the biocide effect is greatly dependent on the presence of Ag+ rather than the nanoparticles themselves. The different polymer coatings are supposed to provide particular Ag+ release kinetics to the resulting hybrid materials. Presumably, the coating provided by polyethyleneimine embraces great stability and hampers Ag+ release from polyethyleneimine-capped AgNPs thereby hindering their biological activity. Instead, the outstanding ionic transport property of PVP and PEO-b-P2VP apparently enables substantial amounts of Ag+ release thus providing the biocide effect to the polymer-stabilized silver colloids.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The investigations were sponsored by FAPESP (Grant no. 2017/00459-4). F.C.G acknowledges the productivity research fellowship granted by CNPq (Grant No. 302467/2014-9) and L.J.C.A thanks the fellowship granted by FAPESP (Grant no. 2016/23844-8). The CEM at UFABC and LCME at UFSC are acknowledged for the accessibility to the Malvern light scattering and electron microscope, respectively.References
- J. R. Morones, J. L. Elechiguerra, A. Camacho, K. Holt, J. B. Kouri, J. T. Ramírez and M. J. Yacaman, Nanotechnology, 2005, 16, 2346–2353 CrossRef CAS PubMed.
- C. Marambio-Jones and E. Hoek, J. Nanopart. Res., 2010, 12, 1531–1551 CrossRef CAS.
- A. Panáček, M. Kolář, R. Večeřová, R. Prucek, J. Soukupová, V. Kryštof, P. Hamal, R. Zbořil and L. Kvítek, Biomaterials, 2009, 30, 6333–6340 CrossRef PubMed.
- S. Galdiero, A. Falanga, M. Vitiello, M. Cantisani, V. Marra and M. Galdiero, Molecules, 2011, 16, 8894–8918 CrossRef CAS PubMed.
- R. R. Arvizo, S. Bhattacharyya, R. a. Kudgus, K. Giri, R. Bhattacharya and P. Mukherjee, Chem. Soc. Rev., 2012, 41, 2943 RSC.
- R. W.-Y. Sun, R. Chen, N. P.-Y. Chung, C.-M. Ho, C.-L. S. Lin and C.-M. Che, Chem. Commun., 2005, 5059 RSC.
- D. M. Eby, H. R. Luckarift and G. R. Johnson, ACS Appl. Mater. Interfaces, 2009, 1, 1553–1560 CAS.
- K. Chaloupka, Y. Malam and A. M. Seifalian, Trends Biotechnol., 2010, 28, 580–588 CrossRef CAS PubMed.
- T. V. Duncan, J. Colloid Interface Sci., 2011, 363, 1–24 CrossRef CAS PubMed.
- E. Lombi, E. Donner, K. G. Scheckel, R. Sekine, C. Lorenz, N. Von Goetz and B. Nowack, Chemosphere, 2014, 111, 352–358 CrossRef CAS PubMed.
- B. Nowack, H. F. Krug and M. Height, Environ. Sci. Technol., 2011, 45, 1177–1183 CrossRef CAS PubMed.
- V. Dal Lago, L. França de Oliveira, K. de Almeida Gonçalves, J. Kobarg and M. Borba Cardoso, J. Mater. Chem., 2011, 21, 12267 RSC.
- S. Pal, Y. K. Tak and J. M. Song, J. Biol. Chem., 2015, 290, 1712–1720 CrossRef PubMed.
- A. M. El Badawy, R. G. Silva, B. Morris, K. G. Scheckel, M. T. Suidan and T. M. Tolaymat, Environ. Sci. Technol., 2011, 45, 283–287 CrossRef CAS PubMed.
- a R. Gliga, S. Skoglund, I. O. Wallinder, B. Fadeel and H. L. Karlsson, Part. Fibre Toxicol., 2014, 11, 11 CrossRef PubMed.
- Z. Chen, P. Yang, Z. Yuan and J. Guo, Sci. Rep., 2017, 7, 1–8 CrossRef PubMed.
- C. Beer, R. Foldbjerg, Y. Hayashi, D. S. Sutherland and H. Autrup, Toxicol. Lett., 2012, 208, 286–292 CrossRef CAS PubMed.
- W. K. Jung, H. C. Koo, K. W. Kim, S. Shin, S. H. Kim and Y. H. Park, Appl. Environ. Microbiol., 2008, 74, 2171–2178 CrossRef CAS PubMed.
- T. C. Dakal, A. Kumar, R. S. Majumdar and V. Yadav, Front. Microbiol., 2016, 7, 1–17 Search PubMed.
- S. Sanyasi, R. K. Majhi, S. Kumar, M. Mishra, A. Ghosh, M. Suar, P. V. Satyam, H. Mohapatra, C. Goswami and L. Goswami, Sci. Rep., 2016, 6, 1–16 CrossRef PubMed.
- M. M. Stevanović, S. D. Škapin, I. Bračko, M. Milenković, J. Petković, M. Filipič and D. P. Uskoković, Polymer, 2012, 53, 2818–2828 CrossRef.
- M. Qasim, N. Udomluck, J. Chang, H. Park and K. Kim, Int. J. Nanomed., 2018, 13, 235–249 CrossRef PubMed.
- H. J. Lee, S. G. Lee, E. J. Oh, H. Y. Chung, S. I. Han, E. J. Kim, S. Y. Seo, H. Do Ghim, J. H. Yeum and J. H. Choi, Colloids Surf., B, 2011, 88, 505–511 CrossRef CAS PubMed.
- M. Stevanović, I. Savanović, V. Uskoković, S. D. Škapin, I. Bračko, U. Jovanović and D. Uskoković, Colloid Polym. Sci., 2012, 290, 221–231 Search PubMed.
- C. A. dos Santos, A. F. Jozala, A. Pessoa and M. M. Seckler, J. Nanobiotechnol., 2012, 10, 1–6 CrossRef PubMed.
- R. C. B. Scaravelli, R. L. Dazzi, F. C. Giacomelli, G. Machado, C. Giacomelli and V. Schmidt, J. Colloid Interface Sci., 2013, 397, 114–121 CrossRef CAS PubMed.
- C. Batista, L. Albuquerque, C. Ribeiro, C. de Castro, E. Miranda, I. Nantes, B. Albuquerque, M. Cardoso and F. Giacomelli, J. Braz. Chem. Soc., 2017, 28(9), 1608–1618 CAS.
- P. Stepánek, Dynamic Light Scattering: The Method and Some Applications, Oxford Science Publications, Oxford, 1993 Search PubMed.
- D. Paramelle, A. Sadovoy, S. Gorelik, P. Free, J. Hobley and D. G. Fernig, Analyst, 2014, 139, 4855 RSC.
- K. Kalishwaralal, S. BarathManiKanth, S. R. K. Pandian, V. Deepak and S. Gurunathan, Colloids Surf., B, 2010, 79, 340–344 CrossRef CAS PubMed.
- S. Vaidyanathan, B. G. Orr and M. M. Banaszak Holl, Acc. Chem. Res., 2016, 49, 1486–1493 CrossRef CAS PubMed.
- Y. Ma, B. Chen, N. He, G. Chen, L. Li and C. Wu, Macromol. Biosci., 2014, 14, 1807–1815 CrossRef CAS PubMed.
- S. Taranejoo, J. Liu, P. Verma and K. Hourigan, J. Appl. Polym. Sci., 2015, 132, 42096 CrossRef.
- C. N. Pedron, M. D. T. Torres, J. A. da S. Lima, P. I. Silva, F. D. Silva and V. X. Oliveira, Eur. J. Med. Chem., 2017, 126, 456–463 CrossRef CAS PubMed.
- R. M. Epand and R. F. Epand, Biochim. Biophys. Acta, Biomembr., 2009, 1788, 289–294 CrossRef CAS PubMed.
- Y. Zhang, H. Peng, W. Huang, Y. Zhou and D. Yan, J. Colloid Interface Sci., 2008, 325, 371–376 CrossRef CAS PubMed.
- B. Perito, E. Giorgetti, P. Marsili and M. Muniz-Miranda, Beilstein J. Nanotechnol., 2016, 7, 465–473 CrossRef CAS PubMed.
- C. Ortiz, R. Torres and D. Paredes, Int. J. Nanomed., 2014, 9, 1717 CrossRef PubMed.
- P. M. Oliver, J. A. Crooks, M. Leidl, E. J. Yoon, A. Saghatelian and D. B. Weibel, J. Bacteriol., 2014, 196, 3386–3398 CrossRef PubMed.
- P. L. Yeagle, The Structure of Biological Membranes, Third Edition, CRC Press, 2011 Search PubMed.
- K. E. Langley, M. P. Yaffe and E. P. Kennedy, J. Bacteriol., 1979, 140, 996–1007 CAS.
- S. Jahan, Y. B. Alias, A. F. Bin Abu Bakar and I. Bin Yusoff, Colloid Polym. Sci., 2017, 295, 1961–1971 CAS.
- M. Connolly, M.-L. Fernandez-Cruz, A. Quesada-Garcia, L. Alte, H. Segner and J. Navas, Int. J. Environ. Res. Public Health, 2015, 12, 5386–5405 CrossRef CAS PubMed.
- R. Zouzelka, P. Cihakova, J. Rihova Ambrozova and J. Rathousky, Environ. Sci. Pollut. Res., 2016, 23, 8317–8326 CrossRef CAS PubMed.
- Z.-M. Xiu, J. Ma and P. J. J. Alvarez, Environ. Sci. Technol., 2011, 45, 9003–9008 CrossRef CAS PubMed.
- G. A. Sotiriou and S. E. Pratsinis, Environ. Sci. Technol., 2010, 44, 5649–5654 CrossRef CAS PubMed.
- Z. M. Xiu, Q. B. Zhang, H. L. Puppala, V. L. Colvin and P. J. J. Alvarez, Nano Lett., 2012, 12, 4271–4275 CrossRef CAS PubMed.
- X. Yang, A. P. Gondikas, S. M. Marinakos, M. Auffan, J. Liu, H. Hsu-Kim and J. N. Meyer, Environ. Sci. Technol., 2012, 46, 1119–1127 CrossRef CAS PubMed.
- D. Xu, Q. Wang, T. Yang, J. Cao, Q. Lin, Z. Yuan and L. Li, Int. J. Environ. Res. Public Health, 2016, 13, 334 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra13597a |
| This journal is © The Royal Society of Chemistry 2018 |