Open Access Article
Open Access ArticleSemi-rational screening of the inhibitors and β-lactam antibiotics against the New Delhi metallo-β-lactamase 1 (NDM-1) producing E. coli
Juan Wang†
,
Yang Li†,
Haizhong Yan,
Juan Duan,
Xihua Luo,
Xueqin Feng,
Lanfen Lu and
Weijia Wang *
*
Laboratory Medicine Department, Zhongshan People's Hospital, The Affiliated Hospital of Sun Yat-Sen University, Guangdong Province, No. 2 Sun Wen East Road, Zhongshan, Guangdong 528403, China. E-mail: wangweijia2017@sina.cn
First published on 7th February 2018
Abstract
Bacteria containing blaNDM-1 gene are a growing threat to almost all clinically β-lactam antibiotics. Especially, the New Delhi metallo-β-lactamase (NDM-1) has become a potential public survival risk. In this study, a novel and efficient strategy for inhibitors and β-lactam antibiotics screening using recombinant New Delhi metallo-beta-lactamase (NDM-1) was developed. First, the gene of blaNDM-1 were identified and cloned from multi-drug resistance of Acinetobacter baumannii isolate; by the means of protein expression and purification, recombinant NDM-1 activity was up to 68.5 U ml−1, and high purity NDM-1 protein with activity of 347.4 U mg−1 was obtained. Finally, for NDM-1, the inhibitors (aspergillomarasmine A (AMA) and EDTA) with high affinity (HI) and the β-lactam antibiotics (imipenem) with low affinity (LA) were screened out. Surprisingly, the inhibition of the NDM-1 was enhanced by the use of inhibitor combinations (AMA–EDTA (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2)), where the IC50 of AMA–EDTA was reduced by 88% and 95%, respectively, comparing to the AMA and EDTA alone. More interesting, AMA–EDTA could restore the activity of imipenem when tested against NDM-1 expressing strains (E. coli and Acinetobacter baumannii), with a working time of 120 min and 330 min, respectively. This method is expected to be used in high-throughput screening, drug redesign (including new inhibitors and drugs) and “old drug new use”.
2)), where the IC50 of AMA–EDTA was reduced by 88% and 95%, respectively, comparing to the AMA and EDTA alone. More interesting, AMA–EDTA could restore the activity of imipenem when tested against NDM-1 expressing strains (E. coli and Acinetobacter baumannii), with a working time of 120 min and 330 min, respectively. This method is expected to be used in high-throughput screening, drug redesign (including new inhibitors and drugs) and “old drug new use”.
1. Introduction
Antibiotics play an important role in disease treatment of animals and humans in the past few years. Tens of thousands of lives have been saved by the high efficacy and low toxicity of β-lactam antibiotics. However, as a large number of antibiotic abuse and resistance screening, pathogens equipped with drug resistance weapons fight against mankind. Nowadays, multidrug-resistant (MDR) pathogens infections are becoming a real threat to international public health.1 Since the emergence of multidrug-resistant strains, the difficulty of clinical treatment and medication are increasing, the health of animal and humans are in risk seriously. Therefore, mechanism of drug resistance of the MDR have been revealed. Research shows, the expression of β-lactamase is the most common mechanism of resistance in drug-resistant bacteria.2β-Lactamases has the ability of hydrolysis the C–N bond of lactam ring of lactam antibiotics, leading the antibiotics inactive.3 β-Lactamases has been classified into two categories: serine-β-lactamases (SBLs) and metallo-β-lactamases (MBLs).4 Of those, serine-β-lactamases (SBLs) can be inhibited by many inhibitors, including the clavulanic acid, sulbactam, tazobactam and avibactam in clinical trials.5 Due to their extensive substrate spectrum and high activity, MBLs is more dangerous to human beings and pose an increasing public health risk.6 Bacterial equipped with MBLs can degrade a large number of β-lactams, such as penicillin, cephalosporin, carbapenems, and aztreonam.7 Bacteria with MBLs infections become a major public health problem worldwide.
The New Delhi metallo-β-lactamase 1 (NDM-1) is a new member of MBLs, and has rapidly emerged as the leading threat to the treatment of infections. The NDM-1 encoded by the blaNDM-1 gene was initially identified in a Klebsiella pneumonia isolate in 2008.8 Since then, NDM-1 has been found at least 180 variants in different strains, including Escherichia coli,9 Acinetobacter baumannii,10 Vibrio cholera, Stenotrophomonas maltophilia and Pseudomonas putida.11 What's worse, few antibiotics can be used for the treatment of patients, who was infected by the bacteria carrying of NDM-1.12
Currently, there are two strategies to preserve the efficacy of β-lactam antibiotics for the NDM-1-producing strains: (I) discover or redesign new β-lactam antibiotics, that can avoid to be hydrolyzed by NDM-1, however, it will takes a long time; (II) inhibit NDM-1 activity, resulting the partner β-lactam antibiotics can work properly interfering with the formation of bacterial cell walls, which has been regarded as the quick and efficient route. In order to ensure activity of antibiotics (efficacy), efficient and safe inhibitors are urgently needed to overcome the activity of NDM-1. However, NDM-1 are not susceptible to the SBLs' inhibitors, various structurally types of inhibitors have been described with no effect, including carboxylic/succinic acids,13 triazoles/tetrazoles,14 thiols,15 trifluoromethyl ketones and others.16 NDM-1-mediated resistance to β-lactam antibiotics emerged as a clinical threat to life-saving.
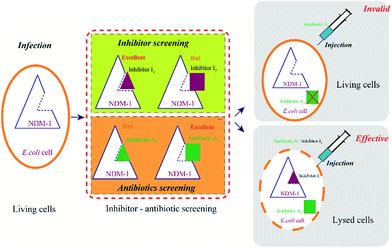
Therefore, the key issue is how to obtain the inhibitors for NDM-1 inhibition, assisting the antibiotics against the multidrug-resistant strains harboring blaNDM-1 gene, synergistically. In the present study, the NDM-1-encoding gene from Acinetobacter baumannii was overexpressed in Escherichia coli, optimal expression conditions and the physical and chemical properties of NDM-1 were then identified. The aim is to obtain a best inhibitor to inhibit NDM-1 activity and combine the inhibitor with excellent antibiotics to against NDM-1 producing strains. A novel and efficient strategy for inhibitors and β-lactam antibiotics screening using recombinant New Delhi metallo-beta-lactamase (NDM-1) was developed.
2. Materials and Methods
2.1. Materials bacterial strains, plasmids, and enzymes
Chemicals and solvents were purchased from standard suppliers and used without further purification. Ampicillin, penicillin G and piperacillin were purchased from Sangon Biotechnology Co. Ltd (Shanghai, China); cefuroxime, ceftizoxime, cefpiramide, ceftazidime, cefoxitin, cefaclor, cephalexin, nitrocefin, cefepime, cephalothin, cefotaxime, imipenem and meropenem were purchased from TCI (Shanghai, China); inhibitors were purchased from Sigma-Aldrich Corporation (USA). The host strain Escherichia coli BL21 (DE3), E. coli JM109 [recA supE hsR Δ(lac-pro)] and the expression plasmid pET-28a (+) were obtained from Novagen (Madison, WI, USA). LATaq DNA and PrimeSTAR DNA polymerase were from TakaRa (Dalian, China). The T-vector, restriction enzymes, T4-ligase, plasmid miniprep kit, and agarose gel DNA purification kit were supplied by TaKaRa Biotechnology (Otsu, Japan).2.2. Expression of the New Delhi metallo-β-lactamase 1 (NDM-1) in E. coli BL21 (DE3)
Acinetobacter baumannii isolates from four general hospitals in the Guangzhou area of China during June 2012 and June 2016. Complete DNA of Acinetobacter baumannii S002 was used as template. The blaNDM-1 gene was amplified by polymerase chain reaction (PCR) using the following primers: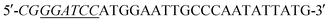 (forward) and
(forward) and 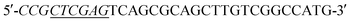 (reverse; the underlined regions are BamHI and XhoI restriction sites, respectively). PCR parameters: initial denaturation at 95 °C for 5 min; denaturation at 94 °C for 1 min, annealing at 60 °C for 1 min, extension at 72 °C for 1.2 min, repeated for 30 cycles; final extension step at 72 °C for 2 min. The resulting PCR product was purified with a SanPrep Column PCR Product Purification Kit (Sangon Biotech Co. Ltd., Shanghai, China). The purified PCR product and the T7-based expression vector pET28 a (+) (Novagen, Madison, Wisconsin) were digested with BamHI and XhoI for 4 h at 37 °C, respectively. The digested product purified using a SanPrep Column PCR product purification Kit, and ligated using T4-DNA ligase at 16 °C 24 h. The ligation mixture was transformed to E. coli JM109 (Novagen, Madison, WI), and transformants were selected for growth on solid Luria–Beltane (LB) agar plates containing kanamycin (50 μg ml−1) and 0.05 M Zn2+. Positive clones were validated by PCR using the primers 5′-ATCGTCAGGGATGGCGGCCGCG-3′ (forward) and 5′-TGCGACATCGTATAACGTTACTGGT-3′ (reverse). The final plasmid, pET28a-NDM-1, purified from a single colony was identified by sequencing (Sangon Biotech, Shanghai). The correct pET28a-NDM-1 was transformed into E. coli BL21 (DE3).
(reverse; the underlined regions are BamHI and XhoI restriction sites, respectively). PCR parameters: initial denaturation at 95 °C for 5 min; denaturation at 94 °C for 1 min, annealing at 60 °C for 1 min, extension at 72 °C for 1.2 min, repeated for 30 cycles; final extension step at 72 °C for 2 min. The resulting PCR product was purified with a SanPrep Column PCR Product Purification Kit (Sangon Biotech Co. Ltd., Shanghai, China). The purified PCR product and the T7-based expression vector pET28 a (+) (Novagen, Madison, Wisconsin) were digested with BamHI and XhoI for 4 h at 37 °C, respectively. The digested product purified using a SanPrep Column PCR product purification Kit, and ligated using T4-DNA ligase at 16 °C 24 h. The ligation mixture was transformed to E. coli JM109 (Novagen, Madison, WI), and transformants were selected for growth on solid Luria–Beltane (LB) agar plates containing kanamycin (50 μg ml−1) and 0.05 M Zn2+. Positive clones were validated by PCR using the primers 5′-ATCGTCAGGGATGGCGGCCGCG-3′ (forward) and 5′-TGCGACATCGTATAACGTTACTGGT-3′ (reverse). The final plasmid, pET28a-NDM-1, purified from a single colony was identified by sequencing (Sangon Biotech, Shanghai). The correct pET28a-NDM-1 was transformed into E. coli BL21 (DE3).
2.3. Media and culture conditions
E. coli containing pET28a (+)-blaNDM-1 was cultured in the following medium. Lysogeny broth (LB): 10 g L−1 peptone, 5 g L−1 yeast extract, and 10 g L−1 NaCl, pH 7.0. Lysogeny broth auto-induction medium (LBA): 0.5 g L−1 glucose, 2 g L−1 lactose, 5 g L−1 glycerol, 10 g L−1 peptone, 5 g L−1 yeast extract, 10 g L−1 NaCl, 6.5 g L−1 KH2PO4, 7.0 g L−1 Na2HPO4, 3.3 g L−1 (NH4)2SO4, and 0.15 g L−1 MgSO4; pH 7.0. Terrific broth (TB): 5 ml L−1 glycerol, 12 g L−1 tryptone, 25 g L−1 yeast extract, 2.31 g L−1 KH2PO4, and 12.54 g L−1 K2HPO4; pH 7.0. Super broth (SB): 35 g L−1 tryptone, 20 g L−1 yeast extract, 5 g L−1 glycerol, 2.31 g L−1 KH2PO4, and 12.54 g L−1 K2HPO4; pH 7.0. SOC medium: 20 g L−1 peptone, 5.0 g L−1 yeast extract, 0.5 g L−1 NaCl, 0.2 g L−1 KCl, 0.01 g L−1 ZnCl2, 1.0 g L−1 MgCl2 and 4 g L−1 glucose, pH 7.0. The seed culture was incubated on a reciprocal shaker (200 rpm) at 37 °C in a 500 ml flask containing 50 ml of LB medium (50 μg ml−1 kanamycin) for 16 h. The prepared seed culture was inoculated (5%, v/v) into the fresh medium. Acinetobacter baumannii was grown in nutrient broth (2 g yeast extract, 1 g meat extract, 5 g peptone and 5 g of NaCl in 1 L of sterile distilled water, 15.0 g agar).17 E. coli JM109 was used as the host for plasmid amplification. E. coli was incubated in lysogeny broth (LB) medium at 37 °C, 200 rpm, for 24 h.2.4. Purification of recombinant NDM-1
NDM-1 were produced in E. coli BL21 (DE3)-pET28a (+)-blaNDM-1 cells at 37 °C using SOC medium supplemented with 50 μg ml−1 kanamycin. NDM-1 expression was induced by IPTG (0.65 mM final concentration), while the OD600 reached 0.6. At this point the temperature was adjusted to 25 °C and the cells were further incubated for 20 h. The culture broth was centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 15 min. The cells were suspended in 0.1 M phosphate buffer (PBS, pH 7.2), and sonicated by an Ultrasonic Cell Disruptor (power 400 W, working 4 s, pause 8 s, total 40 min). The supernatant was concentrated by ultrafiltration with an Amicon Ultra-15 10 K device (Millipore, USA), those proteins with a molecular mass below 10 kDa has been removed. Recombinant NDM-1 were purified using nickel–nitrilotriacetic acid (Ni–NTA) agarose (Qiagen, Hilden, Germany). His tags were digested with TurboTEV protease (Accelagen, San Diego, CA). The purities of NDM-1 was estimated by SDS-PAGE. The purified recombinant NDM-1 was prepared for the further study of the activity, inhibitor screening. The protein concentration was determined using Bradford's method, with bovine serum albumin (BSA) as the standard.18
000 rpm for 15 min. The cells were suspended in 0.1 M phosphate buffer (PBS, pH 7.2), and sonicated by an Ultrasonic Cell Disruptor (power 400 W, working 4 s, pause 8 s, total 40 min). The supernatant was concentrated by ultrafiltration with an Amicon Ultra-15 10 K device (Millipore, USA), those proteins with a molecular mass below 10 kDa has been removed. Recombinant NDM-1 were purified using nickel–nitrilotriacetic acid (Ni–NTA) agarose (Qiagen, Hilden, Germany). His tags were digested with TurboTEV protease (Accelagen, San Diego, CA). The purities of NDM-1 was estimated by SDS-PAGE. The purified recombinant NDM-1 was prepared for the further study of the activity, inhibitor screening. The protein concentration was determined using Bradford's method, with bovine serum albumin (BSA) as the standard.18
2.5. Measurement of NDM-1 activity and antimicrobial susceptibility testing
NDM-1 activity was measured based on the method reported by O'Callaghan et al.19 The reaction mixture containing 200 μM nitrocefin (Sigma-Aldrich) and enzymes 100 μL were incubated with 50 mM Bis–Tris buffer (pH 6.5) containing 50 μM Zn2+, 1 mM 1,4-dithio-DL-threitol (DTT) and 10% (v/v) glycerol in a final volume of 2 ml at 40 °C, 30 min. The initial rate of hydrolysis of nitrocefin was determined by an UV-visible spectrophotometer (Shimadzu, UV-1800, Japan) at 492 nm. Colour changes from yellow to red when nitrocefin hydrolyzed by NDM-1 activity. 1 U corresponds to the amount of enzyme that could generate 1 μM hydrolyzed nitrocefin per minute by hydrolysis. The susceptibility of A. baumannii and recombinant E. coli (containing pET28a-NDM-1) plasmids to imipenem, meropenem, cefepime, cefotaxime, ceftazidime, aztreonam, ampicillin, piperacillin, tigecycline, gentamicin, colistin and amikacin were determined using the E-test method (bioMérieux, Basingstoke, UK) on Mueller–Hinton agar according to the manufacturer's instructions.2.6. Steady-state kinetics, IC50 determination and molecular docking
The kinetic parameters were obtained by measuring the initial rate of the reaction at different substrate concentrations. Hydrolysis of the β-lactam antibiotics by NDM-1 was followed by observing changes in absorption resulting from the opening of the β-lactam ring at specific wavelengths for each antibiotics. The kinetic parameters assays were performed at 30 °C using a UV21800 spectrophotometer (Mapada, Shanghai, China). Assays were performed in 200 μL of assay buffer (30 mM HEPES, 10 μM ZnCl2, and 2 mM DTT, 0.01% Triton X-100, pH = 7.0). The steady-state kinetic parameters (Km) were determined three times to evaluate the standard deviation. The Km was analyzed using in the program of origin 8.0 (OriginLab Corporation, Northampton, MA) to generate Michaelis–Menten and substrate inhibition curves. For the IC50 of these compounds determination, NDM-1 (5 nM) supplemented with 10 μM ZnSO4, was pre-incubated with EDTA and AMA (dissolved in DMSO) 20 minutes, then, was mixed with 60 μM imipenem at 30 °C, and the final concentration of DMSO in the assay was <0.5% (v/v), linear portions of curves were used to analyze data. Assays were read in 96-well microplate format at 293 nm using a Spectramax reader at 37 °C.The structures of the NDM-1 was obtained by homology modeling (SWISS-MODEL workspace, https://www.swissmodel.expasy.org/).20 The three-dimensional structures of the inhibitors were generated using the Chem-office Ultra 11.0 program (Cambridge Soft Corporation, Cambridge, MA, USA).20b Molecular docking was carried out using the Auto-dock 4.0 software. Only ligand molecules were considered to be flexible during the docking, and only the free energy of their best orientation was used to compute the docking free energy.
3. Results
3.1. Emerging of NDM-1 from Acinetobacter baumannii
A total of 12 clinical isolates of Gram-negative bacteria, were collected from the sputum of ten patients suffering from pneumonia. Of these, three isolates identified as Acinetobacter baumannii. All these A. baumannii were resistant to the tested antibiotics, including cephalosporins (Table 1). The A. baumannii were resistant to most of β-lactam antibiotics, including ceftazidime, imipenem; meropenem, cefepime, cefotaxime, ceftazidime, aztreonam, ampicillin, piperacillin, and susceptible to colistin, tigecycline, gentamicin; amikacin (Table 1). A. baumannii isolates (S001, S002, S003) were positive by the MBL E-test. All these A. baumannii carries blaNDM-1 gene validated by PCR screening and sequencing. The sequences of the blaNDM-1 genes was showing 99.5%, 99.9%, 99.2% (S001, S002, S003), respectively, compared with previously reported genes (NDM-1).8| Isolates | IMP | MEM | FEP | CTX | CAZ | ATM | SMP | PIP | TGC | GEN | CST | AMK |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| a IMP, imipenem; MEM, meropenem; FEP, cefepime; CTX, cefotaxime; CAZ, ceftazidime; ATM, aztreonam; SMP, ampicillin; PIP, piperacillin; TGC, tigecycline; GEN, gentamicin; CST, colistin; AMK, amikacin. | ||||||||||||
| S001 | >32 | >32 | 256 | 256 | 256 | 256 | >256 | >256 | 0.5 | 1 | 0.5 | 8 |
| S002 | >32 | >32 | >256 | 256 | 256 | 256 | >256 | >256 | 1 | 1 | 0.5 | 16 |
| S003 | >32 | >32 | 256 | 256 | 256 | 64 | 256 | >256 | 1 | >256 | 0.5 | 8 |
3.2. Overexpression and biochemical characterization of the recombinant NDM-1
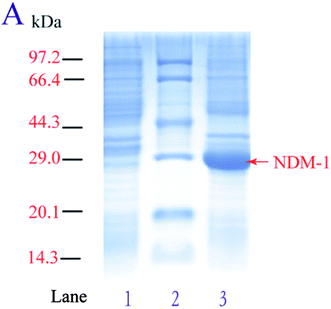
In order to achieve high-level overexpression of NDM-1 in E. coli, 830-bp DNA fragment including the termination codon and the native signal peptide, encoding NDM-1, was amplified by PCR from DNA of Acinetobacter baumannii S002 and cloned into the expression plasmid pET28a (+). Plasmid pET-28a (+)-NDM-1 was confirmed by DNA sequencing and transformed into E. coli BL21 (DE3). Recombinant NDM-1 is intracellular, and crude extract was obtained after expressing cells were grown, harvested, and lysed using a cell disrupter. The recombinant NDM-1 protein is an intracellular protein in a soluble and active form. After induction with 0.5 mM IPTG at 30 °C, proteins from control, purified and over-expressing cells were analyzed by SDS-PAGE, and an intense band with a molecular mass of approximately 29 kDa that corresponds to NDM-1 was observed (Fig. 1). The specific activity of the over-expressing cell extract was 17.8 U mg−1, which was 3.8-fold higher than that of the Acinetobacter baumannii cells.Different medium were investigated to determine the NDM-1 activity. Fig. 2A shows the NDM-1 activity and biomass reached maximum values (14.3 U ml−1 and 1.32 g L−1, respectively) when SOC was used as the medium source. NDM-1 activity improved by 69.7%, as compared with that in LB medium. The optimal induction phase was carried out during the exponential growth phase (OD600 = 0.60, Fig. 2B). We found that 0.65 mM IPTG were required and the maximum recombinant NDM-1 activity (25.8 U ml−1) was observed with 20 h of induction (Fig. 2C and D). An orthogonal L9 (3) array experiment was performed to optimize NDM-1 over-expression. The order of the effect of the factors on NDM-1 production was initial OD600 > IPTG concentration > induction period. The initial cell density was the main factor for NDM-1 production. In this study, the optimal conditions for NDM-1 over-expression were as follows: OD600 = 0.60, IPTG 0.65 mM, and induction 20 h (Table 2). The NDM-1 activity was up to maximum values 68.5 U ml−1 under the optimal condition, improved by 92.3% as compared with that before optimization (LB medium, 5.3 U ml−1).
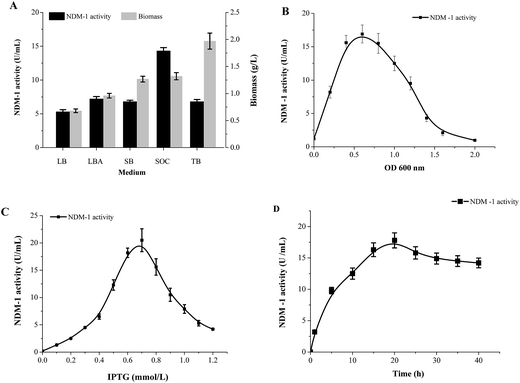 | ||
| Fig. 2 Expression condition optimized on recombinant NDM-1 activity. (A) Medium, (B) OD600, (C) IPTG, (D) time. | ||
| Run | Factors | A initial cell density | B induction period (h) | C IPTG (mM) | NDM-1 activity (U ml−1) | ||
|---|---|---|---|---|---|---|---|
| A | B | C | |||||
| 1 | 1 | 1 | 1 | 0.5 | 15 | 0.55 | 18 |
| 2 | 1 | 2 | 2 | 0.5 | 20 | 0.65 | 67 |
| 3 | 1 | 3 | 3 | 0.5 | 25 | 0.75 | 28 |
| 4 | 2 | 1 | 2 | 0.6 | 15 | 0.65 | 43 |
| 5 | 2 | 2 | 3 | 0.6 | 20 | 0.75 | 52 |
| 6 | 2 | 3 | 1 | 0.6 | 25 | 0.55 | 60 |
| 7 | 3 | 1 | 3 | 0.7 | 15 | 0.75 | 48 |
| 8 | 3 | 2 | 1 | 0.7 | 20 | 0.55 | 23 |
| 9 | 3 | 3 | 2 | 0.7 | 25 | 0.65 | 34 |
| k1 | 37.67 | 36.33 | 33.67 | ||||
| k2 | 51.67 | 47.33 | 48.00 | ||||
| k3 | 35.00 | 40.67 | 42.67 | ||||
| Delt | 16.67 | 11.00 | 14.33 | ||||
| Ran | 1 | 3 | 2 | ||||
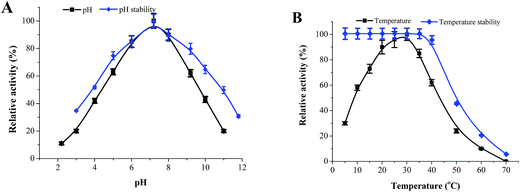
3.3. Purification and biochemical characterization of recombinant NDM-1
The recombinant NDM-1 was purified by HisTrap™ FF affinity chromatography, and the specific activity of purified NDM-1 was up to 347.35 U mg−1 (Table 3). The optimal pH and pH stability of recombinant NDM-1 were determined within a pH range of 2.0–10.0 (Fig. 3A). The results show that the recombinant NDM-1 had an optimal pH of 7.2. Interestingly, NDM-1 was stable at high-pH, the relative activity of NDM-1 retained 45% at pH = 11. Fig. 3B shows the NDM-1 activity increased with the temperature from 20 to 30 °C and decreased from 30 to 60 °C. The maximum activity of the recombinant NDM-1 was observed at 30 °C. The thermos-stability experiments of the recombinant NDM-1 showed the enzyme was stable at 10–37 °C, and became instable when the temperature was above 42 °C (Fig. 3B).| Steps | Total protein (mg) | Total activity (U) | Specific activity (U mg−1) | Yield (%) | Purification fold |
|---|---|---|---|---|---|
| Crude (0.5 L) | 1058.70 | 33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 946.50 946.50 |
32.06 | 100.00 | 1.00 |
| Precipitation | 589.63 | 28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 250.00 250.00 |
47.91 | 83.22 | 1.49 |
| Amicon Ultra-15 10K | 116.40 | 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 024.65 024.65 |
94.71 | 32.48 | 2.95 |
| HisTrap | 15.75 | 5470.77 | 347.35 | 16.12 | 10.83 |
3.4. Inhibitors and β-lactam antibiotic screening
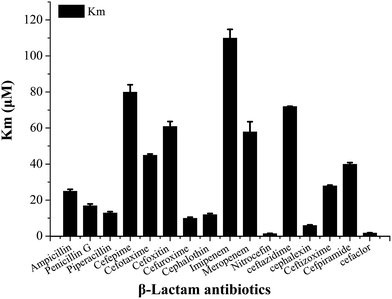
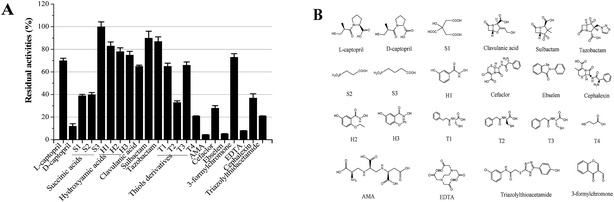
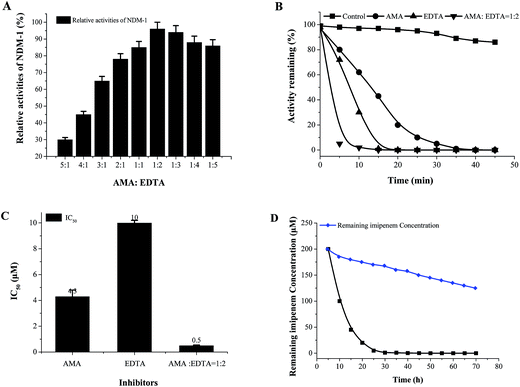
In order to select an excellent antibiotic for inhibitors evaluation and synergistic against the NDM-1 producing Acinetobacter baumannii, kinetic parameters of a representative set of carbapenem, penicillin and cephalosporin β-lactam antibiotic substrates were determined for NDM-1. The results showed that the NDM-1 hydrolyzed all the tested carbapenems (imipenem, meropenem), penicillin (ampicillin, penicillin G, piperacillin); cephalosporins (cefuroxime, ceftizoxime, cefpiramide, ceftazidime, cefoxitin, cefaclor, cephalexin, nitrocefin, cefepime, cephalothin, cefotaxime) (Fig. 4). Of these, the penicillin substrate was all hydrolyzed by the NDM-1. For the tested cephalosporins, there are significant differences in kinetic parameters. Cefepime has higher Km (80 μM), in contrast; the NDM-1 have low Km values for nitrocefin (1.5 μM), therefore, nitrocefin has been used for NDM-1 activity determination. Only a small amount of β-lactam antibiotics have poor affinity towards NDM-1, the Km of cefepime, cefoxitin, imipenem, ceftazidime, meropenem were >60 μM. Imipenem with a Km value of 128 μM, which was higher than the others, was selected as the excellent antibiotic.Our long-term goals is to search for clinically applicable NDM-1-inhibitory agents that interacted with the residue at the active site with the potential of inhibition of all NDMs (NDM-1 and its variants). In order to assess the various classes of metallo-enzyme inhibitors, succinic acid analogues,13b hydroxamic acids,21 ebselen,22 bisthiazolidines,23 captopril analogues,24 thiols,25 and N-hydroxythiazoles,26 triazolylthioacetamide,27 EDTA and others were selected as the potential metallo-enzyme inhibitors for screening. The residual activities of NDM-1 were determined and shown in Fig. 5.
Unfortunately, these commercially available β-lactamase inhibitor (clavulanic acid, sulbactam, tazobactam) do not inhibit the NDM-1 effectively. Moreover, it indicated that the majority of the tested inhibitors had residual activities >30%, including succinic acids (S1, S2, S3), hydroxamic acids (H1–H3) and thiols derivatives (T1–T3), captopril, cefaclor, cephalexin, 3-formylchromone. On the other hand, L-captopril, ebselen, aspergillomarasmine A (AMA), EDTA, D-captopril gave residual activities of <20%, especially, ebselen, aspergillomarasmine A (AMA) had residual activities <5% (Fig. 5). Consequently, EDTA, AMA, L-captopril, ebselen, D-captopril were screened by molecule docking, surprisingly, EDTA and AMA had high affinity for NDM-1 with docking energy of −98.41 kcal mol−1 and −109.19 kcal mol−1 (Table 4). Herein, EDTA and AMA were further selected as excellent NDM-1 inhibitor. Next, the synergistic inhibition of EDTA and AMA was investigated. The result showed that NDM-1 inhibition increases according to the molar ratio of AMA to EDTA (5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1–
1–![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1:
1:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2). Of great interest, when the molar ratio of AMA to EDTA reached 1
2). Of great interest, when the molar ratio of AMA to EDTA reached 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, the inhibition of NDM-1 was up to 98.5% (Fig. 6A). The working time of AMA–EDTA, AMA and EDTA were 5 min, 20 and 35 min, respectively (Fig. 6B).
2, the inhibition of NDM-1 was up to 98.5% (Fig. 6A). The working time of AMA–EDTA, AMA and EDTA were 5 min, 20 and 35 min, respectively (Fig. 6B).
| Inhibitors | Structures | Docking energy (kcal mol−1) | Inhibitors | Structures | Docking energy (kcal mol−1) |
|---|---|---|---|---|---|
| a Molecular docking was carried out using the Auto-dock 4.0 software. Only ligand molecules were considered to be flexible during the docking, and only the free energy of their best orientation was used to compute the docking free energy. | |||||
| AMA | 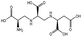 |
−98.41 ± 1.23 | D-Captopril | 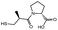 |
−61.76 ± 0.69 |
| Ebselen | 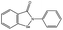 |
−78.03 ± 0.92 | EDTA | 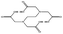 |
−109.19 ± 1.58 |
AMA, EDTA, and their combination (AMA–EDTA = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) were subsequently selected for IC50 determinations. AMA–EDTA was found to be the most potent inhibitor for NDM-1 tested, with IC50 values ranging from 1 to 10 μM; compared to AMA and EDTA alone, the IC50 of AMA–EDTA (1
2) were subsequently selected for IC50 determinations. AMA–EDTA was found to be the most potent inhibitor for NDM-1 tested, with IC50 values ranging from 1 to 10 μM; compared to AMA and EDTA alone, the IC50 of AMA–EDTA (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) decreased by 88% and 95%, respectively (Fig. 6C). The effect of AMA–EDTA on NDM-1 inhibition has been investigated in presence of imipenem, imipenem concentration was reduced 99.5% in the absence of inhibitor (AMA
2) decreased by 88% and 95%, respectively (Fig. 6C). The effect of AMA–EDTA on NDM-1 inhibition has been investigated in presence of imipenem, imipenem concentration was reduced 99.5% in the absence of inhibitor (AMA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EDTA = 1
EDTA = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2), while the imipenem was only reduced by 16% with addition of AMA–EDTA (1
2), while the imipenem was only reduced by 16% with addition of AMA–EDTA (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
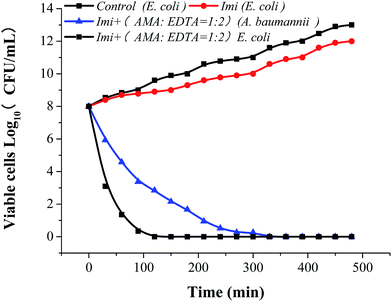
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) within 30 min (Fig. 6D). Most interesting, AMA–EDTA could restore the activity of carbapenem (imipenem) when tested against E. coli expressing NDM-1 and NDM-1-producing Acinetobacter baumannii, with a working time of 120 min and 330 min, respectively (Fig. 7).
2) within 30 min (Fig. 6D). Most interesting, AMA–EDTA could restore the activity of carbapenem (imipenem) when tested against E. coli expressing NDM-1 and NDM-1-producing Acinetobacter baumannii, with a working time of 120 min and 330 min, respectively (Fig. 7).
4. Discussion
Great health risks to human and animals were posed by emerging multi-drug resistant (MDR) bacteria-Acinetobacter baumannii. We found Acinetobacter baumannii was susceptible to colistin, and colistin has been a potent antibiotic to control the NDM-1 bacterial infections. However, it could not be always work. Colistin is destroyed and inactivated by the co-producing NDM-9 and MCR-1 E. coli.28 The New Delhi metallo-β-lactamase 1 (NDM-1) and its variants were reported to be resistant to most of the β-lactam antibiotics.29 Although the mechanism and pathways of cephalosporin hydrolysis catalyzed by NDM-1 has been resolved,30 lack of efficient inhibitors to NDM-1 still is a big clinical concern. Therefore, it is meaningful to develop a rapid and efficient method for inhibitors screening to against NDM-1 mutants.There are two routes for discovering efficient NDM-1 inhibitors. One is rational design of new inhibitors, which is not exist in nature currently, however, new synthesized compounds or newly-discovered drugs (without clinical trials), were lack of the support of clinical trial data, safety of the new inhibitors need to be studied, moreover, R & D period is very long. It is difficult to meet the urgent needs of the current war to against MDR bacteria. The other route is “new use for old drugs”, the old drug has sufficient clinical trial data, high safety and mature synthetic process. The method of inhibiting NDM-1 by adding inhibitors can be rapidly responded to the new NDM-1 variants in a short term, leading to reduce the patient pain and mortality. Towards NDM-1, the inhibitors with high affinity (HI) and the β-lactam antibiotics with low affinity (LA) were selected as the excellent combinations (Fig. 8). This method is expected to be used in drug design (including new inhibitors and drugs) and high-throughput screening.
Aspergillomarasmine A (AMA) was able to completely reserve the antibacterial activity of meropenem against NDM-1.31 NDM-1 was inactive while EDTA covalently binded with the cysteine residue at the active site.22 In this context, the two major advantages of EDTA and AMA has been integrated. AMA combined with EDTA can completely reserve the imipenem to fight against NDM-1-producing E. coli and Acinetobacter baumannii. Imipenem combined with inhibitors (AMA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EDTA = 1
EDTA = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) can kill the NDM-1-producing E. coli efficiently. We found NDM-1-producing Acinetobacter baumannii is harder to be killed than that of E. coli, and the working time of NDM-1-producing Acinetobacter baumannii was 2.75 times of expressing NDM-1 E. coli. It may be caused by the difference of the outer membrane permeability. The outer membrane in A. baumannii is less permeable to antimicrobial agents than that in Escherichia coli.32,33 Outer membrane permeability of A. baumannii decreased by its small number and size of porins.34
2) can kill the NDM-1-producing E. coli efficiently. We found NDM-1-producing Acinetobacter baumannii is harder to be killed than that of E. coli, and the working time of NDM-1-producing Acinetobacter baumannii was 2.75 times of expressing NDM-1 E. coli. It may be caused by the difference of the outer membrane permeability. The outer membrane in A. baumannii is less permeable to antimicrobial agents than that in Escherichia coli.32,33 Outer membrane permeability of A. baumannii decreased by its small number and size of porins.34
Moreover, AMA is a peptide, which can be degraded by biological enzymes. AMA can be obtained by fermentation with a low production cost and more benefit to human beings. Furthermore, efficient and low toxicity new inhibitors (new structures) from the extract of natural plants and marine microorganisms and AMA remodeling will be tried in subsequent research. In the present study, the NDM-1 from Acinetobacter baumannii was overexpressed in E. coli BL21 (DE3), however, the activity of NDM-1 was very low. Then, conditions were further optimized by an orthogonal array experiment, and high purity of recombinant NDM-1 was obtained. NDM-1 was stable (5 < pH < 11, temperature < 45 °C), which is expected to be applied to resolve the problem of antibiotics residues, and antibiotic leakage, antibiotic testing, biological sensor. The NDM-1 variants from A. baumannii S001 and S003, may have some new properties, will be investigated in future.
Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This work was supported by Key Technology Program of Zhongshan (No. 2015B1028).References
- (a) S. N. Richter, I. Frasson, E. Franchin, C. Bergo, E. Lavezzo, L. Barzon, A. Cavallaro and G. Palù, Gut Pathog., 2012, 4, 1 CrossRef PubMed; (b) A. U. Khan, L. Maryam and R. Zarrilli, BMC Microbiol., 2017, 17, 101 CrossRef PubMed.
- S. M. Drawz and R. A. Bonomo, Clin. Microbiol. Rev., 2010, 23, 160 CrossRef CAS PubMed.
- T. Palzkill, Ann. N. Y. Acad. Sci., 2013, 1277, 91 CrossRef CAS PubMed.
- C. Bebrone, Biochem. Pharmacol., 2007, 74, 1686 CrossRef CAS PubMed.
- S. D. Lahiri, S. Mangani, T. Durandreville, M. Benvenuti, F. D. Luca, G. Sanyal and J. D. Docquier, Antimicrob. Agents Chemother., 2013, 57, 2496 CrossRef CAS PubMed.
- C. Bebrone, P. Lassaux, L. Vercheval, J. S. Sohier, A. Jehaes, E. Sauvage and M. Galleni, Drugs, 2010, 70, 651 CrossRef CAS PubMed.
- (a) K. Bush, J. Infect. Chemother., 2013, 19, 549 CrossRef CAS PubMed; (b) N. Tijet, D. Boyd, S. N. Patel, M. R. Mulvey and R. G. Melano, Antimicrob. Agents Chemother., 2013, 57, 4578 CrossRef CAS PubMed.
- D. Yong, M. A. Toleman, C. G. Giske, H. S. Cho, K. Sundman, K. Lee and T. R. Walsh, Antimicrob. Agents Chemother., 2009, 53, 5046 CrossRef CAS PubMed.
- G. Bushnell, F. Mitrani-Gold and L. M. Mundy, Int. J. Infect. Dis., 2013, 17, e325 CrossRef CAS PubMed.
- (a) H. Grundmann, D. M. Livermore, C. G. Giske, R. Canton, G. M. Rossolini, J. Campos, A. Vatopoulos, M. Gniadkowski, A. Toth and Y. Pfeifer, Eurosurveillance, 2010, 15, 22 CrossRef; (b) S. Chatterjee, A. Mondal, S. Mitra and S. Basu, J. Antimicrob. Chemother., 2017, dkx131 Search PubMed; (c) K. Karthikeyan, M. A. Thirunarayan and P. Krishnan, J. Antimicrob. Chemother., 2010, 65, 2253 CrossRef CAS PubMed.
- T. R. Walsh, J. Weeks, D. M. Livermore and M. A. Toleman, Lancet Infect. Dis., 2011, 11, 355 CrossRef PubMed.
- A. Pruden, D. G. Larsson, A. Amézquita, P. Collignon, K. K. Brandt, D. W. Graham, J. M. Lazorchak, S. Suzuki, P. Silley and J. R. Snape, Environ. Health Perspect., 2013, 121, 878 CrossRef PubMed.
- (a) L. B. H. Pinhong Chen, R. L. Mikulski, L. Deng, S. Sundriyal and T. Palzkill, Bioorg. Med. Chem. Lett., 2012, 22, 6229 CrossRef PubMed; (b) J. H. Toney, G. G. Hammond, P. M. D. Fitzgerald, N. Sharma, J. M. Balkovec, G. P. Rouen, S. H. Olson, M. L. Hammond, M. L. Greenlee and Y. Gao, J. Biol. Chem., 2001, 276, 31913 CrossRef CAS PubMed.
- W. T, S. SA, M. D, S. TP, F. JR, S. M, F. JM, B. C, S. KB and H. PS, ACS Med. Chem. Lett., 2010, 1, 150 CrossRef PubMed.
- B. M. R. Liénard, G. Garau, L. Horsfall, A. I. Karsisiotis, C. Damblon, P. Lassaux, C. Papamicael, G. C. K. Roberts, M. Galleni and O. Dideberg, Org. Biomol. Chem., 2008, 6, 2282 Search PubMed.
- (a) M. W. Walter, A. Felici, M. Galleni, R. P. Soto, R. M. Adlington and J. E. Baldwin, Bioorg. Med. Chem. Lett., 1996, 6, 2455 CrossRef CAS; (b) W. Fast and L. D. Sutton, Biochim. Biophys. Acta, Proteins Proteomics, 2013, 1834, 1648 CrossRef CAS PubMed.
- D. S. Uppu, S. Samaddar, C. Ghosh, K. Paramanandham, B. R. Shome and J. Haldar, Biomaterials, 2016, 74, 131 CrossRef CAS PubMed.
- M. M. Bradford, Anal. Biochem., 1976, 72, 248 CrossRef CAS PubMed.
- C. H. O'Callaghan, A. Morris, S. M. Kirby and A. H. Shingler, Antimicrob. Agents Chemother., 1972, 1, 283 CrossRef.
- (a) J. Wu, H. Wang, B. Yang, W. Song, C. Liang and L. Liu, RSC Adv., 2017, 7, 38264 RSC; (b) M. Biasini, S. Bienert, A. Waterhouse, K. Arnold, G. Studer, T. Schmidt, F. Kiefer, T. C. Gallo, M. Bertoni and L. Bordoli, Nucleic Acids Res., 2014, 42, W252 CrossRef CAS PubMed.
- B. M. Liénard, L. E. Horsfall, M. Galleni, J. M. Frère and C. J. Schofield, Bioorg. Med. Chem. Lett., 2007, 17, 964 CrossRef PubMed.
- J. Chiou, S. Wan, K. F. Chan, P. K. So, D. He, E. W. Chan, T. H. Chan, K. Y. Wong, J. Tao and S. Chen, Chem. Commun., 2015, 51, 9543 RSC.
- M. M. González, M. Kosmopoulou, M. F. Mojica, V. Castillo, P. Hinchliffe, I. Pettinati, J. Brem, C. J. Schofield, G. Mahler and R. A. Bonomo, ACS Infect. Dis., 2015, 1, 544 CrossRef PubMed.
- (a) G. Yu, W. Jing, G. Niu, W. Shui, Y. Sun, H. Zhou, Y. Zhang, Y. Cheng, Z. Lou and Z. Rao, Protein Cell, 2011, 2, 384 CrossRef PubMed; (b) N. Li, Y. Xu, X. Qiang, C. Bai, T. Wang, W. Lei, D. He, N. Xie, L. Li and W. Jing, Bioorg. Med. Chem. Lett., 2014, 24, 386 CrossRef CAS PubMed; (c) D. T. King, L. J. Worrall, R. Gruninger and N. C. Strynadka, J. Am. Chem. Soc., 2012, 134, 11362 CrossRef CAS PubMed.
- S. Siemann, A. J. Clarke, T. Viswanatha and G. I. Dmitrienko, Biochemistry, 2003, 42, 1673 CrossRef CAS PubMed.
- C. M. Tegley, V. N. Viswanadhan, K. Biswas, M. J. Frohn, T. A. Peterkin, C. Chang, R. W. Bürli, J. H. Dao, H. Veith and N. Rogers, Bioorg. Med. Chem. Lett., 2008, 18, 3925 CrossRef CAS PubMed.
- (a) L. Zhai, Y. L. Zhang, J. S. Kang, P. Oelschlaeger, L. Xiao, S. S. Nie and K. W. Yang, ACS Medicinal Chemistry Letters, 2016, 7, 413 CrossRef CAS PubMed; (b) P. W. Thomas, T. S. Michael Cammarata, J. S. Brodbelt, P. Hodder and W. Fast, Bioorg. Med. Chem., 2013, 21, 3138 CrossRef CAS PubMed.
- X. Yao, Y. Doi, L. Zeng, L. Lv and J. H. Liu, Lancet Infect. Dis., 2016, 16, 288 CrossRef CAS PubMed.
- K. K. Kumarasamy, M. A. Toleman, T. R. Walsh, J. Bagaria, F. Butt, R. Balakrishnan, U. Chaudhary, M. Doumith, C. G. Giske and S. Irfan, Lancet Infect. Dis., 2010, 10, 578 CrossRef.
- H. Feng, J. Ding, D. Zhu, X. Liu, X. Xu, Y. Zhang, S. Zang, D. C. Wang and W. Liu, J. Am. Chem. Soc., 2014, 136, 14694 CrossRef CAS PubMed.
- A. M. King, S. A. Reid-Yu, W. Wang, D. T. King, G. De Pascale, N. C. Strynadka, T. R. Walsh, B. K. Coombes and G. D. Wright, Nature, 2014, 510, 503 CrossRef CAS PubMed.
- J. Hrabák, E. Chudáčková and R. Walková, Clin. Microbiol. Rev., 2013, 26, 103 CrossRef PubMed.
- J. Vila, S. Martí and J. Sánchez-Céspedes, Journal of Antimicrobial Chemotherapy, 2007, 59, 1210 CrossRef CAS PubMed.
- M. Obara and T. Nakae, J. Antimicrob. Chemother., 1991, 28, 791 CrossRef CAS PubMed.
Footnote |
| † The co-first authors. |
| This journal is © The Royal Society of Chemistry 2018 |