Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Eugenol triggers CD11b+Gr1+ myeloid-derived suppressor cell apoptosis via endogenous apoptosis pathway
Ying Ding†
 a,
Zecheng Yang†b,
Wensheng Zhang
a,
Zecheng Yang†b,
Wensheng Zhang c,
Yuwei Xu
c,
Yuwei Xu b,
Yuanyuan Wang
b,
Yuanyuan Wang d,
Minghua Hu
d,
Minghua Hu d,
Fangli Ma
d,
Fangli Ma d,
Hanan Long*fg,
Ning Tao
d,
Hanan Long*fg,
Ning Tao *e and
Zhihai Qin
*e and
Zhihai Qin *ae
*ae
aSchool of Basic Medical Sciences of Southwest Medical University, Luzhou, China
bCollege of Life Science, University of the Chinese Academy of Sciences, Beijing, China
cDepartment of Microbiology and Immunology, Shanxi Medical University, Taiyuan, China
dInfinitus Chinese Herbal Immunity Research Centre, Infinitus China Company Ltd, Guangzhou, China
eKey Laboratory of Protein and Peptide Pharmaceuticals, Institute of Biophysics, Chinese Academy of Sciences, Datun Road No. 15, Chaoyang District, Beijing, China. E-mail: tao@ibp.ac.cn; zhihai@ibp.ac.cn
fDepartment of Pathology, The Affiliated Hospital of Southwest Medical University, Luzhou, Sichuan, China. E-mail: longhanan@swmu.edu.cn
gDepartment of Science and Technology, Southwest Medical University, Luzhou, Sichuan, China
First published on 19th January 2018
Abstract
To study the effect and underlying molecular mechanism of eugenol on CD11b+Gr1+ myeloid-derived suppressor cells (MDSCs). The effect of eugenol on the inhibition of immortalized MDSC cell line MSC-2 and murine peritoneal macrophages was detected by MTT. Flow cytometry was used to detect the pro-apoptosis effect of eugenol on MDSCs. The expression levels of apoptosis-related proteins were detected by western blot. Eugenol has a selective inhibitory effect on MDSCs in a dose-dependent manner, which activates an endogenous apoptosis pathway, leading to apoptosis. Eugenol promotes the apoptosis of MDSCs via the intrinsic pathway.
1. Introductions
Cancer is a major public health problem worldwide and is the main cause of death in most countries.1–3 However, conventional cancer therapies, including surgery, radiotherapy and chemotherapy, have faced impenetrable challenges due to their side effects and high recurrence rates.4–6 Therefore, safe and effective therapeutic strategies are urgently needed for treating malignant tumours.7,8CD11b+Gr1+ myeloid-derived suppressor cells (MDSCs) are heterogeneous cells within the tumour microenvironment;9,10 they consist of myeloid precursor cells and immature myeloid cells, and they play a major role in the regulation of the immune response to tumours.11,12 Previous studies have demonstrated that MDSCs downregulate the innate and adaptive immune systems.13,14 As the primary immune suppressors of the tumour microenvironment, MDSCs have been shown to be effective targets for cancer therapy.15,16 Many molecules (for example, gemcitabine, docetaxel and 5-fluorouracil) targeting MDSCs have been demonstrated to have therapeutic effects in tumour treatment.17–19
Natural products are important for drug development, and many extractions of natural materials, Taxol and MPSSS, for instance, have strong anti-tumour features.18–20 Eugenol (4-allyl-1-hydroxy-2-methoxybenzene) is the main component of clove oil, which is often used as a preservative, analgesic and antimicrobial agent.21 Eugenol has extensive pharmacological effects, such as inhibiting the NF-κB pathways, participating in immune regulation and the anti-inflammatory response, blocking the activity of xanthine oxidase, affecting temperature-sensitive neuron discharge and antipyretic activities.22–26 Moreover, eugenol is used for animal anaesthesia, mosquito repellent and the promotion of drug permeation absorption. Eugenol shows potential for application in the prevention and treatment of certain tumours.27–29 However, whether eugenol can reduce immune suppression in the tumour microenvironment especially that caused by MDSCs, is unknown.
In this study, we explored the effect of eugenol on MDSCs. We found that eugenol effectively promoted the apoptosis of MDSCs and inhibited their immunosuppressive function. Our research reveals a novel use for eugenol in anti-tumour immunity, suggesting its potential use in anti-tumour therapy.
2. Materials and methods
2.1. Cell lines
Three immortalized cell lines were used in this study. MSC-2, generated from a retrovirus encoding the v-raf and v-myc oncogenes, had a macrophage-like phenotype and was provided by the Francois Ghiringhelli laboratory, Department of Medical Oncology, France. CT-26, a murine colon carcinoma cell line, was provided by Yang xin, Fu research group of the Institute of Biophysics, Chinese Academy of Sciences. MC-38 cells from a weakly immunogenic murine colon adenocarcinoma induced by the SC injection of dimethyl hydrazine in C57BL/6 mice were maintained in our research group. MSC-2, CT-26 and MC-38 cells were routinely cultured in DMEM high sugar medium (HyClone, USA) supplemented with 10% fetal bovine serum (PAN, Co. Germany), 1% penicillin and streptomycin. Cells were cultured at 37 °C in a fully humidified incubator equilibrated with 5% carbon dioxide (CO2).2.2. The preparation of the primary cells
2.3. MTT assay
An MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl-tetra-zolium bromide) assay was used to evaluate the eugenol-induced cytotoxicity. Briefly, cells in the logarithmic growth phase were seeded into a 96-well culture plate at a density of 0.6 × 104 cells per well in 100 μL, incubated with different doses of eugenol for 24 hours. Then 10 μL of the MTT solution (5 mg mL−1) was added and incubated for 4 hours before dissolving. Triple combine buffer (10% SDS, 5% isobutanol, 0.012 mol L−1 HCl, dissolved in distilled water) was added to dissolve the formazan crystal. The absorbance was measured at 570 nm using an absorbance microplate reader (BioTek, USA), and the half-maximal inhibitory concentration (IC50) values were determined.Cell viability was calculated as follows: 100 × (absorbance of eugenol treated cells/absorbance of maximum MTT released control cells). Graph Pad Prism 5.0 was used to analyse the data.
2.4. Flow cytometry
2.5. Western blot analysis of cell proteins
MSC-2 cells treated with a certain concentration. For various durations were harvested from a 6-well plate and then lysed in RIPA buffer (Beyotime, China) to isolate whole cell proteins. The cell extract containing proteins (30 μg) was separated on 12% SDS–polyacrylamide gels and electrophoretic ally transferred onto a nitrocellulose membrane (GE Healthcare, Milwaukee, USA). The membrane was blocked in 3% BSA in PBS-T (0.1% Tween-20) at 4 °C overnight and probed with the primary antibodies as follows: anti-p65 nuclear factor kappa B (NF-κB) monoclonal antibody, anti-β-actin monoclonal antibody, and anti-cytochrome C, anti-bax, anti-caspase 3, anti-BCL-2, anti-caspase 8, and anti-caspase 9 polyclonal antibodies (Cell Signalling Technology, US). After washing three times with PBS-T, HRP-conjugated goat anti-mouse or goat anti-rabbit IgG was used as the secondary antibody. Specific bands were visualized using the Chemiluminescence Imaging System (Clinx Science Instruments Co. Ltd, Shanghai, China).2.6. Statistical analysis
All data are presented as the means ± SD (standard deviation) and evaluated by one-way ANOVA. *p < 0.05 was regarded as statistically significant. All of the experimental procedures were independently repeated at least three times.3. Results
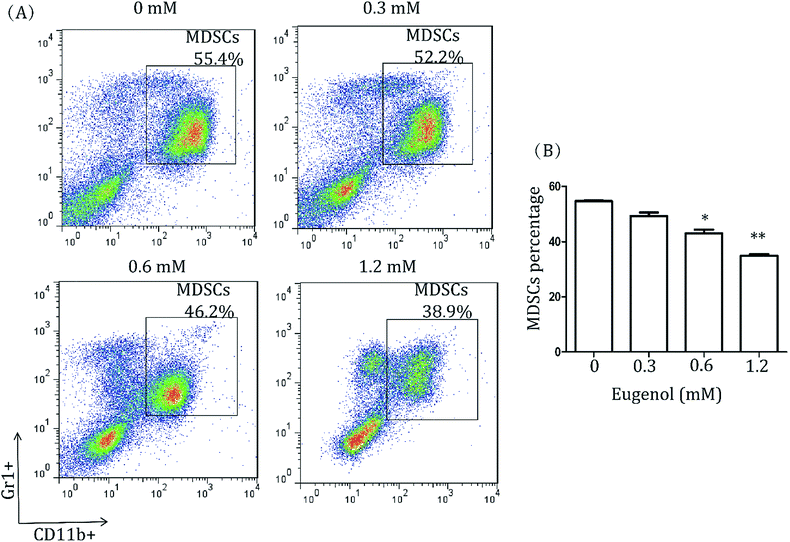
3.1. Eugenol reduces the proportion of MDSCs in splenocytes
To study the effect of eugenol on MDSCs, spleen cells from CT-26 tumour-bearing mice were treated with different concentrations of eugenol and analysed by flow cytometry. As shown in Fig. 1, eugenol reduced the MDSCs (Gr1+, CD11b+) numbers in a dose-dependent manner and significantly suppressed MDSCs at a concentration of 0.60 mM.3.2. Eugenol has an inhibitory effect on immortalized MDSC cell line MSC-2, and eugenol promotes the cell viability of macrophages near the IC50 concentration
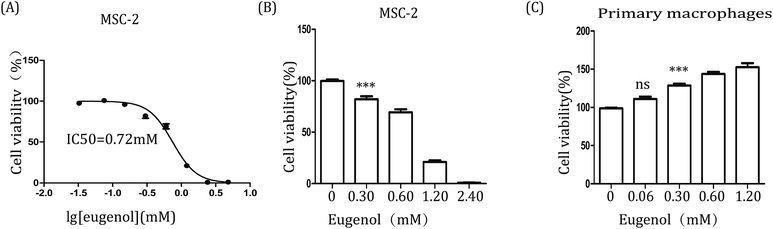
Further, to examine the effect of eugenol on MDSCs, we first investigated the immortalized MDSC cell line MSC-2. After the cells were treated with eugenol at various concentrations for 24 hours, the concentration that resulted in ∼50% cell death (IC50) was determined to be 0.72 mM (Fig. 2A). Eugenol reduced the viability of MSC-2 cells in a dose-dependent manner and significantly affected cell viability at a concentration of 0.30 mM (*p < 0.05) (Fig. 2B). Interestingly, the MTT assay results also showed that the viability of macrophages (isolated from abdominal cavity of male BALB/c mice) was promoted when the cells were stimulated with eugenol (Fig. 2C).3.3. Eugenol induces apoptosis in MSC-2 cells via the intrinsic pathway
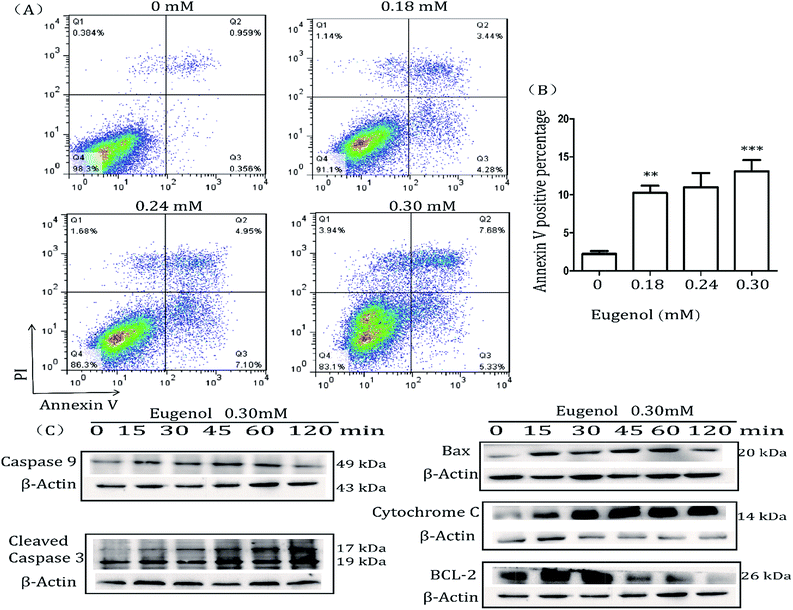
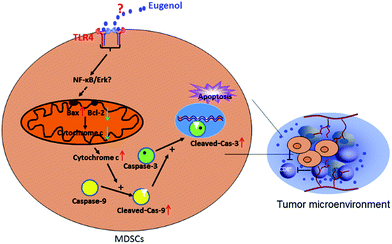
MTT results showed that eugenol decreased the proportion of living MSC-2 cells. We next investigated the mechanism underlying this phenomenon. The Annexin V-FITC/PI double-stained apoptosis detection assay revealed that the apoptosis rate of MSC-2 cells was significantly increased when cells were treated with increasing concentrations of eugenol (Fig. 3A and B). To investigate the molecular mechanism underlying eugenol-induced MSC-2 cell apoptosis, we performed a western blot to investigate the changes in expression of apoptosis-related proteins. As shown in Fig. 3C, the expression levels of caspase-9, cleaved caspase-3 and cytochrome C were upregulated in a time-dependent manner.Caspase-8 was barely expressed, even when cells were treated for 120 min (data not shown). Given that caspase-9, cleaved caspase-3 and cytochrome C are all involved in the intrinsic apoptosis pathway, and caspase-8 is related to the extrinsic pathway, these findings suggest that eugenol induces the apoptosis of MSC-2 cells via the endogenous mitochondrial cytochrome C pathway rather than the extrinsic pathway. Bcl-2 was initially expressed, but expression decreased over time.
3.4. Eugenol could not significantly decrease the percentage of MDSCs in the tumour-bearing mice with TLR4 knockout
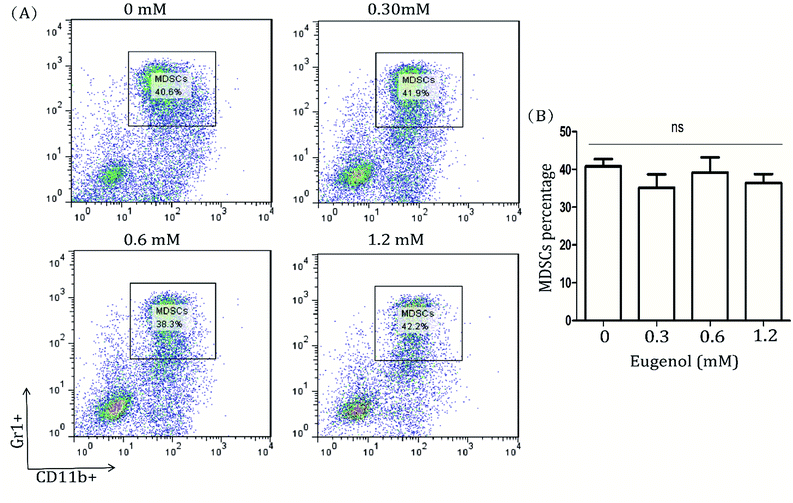
Detecting the spleen cells of other tumour-bearing mice, we found a different data. The results from eugenol-treated MDSCs isolated from TLR4 gene knockout mice indicated that eugenol did not significantly induce apoptosis in these MDSCs (Fig. 4A). That finding, taken together with the results of the wild-type BABL/c mice, indicated TLR was possible the receptor for eugenol on MDSCs (Fig. 1B and 4B).4. Discussion
Many natural compounds have been used in cancer therapy to induce differentiation, inhibit function, and reduce the population of MDSCs.16 We investigated the percentage of MDSCs of splenocytes isolated from tumour-bearing mice. The results showed that the percentage of MDSCs in the spleen was decreased after treatment with eugenol (Fig. 1), suggesting that eugenol reduced the population of MDSCs. The MTT assay performed on the immortalized MDSC cell line MSC-2 confirmed this result, and the IC50 was determined to be 0.72 mM (Fig. 2A and B). Moreover, macrophages isolated from ascitic fluid demonstrated a visible proliferation when treated with eugenol, which may be evidence of an additional anti-tumour function of eugenol (Fig. 2C).Apoptosis has been widely investigated for drug development and for elucidating anti-tumour mechanisms. Programmed cell death is launched from the intrinsic or extrinsic pathway, which are characterized by caspase-8 and caspase-9, respectively.30,31 In addition to the flow cytometry results showing that eugenol induced MSC-2 cell apoptosis (Fig. 3A and B), further experiments showed that the expression level of caspase-9 increased when cells were treated with eugenol (Fig. 3C). Taken together, these results suggested that eugenol activated the intrinsic apoptosis pathway in the immortalized MDSC cell line MSC-2. The loss of mitochondrial membrane potential results in the release of apoptosis-inducing proteins, such as cytochrome C, from the intermembrane space into the cytosol.32,33 The fate of cells undergoing apoptosis primarily depends on the ratio of antagonist molecules (Bcl-2, Bcl-xL, Mcl-1, and A1) to agonist molecules (Bax, Bak, Bcl-xs, and Bad).34 In this study, we found that the expression levels of cytochrome C and Bax increased, whereas the expression level of Bcl-2 decreased in a time depended manner in MSC-2 cells when treated with eugenol (Fig. 3C and D). This finding contributed to our conclusion.
Many studies have shown that the activation of the TLR4/NF-κB signalling pathway triggers apoptotic cascades.35 Our research demonstrated that eugenol did not decrease the ratio of MDSCs in the spleens of TLR4 knockout mice bearing colon tumours, suggesting that TLR4 is a possibly receptor involved in the promotion of apoptosis by eugenol. Further studies focusing on the detailed mechanisms and the role of TLR4 in this process are warranted.
In summary, our study demonstrated a strong anti-MDSC effect of eugenol in tumour immune therapy, suggesting the potential of eugenol as a prototype for designing and developing new anti-tumour medicine (Fig. 5).
5. Conclusions
In conclusion, we found that eugenol induces apoptosis in CD11b+Gr1+ myeloid-derived suppressor cells and promotes the viability of primary macrophages in mice, thereby mobilizing the body's ability to suppress tumour growth.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 31370910 and 81373327).References
- W. Chen, R. Zheng, P. D. Baade, S. Zhang, H. Zeng and F. Bray, et al., Ca-Cancer J. Clin., 2016, 66, 115–132 CrossRef PubMed.
- R. L. Siegel, K. D. Miller and A. Jemal, Ca-Cancer J. Clin., 2015, 65, 5–29 CrossRef PubMed.
- R. L. Siegel, K. D. Miller and A. Jemal, Ca-Cancer J. Clin., 2016, 66, 7–30 CrossRef PubMed.
- G. Kroemer, L. Galluzzi, O. Kepp and L. Zitvogel, Annu. Rev. Immunol., 2013, 31, 51–72 CrossRef CAS PubMed.
- G. Kroemer, L. Senovilla, L. Galluzzi, F. Andre and L. Zitvogel, Nat. Med., 2015, 21, 1128–1138 CrossRef CAS PubMed.
- M. Obeid, A. Tesniere, F. Ghiringhelli, G. M. Fimia, L. Apetoh and J. L. Perfettini, et al., Nat. Med., 2007, 13, 54–61 CrossRef CAS PubMed.
- M. Michaud, I. Martins, A. Q. Sukkurwala, S. Adjemian, Y. T. Ma and P. Pellegatti, et al., Science, 2011, 334, 1573–1577 CrossRef CAS PubMed.
- O. Kepp, L. Galluzzi, I. Martins, F. Schlemmer, S. Adjemian and M. Michaud, et al., Cancer Metastasis Rev., 2011, 30, 61–69 CrossRef CAS PubMed.
- D. I. Gabrilovich and S. Nagaraj, Nat. Rev. Immunol., 2009, 9, 162–174 CrossRef CAS PubMed.
- K. H. Parker, D. W. Beury and S. Ostrand-Rosenberg, Adv. Cancer Res., 2015, 128, 95–139 CrossRef PubMed.
- V. Kumar, S. Patel, E. Tcyganov and D. I. Gabrilovich, Trends Immunol., 2016, 37, 208–220 CrossRef CAS PubMed.
- Y. Zhao, T. Wu, S. Shao, B. Shi and Y. Zhao, OncoImmunology, 2016, 5, e1004983 CrossRef PubMed.
- P. Trikha and W. E. Carson III, Biochim. Biophys. Acta, 2014, 1846, 55–65 CAS.
- L. Yang, L. M. DeBusk, K. Fukuda, B. Fingleton, B. Green-Jarvis and Y. Shyr, et al., Cancer Cell, 2004, 6, 409–421 CrossRef CAS PubMed.
- J. Medina-Echeverz, F. Aranda and P. Berraondo, OncoImmunology, 2014, 3, e28398 CrossRef PubMed.
- T. J. Waldron, J. G. Quatromoni, T. A. Karakasheva, S. Singhal and A. K. Rustgi, OncoImmunology, 2013, 2, e24117 CrossRef PubMed.
- K. S. Atretkhany, M. A. Nosenko, V. S. Gogoleva, R. V. Zvartsev, Z. Qin and S. A. Nedospasov, et al., Front. Immunol., 2016, 7, 147 Search PubMed.
- H. Qin, B. Lerman, I. Sakamaki, G. Wei, S. C. Cha and S. S. Rao, et al., Nat. Med., 2014, 20, 676–681 CrossRef CAS PubMed.
- H. Wu, N. Tao, X. Liu, X. Li, J. Tang and C. Ma, et al., PLoS One, 2012, 7, e51751 CAS.
- P. L. Raber, P. Thevenot, R. Sierra, D. Wyczechowska, D. Halle and M. E. Ramirez, et al., Int. J. Cancer, 2014, 134, 2853–2864 CrossRef CAS PubMed.
- S. Fujisawa and Y. Murakami, Adv. Exp. Med. Biol., 2016, 929, 45–66 CrossRef CAS PubMed.
- T. F. Bachiega, J. P. de Sousa, J. K. Bastos and J. M. Sforcin, J. Pharm. Pharmacol., 2012, 64, 610–616 CrossRef CAS PubMed.
- E. Dervis, A. Yurt Kilcar, E. I. Medine, V. Tekin, B. Cetkin and E. Uygur, et al., Cancer Biother.Radiopharm., 2017, 32, 75–81 CrossRef CAS PubMed.
- X. Han and T. L. Parker, Pharm. Biol., 2017, 55, 1619–1622 CrossRef CAS PubMed.
- A. Hussain, K. Brahmbhatt, A. Priyani, M. Ahmed, T. A. Rizvi and C. Sharma, Cancer Biother.Radiopharm., 2011, 26, 519–527 CrossRef CAS PubMed.
- P. L. Junior, D. A. Camara, A. S. Costa, J. L. Ruiz, D. Levy and R. A. Azevedo, et al., Phytomedicine, 2016, 23, 725–735 CrossRef CAS PubMed.
- S. K. Jaganathan and E. Supriyanto, Molecules, 2012, 17, 6290–6304 CrossRef CAS PubMed.
- M. Ma, Y. Ma, G. J. Zhang, R. Liao, X. F. Jiang and X. X. Yan, et al., Oncotarget, 2017, 8, 56296–56310 Search PubMed.
- A. Sarkar, S. Bhattacharjee and D. P. Mandal, Asian Pac. J. Cancer Prev. APJCP, 2015, 16, 6753–6759 CrossRef PubMed.
- M. H. Wu, X. K. Jin, A. Q. Yu, Y. T. Zhu, D. Li and W. W. Li, et al., Fish Shellfish Immunol., 2014, 41, 625–632 CrossRef CAS PubMed.
- S. Kumar, Cell Death Differ., 2007, 14, 32–43 CrossRef CAS PubMed.
- M. Madesh, B. Antonsson, S. M. Srinivasula, E. S. Alnemri and G. Hajnoczky, J. Biol. Chem., 2002, 277, 5651–5659 CrossRef CAS PubMed.
- G. van Loo, P. Schotte, M. van Gurp, H. Demol, B. Hoorelbeke and K. Gevaert, et al., Cell Death Differ., 2001, 8, 1136–1142 CrossRef CAS PubMed.
- J. M. Adams and S. Cory, Science, 1998, 281, 1322–1326 CrossRef CAS PubMed.
- X. Wang, Y. Sun, H. Yang, Y. Lu and L. Li, Sci. Rep., 2016, 6, 27866 CrossRef CAS PubMed.
Footnote |
| † These authors contributed equally to the manuscript. |
| This journal is © The Royal Society of Chemistry 2018 |