 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Isolation, differentiation and biodiversity of ureolytic bacteria of Qatari soil and their potential in microbially induced calcite precipitation (MICP) for soil stabilization
Shazia Bibia,
Meriam Oualhaa,
Mohammad Yousaf Ashfaqa,
Muhannad T. Suleimanb and
Nabil Zouari *a
*a
aDepartment of Biological and Environmental Sciences, College of Arts and Sciences, Qatar University, PO Box 2713, Doha, Qatar. E-mail: Nabil.Zouari@qu.edu.qa; Tel: +974-4403-4559
bDepartment of Civil and Environmental Engineering, Lehigh University, USA
First published on 6th February 2018
Abstract
Biomineralization plays a key role in modifying the geological properties of soil, thereby stabilizing it against wind erosion, especially in areas characterized by harsh weather and harsh soil (calcareous and arid); i.e. Arabic Gulf region. Among soil microorganisms, ureolytic bacteria are capable of modifying soil characteristics and thus, inducing biomineralization. This research investigated the occurrence and diversity of ureolytic bacteria in Qatari soils, specifically to study their acquired potential to adapt to harsh conditions exhibiting ureolytic activity. Soil samples were collected from various locations in Qatar and were used to isolate the indigenous ureolytic bacteria. It was noticed that most of the ureolytic bacteria in Qatari soil belong to the genus Bacillus mainly Bacillus cereus. Identification and differentiation of 18 ureolytic isolates were performed using MALDI-TOF MS techniques while ribotyping (16S rRNA) molecular technique was used mainly for 6 selected strains. This study not only shows the diversity of species of ureolytic bacteria in Qatari soil but also shows the diversity in their protein profiles, which confirms that bacteria have adapted well to the harsh environment. In addition, the strains were evaluated based on a newly modified screening method in this work; i.e. production of arbitrary urease activity (AUA). Thus, the strains showing the highest AUA, exhibited the highest capability to produce urease enzymes induced by urea. Analysis of calcium carbonate precipitation utilizing SEM-EDX showed that the ureolytic bacteria also play a significant role in the precipitation of minerals such as CaCO3, in the presence of urea in soil. Therefore, this research showed a high occurrence of indigenous Bacillus bacteria in Qatari soil that can perform biomineralization and thus can be helpful, if properly stimulated, in enhancing soil stabilization, and for other local applications as well, since they are adapted to these soil and weather conditions.
1. Introduction
The soil stabilization techniques currently available to minimize or avoid airborne particles due to wind erosion use significant amounts of fossil fuel energy and/or hazardous materials such as mixing techniques using bituminous materials as stabilizing agents. Investigating sustainable biological soil stabilization methods to resist soil erosion due to wind is more appealing, in particular, if the biological activities are delivered by indigenous soil bacteria living under the corresponding environmental conditions. Bio-modification methods are innovative soil stabilization improvement solutions that use indigenous bacteria in the soil to mimic and engineer naturally occurring processes.Microorganisms play an important role in inducing the precipitation of minerals, hence, promoting a change in the geological properties of the soil or any earth component. Biomineralization can help to improve soil quality by increasing soil stiffness through the production of calcite precipitates using bacterial activities.21 Bacteria are important in biomineralization processes with, approximately, 200 different types known to induce calcium carbonate formation.4 There are two mechanisms known through which biomineralization occurs.3 Either, the bacterial cells provide nucleation site for the precipitation of calcium carbonates or ureolysis increases alkalinity of the surrounding environment resulting in precipitation. An alternative mechanism involves extracellular macromolecules; extracellular polymeric substances (EPS).19 EPS assists the microbial cell to adhere to different substrates while providing additional protection, nutrition and adsorption. EPS helps bacterial cells trap and precipitate sediments and minerals. After adhering to different surfaces, the bacterial cells start to dissolve the nutrients to obtain energy, resulting in precipitation of minerals as by-product. Ureolytic bacteria increases the pH through hydrolysis of urea and thus precipitates minerals.15 Bacillus is the most common genus of bacteria known to perform biomineralization by implementing ureolytic activities. Some specific species are B. sphaericus,7 B. lentus,17 B. pasteurii30 and B. licheniformis.16
Bacteria involved in biomineralization are important in stabilizing soil, especially in regions and cities with windy and desert-like conditions such as in the Gulf region. Among large number of applications of ureolytic bacteria, the stabilization of soils, especially calcareous ones, is highly recommended. At such conditions, calcification may be favorable due to the stress by the elevated temperature and high calcium content in the soil. To assist biomineralization of calcite, bacterial cells must produce either constitutive or inducible urease, however, very few bacterial species produce constitutive urease.5 Only one bacterium, Sporosarcina koreensis, was isolated from the Qatari desert dunes and is shown to harbor the urease gene, as per its whole-genome sequencing.1 The use of nitrogen fertilizers like urea; a product of petroleum industry is also readily available in some countries, especially in the Arabic Gulf countries.12,13 Urea remains the solid nitrogen fertilizer in common use with the highest nitrogen content and with the lowest transportation costs per unit of nitrogen nutrient. Its quality and ability to fertilize soil for crop cultivation make it one of the most widely used nitrogen fertilizer showing an increase in demand in agriculture. Bioaugmentation/biostimulation of soils with ureolytic bacteria in areas with nitrogen-poor soils has a beneficial side effect of biomineralization.
The aim of the research presented in this paper is to characterize the soil bacterial populations exhibiting ureolytic activity and investigate their diversity and distribution in a region with harsh weather conditions. The innovation resides in the establishment of a screening program of isolated bacteria based on rapid estimation of urease activity and bio-modification of soil. Furthermore, this study addresses the current knowledge gaps related to the microscale response of bio-mediated soils. The objectives of this research include (1) isolation of ureolytic bacteria from the soil in Qatar; (2) identification and differentiation of the isolated strains, using molecular techniques (16S rRNA sequencing) and MALDI-TOF MS (proteins profiling), to study their biodiversity; (3) quantification of the specific production of urease enzyme produced by ureolytic bacteria and (4) analysis of their role in microbially-induced calcite precipitation (MICP) using SEM-EDS technique.
2. Methodology
2.1. Soil sampling
Soil samples were collected from six locations around Qatar; Hamad Airport area, Duhail, Al-Wakra, Mesaieed, Dukhan and Abu hamur. The samples were stored in ice for transporting to the lab.2.2. Enrichment and isolation of ureolytic bacteria
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 5 min, pellets were obtained. The pellets were washed with cold sterile phosphate-buffered saline solution (g L−1) (8 NaCl, 0.2 KCl, 1.44 Na2HPO4 and 0.24 KH2PO4) thrice followed by centrifugation. The resulting pellets were suspended in 5 mL sterile PBS solution.
000 rpm for 5 min, pellets were obtained. The pellets were washed with cold sterile phosphate-buffered saline solution (g L−1) (8 NaCl, 0.2 KCl, 1.44 Na2HPO4 and 0.24 KH2PO4) thrice followed by centrifugation. The resulting pellets were suspended in 5 mL sterile PBS solution.2.3. Identification of isolates by MALDI-TOF-MS
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 2 min. The pellet was suspended in 70% formic acid and 100% acetonitrile. The supernatant was transferred to Biotarget plate and was overlaid with 1 μL of HCCA (α-cyanohydroxycinnamic acid) matrix solution.
000 rpm for 2 min. The pellet was suspended in 70% formic acid and 100% acetonitrile. The supernatant was transferred to Biotarget plate and was overlaid with 1 μL of HCCA (α-cyanohydroxycinnamic acid) matrix solution.2.4. Molecular identification of ureolytic bacteria
Pure cultures of isolates were obtained after overnight growth on Luria Broth (LB) agar medium. Thermal lysis (suspension in 0.5 mL H2O, boiling for 10 min, centrifugation for 10 min at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm) was performed on the cells for DNA extraction. The resulting supernatant was used for PCR amplification. Primer RibS73 and RibS74 were used to amplify 16S rRNA fragment.2 Purification was done after successful amplification and the amplicons were sequenced by Applied Biosystems 3500 Series Genetic Analyzer System.
000 rpm) was performed on the cells for DNA extraction. The resulting supernatant was used for PCR amplification. Primer RibS73 and RibS74 were used to amplify 16S rRNA fragment.2 Purification was done after successful amplification and the amplicons were sequenced by Applied Biosystems 3500 Series Genetic Analyzer System.
BLAST was used to find identical or related sequences in the Gene Bank database provided at NCBI (National Center for Biotechnology Information).
2.5. Determination of urease activity of isolates
At the time of analysis, samples from each culture were centrifuged for 10 min at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm and the supernatant served for ureolytic activity measurement.
000 rpm and the supernatant served for ureolytic activity measurement.
2.6. Scanning electron microscopy (SEM-EDX)
Scanning electron microscopy (SEM) is a technique that uses beam of electrons with high energy to generate different signals on the surface of specimens. The signals reveal the information about the sample under observation such as its morphology, chemical nature, and material orientation. SEM-EDX was used in our research to analyse the morphology and composition of calcium carbonate precipitates.The precipitates were obtained in 20 mL cultures performed with urea media containing (g L−1): 20 urea, 3.7 CaCl2, inoculated with separate isolates with an initial OD (600 nm) of 0.1, and incubated at 37 °C in a rotary shaker set at 150 rpm for 30 days. Cultures with both soil (0.4 g) and without soil were inoculated with bacteria. Three controls were performed with the same volume of 20 mL: first, urea medium with calcium chloride, second, 0.4 g soil in urea media with calcium chloride and third, 0.4 g soil in water. The soil used was collected from Abu hamur region in Doha City (Qatar). Precipitates were washed thrice with distilled water and the pellet obtained was oven-dried at 40 °C for 72 h before SEM-EDX analysis.
2.7. Statistical analysis
The statistical analysis was done for the results obtained using Microsoft Excel 2016 and ANOVA was also used choosing 95% confidence level.3. Results
3.1. Isolation of ureolytic bacteria from soil samples
The basis of this work was isolation of indigenous ureolytic bacteria and evaluation of their growth and production of ureolytic activity in soil surface to determine their potential viable bio-modification processes in Qatari soil with its environmental conditions. Six regions around Doha city were chosen to investigate and evaluate the occurrence and abundance of bacteria which demonstrate ureolytic activities. The samples were collected and enrichment cultures were performed for boosting growth of the corresponding bacteria as mentioned in the Material and Methods section. Ureolytic bacteria were grown in the media (enrichment medium, urea modified agar, urea broth media). Table 1 shows the origin of the sample, the isolates codes and the positive ureolytic ones, using the selective ureolytic medium. Modified urea agar was used to test qualitatively the isolates for their ureolytic activity. The pinkish/red change in colour indicates secretion of urease enzyme which hydrolyses urea, allowing further growth of the isolate on such medium. Out of 30 isolates, 18 isolates were tested positive for ureolytic activity at the experimental conditions.| Sample origin | Isolate code | Positive for ureolysis |
|---|---|---|
| Airport area | QBB1, QBB2, QBB3, QBB4, QBB5, QBB6 | QBB1, QBB4, QBB5, QBB6 |
| Duhail | QBB7, QBB8, QBB9, QBB10, QBB11, QBB12, QBB13, QBB14 | QBB7, QBB8, QBB11, QBB12, QBB13, QBB14 |
| Al-Wakra | QBB15, QBB16, QBB17, QBB18 | QBB17, QBB18, |
| Mesaieed | QBB19, QBB20, QBB21, QBB22, QBB23, QBB24, QBB25 | QBB20, QBB21, QBB22, QBB24, QBB25 |
| Dukhan | QBB26, QBB27, QBB28, QBB29 | QBB29 |
| Abu hamur | QBB30 | — |
3.2. Identification of isolates by MALDI-TOF MS
MALDI-TOF MS technique was used to identify the strains exhibiting ureolytic activity. The isolate code, reliability score and the corresponding identity are shown in Table 2. The profile matching was expressed on a log scale from 0 to 3 score. The score was interpreted as per the manufacturer's instruction in which 2.3–3.000 score indicates highly probable species-level identification, 2.00–2.299 shows genus identification and probable species-level identification and 1.70–1.999 score is designated as probable genus level identification. The calibration was done using Bacterial Test Standards (Item Catalogue #255343) provided by the manufacturer (Bruker Daltonics, Bremen, Germany).| Isolate code | Score | Identity |
|---|---|---|
| QBB1 | 1.70 | B. subtilis |
| QBB4 | 2.27 | B. cereus |
| QBB5 | 2.20 | B. cereus |
| QBB6 | 2.17 | B. cereus |
| QBB7 | 2.01 | B. cereus |
| QBB8 | 2.00 | B. licheniformis |
| QBB11 | 2.15 | B. cereus |
| QBB12 | 2.21 | B. cereus |
| QBB13 | 2.10 | B. cereus |
| QBB14 | 2.07 | B. cereus |
| QBB17 | 2.10 | B. cereus |
| QBB18 | 2.19 | B. cereus |
| QBB20 | 1.88 | B. licheniformis |
| QBB21 | 2.10 | B. cereus |
| QBB22 | 2.19 | B. cereus |
| QBB24 | 2.18 | B. cereus |
| QBB25 | 2.14 | B. cereus |
| QBB29 | 2.16 | B. cereus |
Most of the isolates were Bacillus cereus, one isolate was identified as Bacillus subtilis (code: QBB1) with a low identification score, and two were identified as Bacillus licheniformis (code: QBB8, QBB20) with high reliability scores. Despite several trials, the identification scores for all the isolates were mostly higher than 2 which can be considered as Probable Species-Level identification.
3.3. Differentiation of the isolated strains using protein profiles
The MADLI-TOF MS technique was also used for subtyping the strains based on their proteins profiles. Although the technique has been widely used for genus and species level identification, its use for subspecies level identification has not yet been fully explored.25 Recently, this technique has been used for subtyping bacteria. The subspecies level identification is quite challenging as the strains (within same species) tend to be very similar in terms of genotype and phenotype.25 Lately, subspecies level typing using MALDI-TOF MS has been reported for bacteria like Yersinia enterocolitica,24 methicillin-resistant Staphylococcus aureus,29 Staphylococcus enterica10 and Streptococcus agalactiae.18 In this research, strain differentiation, that relies on the presence or absence of protein peaks25 was adopted through proteins profiling. An adequate amount of stable mass signals for the ribosomal proteins typically 2000 to 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Da were obtained. The mass signals are used to produce profile spectra which comprises series of peaks that are conserved at genus, species and even subspecies levels.6 The variation among the strains can be observed by the absence or presence of peaks as shown in Fig. 1 for the isolated bacteria. It is evident that some isolates which were identified as same species demonstrated differences at the protein level. The isolates were categorized into five groups (named as category A, B, C, D and un-categorized) based on similarities observed in their protein profiles. The un-categorized group comprises isolates QBB1, QBB8, QBB14, QBB20 and QBB25 that could not qualify for any of the remaining four groups due to the differences in m/z ratio of their proteins. Fig. 1 shows the comparison of all isolated bacteria belonging to dissimilar categories. Category A comprises isolates QBB4, QBB6 and QBB7 grouped together as they share the same protein peaks at m/z of 5618.002, 6430.410, 7772.851, 9212.610 and 10
000 Da were obtained. The mass signals are used to produce profile spectra which comprises series of peaks that are conserved at genus, species and even subspecies levels.6 The variation among the strains can be observed by the absence or presence of peaks as shown in Fig. 1 for the isolated bacteria. It is evident that some isolates which were identified as same species demonstrated differences at the protein level. The isolates were categorized into five groups (named as category A, B, C, D and un-categorized) based on similarities observed in their protein profiles. The un-categorized group comprises isolates QBB1, QBB8, QBB14, QBB20 and QBB25 that could not qualify for any of the remaining four groups due to the differences in m/z ratio of their proteins. Fig. 1 shows the comparison of all isolated bacteria belonging to dissimilar categories. Category A comprises isolates QBB4, QBB6 and QBB7 grouped together as they share the same protein peaks at m/z of 5618.002, 6430.410, 7772.851, 9212.610 and 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 432.820. Similarly, category B isolates includes QBB17, QBB21 and QBB22 sharing same peaks at m/z of 5616.721, 6426.591, 7169.339, 7776.814, 9215.209 and 10
432.820. Similarly, category B isolates includes QBB17, QBB21 and QBB22 sharing same peaks at m/z of 5616.721, 6426.591, 7169.339, 7776.814, 9215.209 and 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 430.346. Furthermore, isolates under category C include QBB5, QBB11, QBB12 and QBB13. These strains showed similarities in proteins peaks with m/z of 5885.522, 6430.672, and 9212.476. Lastly, strains QBB18, QBB24 and QBB29 were categorized under category D (Fig. 1) showing peaks at m/z of 5888.431, 7399.226, 9211.565 and 10
430.346. Furthermore, isolates under category C include QBB5, QBB11, QBB12 and QBB13. These strains showed similarities in proteins peaks with m/z of 5885.522, 6430.672, and 9212.476. Lastly, strains QBB18, QBB24 and QBB29 were categorized under category D (Fig. 1) showing peaks at m/z of 5888.431, 7399.226, 9211.565 and 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 433.256. Variations among the proteins profiles of isolates grouped differently is obvious. For example, presence of peaks at m/z of 5824.645 and 9890.842 from category A are not found in the other groups. From group B, the peak at m/z of 6430.410 is not present in group D and un-categorized isolates. Furthermore, peak at m/z 10
433.256. Variations among the proteins profiles of isolates grouped differently is obvious. For example, presence of peaks at m/z of 5824.645 and 9890.842 from category A are not found in the other groups. From group B, the peak at m/z of 6430.410 is not present in group D and un-categorized isolates. Furthermore, peak at m/z 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 430.346 from group C is not found in groups A, B and D.
430.346 from group C is not found in groups A, B and D.
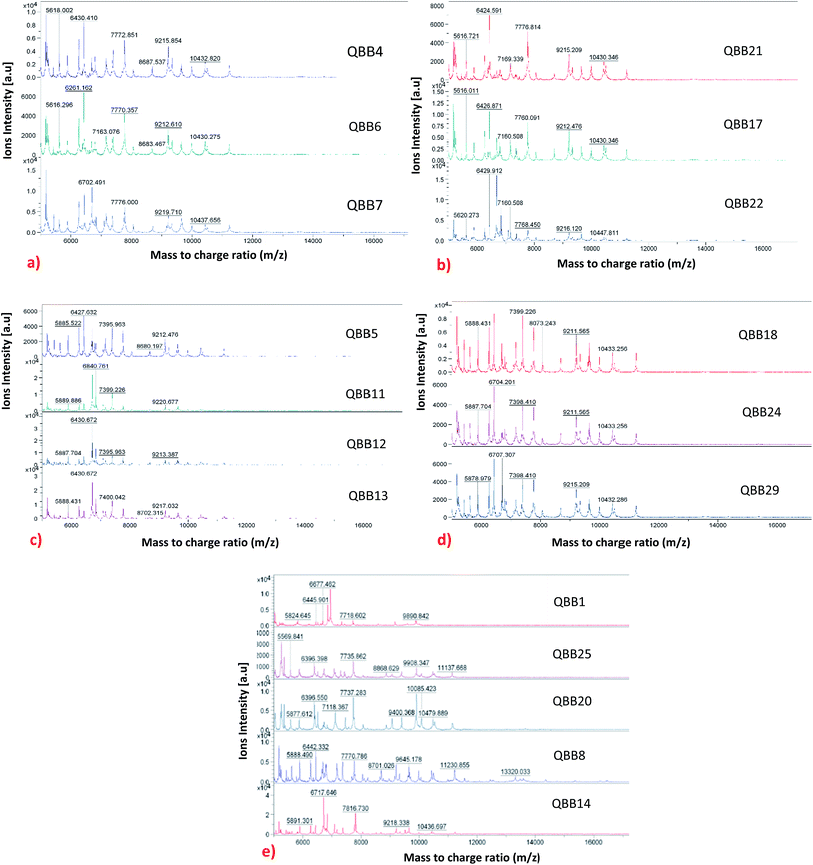 | ||
| Fig. 1 Categorization of isolates into 5 groups based on their similarities and differences in protein profiles: (a) category A; (b) category B; (c) category C; (d) category D and (e) uncategorized. | ||
In term of occurrence and distribution of the isolates based on their corresponding soil sampling site (Table 1 and Fig. 1), isolates from the Hamad Airport soil sample are distributed in the un-categorized group (QBB1), group A (QBB4 and QBB6) and group C (QBB5). Isolates from Duhail soil belong to un-categorized group (QBB8, QBB14 and QBB), group A (QBB7), and group C (QBB11, QBB12 and QBB13). Al-Wakra soil provided isolates belonging to group B (QBB17) and group D (QBB8). Strains isolated from Mesaieed soil are distributed in un-categorized group (QBB20), group B (QBB21 and QBB22), and group D (QBB24). From Dukhan soil, only one isolate exhibited detectable urease activity, which belongs to group D. Isolates from Abu-Hamur soil did not show ureolytic activity. These results show that some strains could be more distributed around the city than others.
3.4. Kinetics of ureolytic activity production and optimization of urea concentration for bacterial growth
In order to compare the isolates for their potential to grow and produce ureolytic activity, the behaviour of each strain as kinetic of growth and activity production should be identified. Since, the optimal conditions, nutrient availabilities and balances are important for efficient microbial activity and may vary from one isolate to another based on its specific metabolism, comparisons between the isolates were performed at the corresponding optimal conditions. All the 18 ureolytic isolates were following a similar pattern of growth, with the highest growth at 20 g L−1 urea after 72 h incubation as clearly shown in Fig. 2(a) for the isolate B. cereus QBB29. A drastic decrease in the counted cells was noticed beyond that incubation time, independently on the urea concentration. The pH in the cultures started to increase from day 2 and a high pH of 8.75 was observed on day 3. This means the hydrolysis of urea was maximum thereby increasing the pH as can be seen in Fig. 2(b). This may explain the inhibition of growth by excess of substrate (urea) or catabolic repression or increase of pH in the microenvironment of the cell.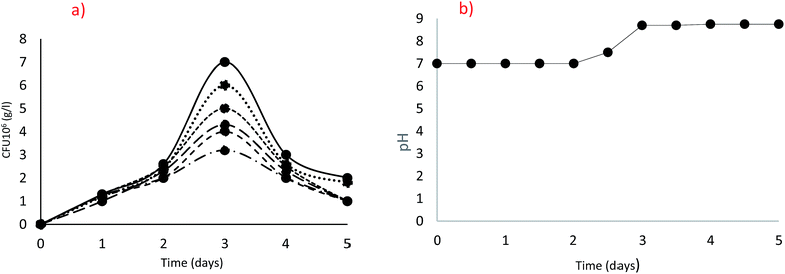 | ||
Fig. 2 Optimization of urea concentration [5 g L−1 (⋯), 10 g L−1 (●), 20 g L−1 (—), 30 g L−1 (![[dash dash, graph caption]](https://www.rsc.org/images/entities/char_e091.gif) ), 40 g L−1 ( ), 40 g L−1 (![[dash dot, graph caption]](https://www.rsc.org/images/entities/char_e090.gif) ), 50 g L−1 (----)]; (a) Colony Forming Units (cfu), (b) pH. ), 50 g L−1 (----)]; (a) Colony Forming Units (cfu), (b) pH. | ||
3.5. Potentialities of production of urease activity by the ureolytic isolates
In order to study the ureolytic bacteria diversity around the studied city, another criterion of differentiation between the isolates belonging to the same group and among the groups differentiated through MALDI-TOF MS analysis, was the determination of their potential to produce ureolytic activity (Table 3). This can be attributed to the level of gene expression and the metabolic regulations in these isolates. Thus, the isolate showing more ureolytic activity than the member of its own group may have expressed the urease gene higher than the others. Similarly, the activity is a result of the total metabolism of the cell, through interactions and regulations occurring in the cell.15| Categories | Sample locations | Isolate | Identity | cfu (106 cfu mL−1) | Arbitrary urease activity (AUA mL−1) | Specific production (AUA/107 cfu) | 95% confidence interval |
|---|---|---|---|---|---|---|---|
| A | Airport | QBB4 | B. cereus | 27 ± 1 | 14.14 ± 0.128 | 5.2388 | 0.630 ± 0.114 |
| Airport | QBB6 | B. cereus | 34 ± 1 | 8.49 ± 0.086 | 2.4984 | 0.378 ± 0.125 | |
| B | Mesaieed | QBB21 | B. cereus | 44 ± 3 | 4.75 ± 0.044 | 1.0796 | 0.211 ± 0.037 |
| Mesaieed | QBB22 | B. cereus | 57 ± 3 | 7.48 ± 0.084 | 1.3133 | 0.333 ± 0.029 | |
| C | Airport | QBB5 | B. cereus | 29 ± 9 | 14.77 ± 0.126 | 5.094 | 0.658 ± 0.204 |
| Duhail | QBB11 | B. cereus | 30 ± 1 | 7.55 ± 0.085 | 2.5176 | 0.336 ± 0.053 | |
| Duhail | QBB12 | B. cereus | 26 ± 5 | 10.92 ± 0.106 | 4.1985 | 0.486 ± 0.078 | |
| Duhail | QBB13 | B. cereus | 35 ± 1 | 11.52 ± 0.110 | 3.2918 | 0.513 ± 0.172 | |
| D | Al-Wakra | QBB18 | B. cereus | 35 ± 6 | 8.13 ± 0.093 | 2.3245 | 0.362 ± 0.009 |
| Mesaieed | QBB24 | B. cereus | 27 ± 5 | 9.73 ± 0.104 | 3.6029 | 0.433 ± 0.132 | |
| Dukhan | QBB29 | B. cereus | 27 ± 6 | 22.75 ± 0.148 | 5.0566 | 1.014 ± 0.667 | |
| Un-categorized | Airport | QBB1 | B. subtilis | 20 ± 1 | 3.05 ± 0.02 | 1.5231 | 0.135 ± 0.012 |
| Duhail | QBB8 | B. licheniformis | 26 ± 2 | 5.74 ± 0.056 | 2.2064 | 0.255 ± 0.044 | |
| Duhail | QBB14 | B. cereus | 37 ± 9 | 8.05 ± 0.086 | 2.1746 | 0.358 ± 0.156 | |
| Mesaieed | QBB20 | B. licheniformis | 45 ± 3 | 5.44 ± 0.053 | 1.21005 | 0.242 ± 0.038 |
Growth and urease activity determination was performed in the cultures using 20 g L−1 urea after 3 days incubation. Statistically significant variations of growth were shown among most of the bacteria as studied with ANOVA analysis with 95% confidence level, showing the diversity of their metabolic requirements and thus metabolites balances which in turn significantly affected the ureolytic activities. Indeed, the highest growth is not necessarily translated into high ureolytic activity in the medium. The cfu of isolate B. cereus QBB22 was the highest (57 ± 3 × 106 cfu mL−1) but the corresponding AUA was one of the lowest (7.48 ± 0.084). The determination of the specific production of AUA by each isolate would be the criterion of selection of the appropriate one for further applications. Consequently, QBB4, QBB5, QBB12, QBB29 and to some extent QBB13 and QBB24 seemed to be appropriate overproducers of urease activity at the experimental conditions. Some isolates such as QBB7, QBB9, QBB17 and QBB25 were not growing efficiently in liquid and solid modified urea media, therefore, they could not qualify for the determination of the specific production of AUA and be compared to the most efficient isolates. These results showed obvious diversity among the collection of the 18 isolates although 16 are identified as B. cereus.
3.6. Identification by 16S rRNA ribotyping
In order to combine techniques of identification, QBB4, QBB5, QBB12 and QBB29 exhibiting high urease activity and QBB11 and QBB18 exhibiting relatively low activity were identified. The objective was to confirm and validate the identification performed by MALDI-TOF MS technique.Table 4 shows the isolate code, its identity, accession number and similarity percentage. There was no discrepancy noted among the results of 16S rRNA and MALDI-TOF MS. Based on these results, it is clear that Bacillus sp., especially B. cereus are the most abundant bacteria exhibiting urease activity in Qatari soils.
| Isolate | Identity | Accession number | Identity percentage | GenBank number for submitted sequences |
|---|---|---|---|---|
| QBB4 | Bacillus cereus | KP743133.1 | 99% | MG745362 |
| QBB5 | Bacillus cereus | KB743133.1 | 100% | MG745365 |
| QBB11 | Bacillus cereus | KY316447.1 | 96% | MG745366 |
| QBB12 | Bacillus cereus | CP011151.1 | 99% | MG745367 |
| QBB18 | Bacillus cereus | LC260003.1 | 100% | MG745332 |
| QBB29 | Bacillus cereus | CP011151.1 | 96% | MG751339 |
3.7. Role of ureolytic bacteria in MICP
All the isolated strains were tested for their potentiality of calcium carbonate precipitation analysed by SEM/EDS. Similar results were obtained with all the ureolytic activity producers, but with incubation periods fluctuating between 1 month and 2 months for the pH to reach a value of 8.5 (results not shown). Table 5 summarizes the EDS analysis results accompanied by the measurement of pH with 60 days incubated cultures of QBB5, which was shown as one of the best AUA producer exceeding the specific production of 5 AUA/107 cfu (Table 3). In the negative controls performed abiotically (without inoculation of any of the isolates) mineral formation was not detected without soil, while no significant differences were observed in abiotic soil-cultures compared to biotic soil-cultures at the time of inoculation.| Medium | QBB5 inoculation | Precipitate type | Composition of precipitates (at%) | pH (after incubation) | |||
|---|---|---|---|---|---|---|---|
| C | O | Ca | P | ||||
| Urea medium + CaCl2 without soil | No | NA | NA | NA | NA | NA | 7.0 |
| Urea medium + CaCl2 without soil | Yes | Amorphous CaCO3 | 16.8 | 65.3 | 16.8 | 1.8 | 8.5 |
| Soil in water | No | No clear amorphous nor crystals of CaCO3 | 7.8 | 57.1 | 2.5 | 0.6 | 7.0 |
| Urea medium + CaCl2 with soil | No | No clear amorphous nor crystals of CaCO3 | 7.8 | 57.1 | 2.5 | 0.6 | 7.0 |
| Urea medium + CaCl2 with soil | Yes | Calcite | 24.0 | 60.5 | 23.9 | 1.1 | 8.8 |
| Yes | Amorphous CaCO3 | 13.9 | 60.8 | 13.4 | 2.6 | 8.4 | |
QBB5 was able to induce formation of amorphous CaCO3 precipitates with equal C and Ca proportions (Atomic (at) %). Oxygen (65.31 at%) in the precipitates was much higher than that should be (50.4 at%) in the formed calcium carbonates normally containing 3 times C at% C than Ca (at%). Phosphorus was of 1.8 at% which may be a result of exopolymeric substances precipitated with the minerals. The EPS are also a source of oxygen in the precipitates, explaining the high oxygen content (at%) in the precipitates. Induction of precipitation was concomitant with the increased pH, to almost 8.5, favourable to biomineralization. Fig. 3(a) shows the amorphous calcium carbonate precipitates obtained in urea medium supplemented with calcium chloride and inoculated with QBB5. When soil was added to the urea medium containing calcium chloride, SEM analysis did not show precipitates or crystals of calcium carbonates in abiotic cultures (not inoculated) (Fig. 3(b)). Similar images were obtained with soil in water. However, EDS analysis showed presence of calcium, carbon, oxygen and phosphorus in the used soil in water or in urea medium supplemented with calcium chloride. They are components of soil, as minerals, but not necessary pure calcium carbonate as amorphous or crystal forms. The isolate QBB5 induced calcium carbonate precipitation since clear calcite crystals were observed in the SEM images (Fig. 3(c)). The carbon, oxygen, calcium and phosphorus with atomic percentages of 24.0%, 60.50%, 23.90% and 1.1% were observed in EDS analysis. Although calcium and carbons are in the same proportion (24 at%), it is not accurate to conclude about the importance of the calcium carbonate formation in the soil. The pH of the biotic culture was almost 8.8, highly appropriate for calcium carbonate precipitation. On the other hand, at a different region within the same culture, the carbon, oxygen, calcium and phosphorus with atomic percentages 13.9%, 60.8%, 13.4% and 2.6% were observed in EDS analysis, however, no regular calcite has been observed instead amorphous calcium carbonate is seen precipitated.
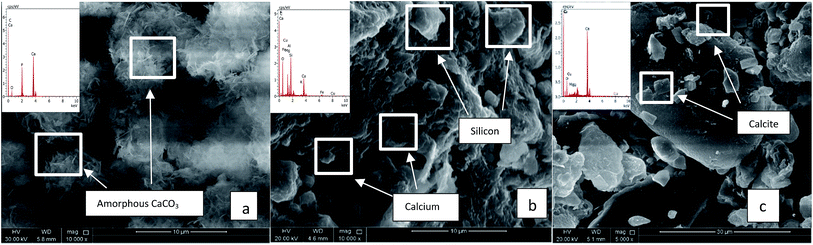 | ||
| Fig. 3 SEM analysis of cultures with urea media. (a) Urea media + CaCl2 + QBB5. (b) Soil + urea media + CaCl2 without QBB5. (c) Soil + urea media + CaCl2 with QBB5. | ||
4. Discussion
Qatar has extreme weather conditions that results in a stressful environment affecting the microbial growth and processes.2 However, certain micro-organisms such as Bacillus, Sporolactobacillus and Sporosarcina are well known to be adapted to harsh conditions.11 In our study, the identified strains exhibiting urease activity were all identified as belonging to the Bacillus genus.It is important to gather information on the bacterial species that are involved in ureolysis and biomineralization specially when there is no preliminary data available within the region. B. licheniformis, B. subtilis and B. cereus were isolated utilizing modified urea agar among which sub-species of B. cereus were abundant. B. cereus isolated from soil and marine environment are known to hydrolyse urea.9,17,27,28 Similarly, B. licheniformis is also involved to precipitate calcium carbonate by ureolysis.16 All the species that tested positive for ureolytic activity could grow on urea as sole carbon and nitrogen source consistently. The growth remained stable throughout the experimental analysis. Hence, it can be said that the screened and identified group of species are not characteristic of ureolytic bacteria in Qatar but rather represent species that are able to express the gene responsible for production of urease enzyme under the experimental conditions used. Based on the results (Table 1), it can be concluded that ureolytic bacteria are abundant in Qatari soil and further in-depth analysis could reveal more species.
All isolated strains followed a similar pattern of growth when subjected to different concentrations of urea (Fig. 2). Based on the uniformity in results, 20 g L−1 of urea was used as a final concentration in further experiments. In the literature, 20 g L−1 of urea is used as well for ureolytic assisted biomineralization experiments.5,14,20,27 Regarding urease activity, a new technique of arbitrary urease activity was used that allows researchers to determine the specific production per colony forming unit. As per the ANOVA analysis, the urease activity between all the isolated species was not significantly different. However, based on the specific production (AUA/106 cfu), each species can be categorized in groups. There is a possibility that species with lower specific production needs more time to hydrolyse urea as compared to species with higher specific production due to a change in their conditions.2
The hydrolysis of urea and the process of calcium carbonate precipitation are directly linked.23 Calcium carbonate precipitation is present in the cultures in its regular (calcite) and irregular (amorphous) precipitate types. The SEM and EDS results provide evidence of the precipitation of calcium carbonate in the cultures by the hydrolysis of urea as well as differences in the morphologies of the soil in cultures with and without the inoculum. Based on the atomic percentages observed, it is evident that carbon and calcium percentages are much lower than the percentages observed for the region with calcite precipitation which means not enough carbon and calcium ions were available for proper crystal formation.
It is important to mention here that urea hydrolysis results in the production of ammonia (NH3) and carbonic acid (H2CO3). The carbonic acid further dissociates and produce carbonate ions (CO3−2). Furthermore, carbonate ions would react with calcium ions (from CaCl2) and precipitate calcium carbonate (CaCO3) at higher pH.8 From this, we can conclude that the precipitation of calcium carbonate in the cultures is the outcome of urea hydrolysis by ureolytic bacteria as no foreign source of carbonate in the cultures was introduced.
The result of this research showed that the abundance of ureolytic Bacillus sp. is high with respect to any other genus. There could be a possibility that all the conditions were favourable for this genus to survive and grow as compared to others during the isolation.
5. Conclusion
The results obtained from this research confirms the presence of ureolytic bacteria in Qatari soil indicating their adaptation to the harsh environment. Using MALDI-TOF MS technique, differentiation of bacteria within the same species such as B. cereus was possible through protein profiles. This showed that the urease activity can be different within the same species, perhaps, due to the differences in the levels of gene expression for urease enzyme. This research also proposed a new method for determining urease activity i.e. arbitrary urease activity (AUA). Hence, the isolates categorized by high AUA values demonstrates higher ureolytic activity. Since, the biodegradation of urea by these bacteria will lead to increase in pH, thereby enhancing minerals precipitation such as calcium carbonate. Therefore, SEM-EDX analysis was performed in order to analyse the morphology and composition of deposits. It was noticed that the presence or absence of ureolytic bacteria significantly affects the composition and morphology of calcium carbonate crystals. Accordingly, it can be concluded that the indigenous ureolytic bacteria present in Qatari soil has the potential to carry out soil stabilization in the presence of urea by enhancing biomineralization. Therefore, introduction of urea into the soil can also be one of the management measure to control the soil erosion caused by winds and to enhance soil stabilization in Qatar.6. Author contributions
Nabil Zouari, conceived and designed the experiments, contributed in analysis of the data and wrote the paper. Shazia Bibi and Meriam Oualha, performed the experiments and contributed in writing of the manuscript. Mohammad Yousaf Ashfaq, performed the MALDI-TOF MS experiments and contributed in writing of the manuscript. Muhannad T. Suleiman, contributed in conceiving the experiments, analysis of the data and review of the manuscript.Conflicts of interest
The authors declare no conflict of interest. All authors decided to publish this work. The funding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.Acknowledgements
This work was made possible by an NPRP 819292766, a grant from the Qatar National Research Fund (a member of the Qatar Foundation). The statements made herein are solely the responsibility of the authors.References
- S. Abdul Majid, M. F. Graw, H. Nguyen and A. G. Hay, Draft Whole-Genome Sequence of Urease-Producing Sporosarcina koreensis, Genome Announc., 2016, 4, e00096-16 CrossRef PubMed.
- Z. Al Disi, S. Jaoua, D. Al-Thani, S. Al-Meer and N. Zouari, Considering the Specific Impact of Harsh Conditions and Oil Weathering on Diversity, Adaptation, and Activity of Hydrocarbon-Degrading Bacteria in Strategies of Bioremediation of Harsh Oily-Polluted Soils, BioMed Res. Int., 2017, 8649350 Search PubMed.
- A. Al Qabany, K. Soga and C. Santamarina, Factors Affecting Efficiency of Microbially Induced Calcite Precipitation, J. Geotech. Geoenviron. Eng., 2012, 138, 992–1001 CrossRef.
- Y. Bai, X. J. Guo, Y. Z. Li and T. Huang, Experimental and visual research on the microbial induced carbonate precipitation by Pseudomonas aeruginosa, AMB Express, 2017, 7, PMC5342990 CrossRef PubMed.
- M. B. Burbank, T. J. Weaver, B. C. Williams and R. L. Crawford, Urease activity of ureolytic bacteria isolation from six soils in which calcite was precipitated by indigenous bacteria, Geomicrobiol. J., 2012, 389–395 CrossRef CAS.
- S. B. Barbuddhe, T. Maier, G. Schwarz, M. Kostrzewa, H. Hof, E. Domann, T. Chakraborty and T. Hainm, Rapid identification and typing of listeria species by matrix-assisted laser desorption ionization-time of flight mass spectrometry, Appl. Environ. Microbiol., 2008, 74, 5402–5407 CrossRef CAS PubMed.
- L. Cheng and R. Cord-Ruwisch, In situ soil cementation with ureolytic bacteria by surface percolation, Ecol. Eng., 2012, 42, 64–72 CrossRef.
- N. K. Dhami, S. M. Reddy and A. Mukherjee, Biofilm and Microbial Applications in Biomineralized Concrete, INTECH Open Access Publisher, 2012 Search PubMed.
- J. Dick, W. De Windt, B. De Graef, H. Saveyn, M. P. Vander, N. De Belie and W. Verstraete, Bio-deposition of a calcium carbonate layer on degraded limestone by Bacillus species, Biodegradation, 2006, 17, 357–367 CrossRef CAS PubMed.
- R. Dieckmann and B. Malorny, Rapid screening of epidemiologically important Salmonella enterica subsp. enterica serovars by whole-cell matrix-assisted laser desorption ionization-time of flight mass spectrometry, Appl. Environ. Microbiol., 2011, 77, 4136–4146 CrossRef CAS PubMed.
- C. F. Ferris, R. H. Melloni, G. Koppel, K. W. Perry, R. W. Fuller and Y. Delville, Vasopressin/serotonin interactions in the anterior hypothalamus control aggressive behavior in golden hamsters, J. Neurosci., 1997, 17, 4331–4340 CAS.
- GRC, Arabesque GRC Qatar – The International Glassfibre Reinforced Concrete Association (GRCA) for all your GRC/GFRC information, 2008, retrieved from http://www.grca.org.uk/grca-members/arabesque.php, accessed on 23 Oct 2017 Search PubMed.
- M. Hvidt, Economic diversification in GCC countries: Past record and future trends, Governance and Globalisation in the Gulf States, 2013 Search PubMed.
- I. Hammad, A. Zoheir and F. Talkhan, Urease activity and induction of calcium carbonate precipitation by Sporosarcina pasteurii NCIMB 8841, J. Appl. Sci. Res., 2013, 9, 1525–1533 CAS.
- F. Hammes, N. Boon, J. De Villiers, W. Verstraete, S. D. Siciliano, J. D. Villiers and J. De, Strain-Specific Ureolytic Microbial Calcium Carbonate Precipitation Strain-Specific Ureolytic Microbial Calcium Carbonate Precipitation, Appl. Environ. Microbiol., 2003, 69, 4901–4909 CrossRef CAS PubMed.
- F. M. Helmi, H. R. Elmitwalli, S. M. Elnagdy and A. F. El-Hagrassy, Calcium carbonate precipitation induced by ureolytic bacteria Bacillus licheniformis, Ecol. Eng., 2016, 90, 367–371 CrossRef.
- G. Kim and H. Youn, Microbially induced calcite precipitation employing environmental isolates, Materials, 2016, 9, 468, DOI:10.3390/ma9060468.
- M. F. Lartigue, E. H. Arnaud, E. Haguenoer, A. S. Domelier, P. O. Schmit, N. V. Marquet, P. Lanotte, L. Mereghetti, M. Kostrzewa and R. Quentin, Identification of Streptococcus agalactiae Isolates from Various Phylogenetic Lineages by Matrix-Assisted Laser Desorption Ionization – Time of Flight Mass Spectrometry, Society, 2009, vol. 47, pp. 2284–2287 Search PubMed.
- B. Lian, Q. Hu, J. Chen, J. Ji and H. H. Teng, Carbonate biomineralization induced by soil bacterium Bacillus megaterium, Geochim. Cosmochim. Acta, 2006, 70, 5522–5535 CrossRef CAS.
- H. Lin, T. M. Suleiman, D. G. Brown and E. Kavazanjian, Mechanical Behavior of Sands Treated by Microbially Induced Carbonate Precipitation, J. Geotech. Geoenviron. Eng., 2016, 142, 04015066 CrossRef.
- D. Mujah, M. A. Shahin and L. Cheng, State-of-the-art review of bio-cementation by microbially induced calcite precipitation (MICP) for soil stabilization, Geomicrobiol. J., 2016, 524–537 Search PubMed.
- M. Nacef, M. Chevalier, S. Chollet, D. Drider and C. Flahaut, MALDI-TOF mass spectrometry for the identification of lactic acid bacteria isolated from a French cheese: The Maroilles, Int. J. Food Microbiol., 2017, 247, 2–8 CrossRef CAS PubMed.
- A. Otlewsk and B. Gutarowska, Environmental parameters conditioning microbially induced mineralization under the experimental model conditions, Acta Biochim. Pol., 2016, 63, 343–351 CrossRef PubMed.
- K. Rizzardi, T. Wahab and C. Jernberg, Rapid subtyping of Yersinia enterocolitica by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI TOF MS) for diagnostics and surveillance, J. Clin. Microbiol., 2013, 51, 4200–4203 CrossRef PubMed.
- T. R. Sandrin, J. E. Goldstein and S. Schumaker, MALDI TOF MS profiling of bacteria at the strain level: a review, Mass Spectrom. Rev., 2012, 32, 188–217 CrossRef PubMed.
- Y. Wang, Q. Zhou, B. Li, B. Liu, G. Wu, M. Ibrahim, G. Xie, H. Li and G. Sun, Differentiation in MALDI-TOF MS and FTIR spectra between two closely related species Acidovorax oryzae and Acidovorax citrulli, BMC Microbiol., 2012, 12, PMC3438124 Search PubMed.
- S. Wei, H. Cui, Z. Jiang, H. Liu, H. He and N. Fang, Biomineralization processes of calcite induced by bacteria isolated from marine sediments, Braz. J. Microbiol., 2015, 46, 455–464 CrossRef PubMed.
- J. Whitaker, Assessing Recombinant Expression of Urease Enzyme from Sporosarcina Ureae as a carbonatogenic method for Strength Enhancement of Loose, Sandy Soils, University of Ottawa, Canada, 2016 Search PubMed.
- M. Wolters, H. Rohde, T. Maier, C. Belmar-Campos, G. Franke, S. Scherpe, M. Aepfelbachera and M. Christner, MALDI-TOF MS fingerprinting allows for discrimination of major methicillin-resistant Staphylococcus aureus lineages, International Journal of Medical Micrbiology, 2011, 301, 64–68 CrossRef CAS PubMed.
- T. Zhu and M. Dittrich, Carbonate Precipitation through Microbial activities in Natural environment and their potential in biotechnology: A Review, Front Bioeng Biotechnol., 2016, 4, 1–21 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2018 |
