Open Access Article
Open Access ArticleN-Acetyl-L-leucine-polyethylenimine-mediated miR-34a delivery improves osteogenesis and bone formation in vitro and in vivo
Yuqin Shena,
Yin Liua,
Han Gaoa,
Hongbo Feia,
Wenwen Yu b,
Tianqi Hua,
Yi Zheng
b,
Tianqi Hua,
Yi Zheng a,
Xueting Bia and
Chongtao Lin*a
a,
Xueting Bia and
Chongtao Lin*a
aDepartment of Periodontics, School and Hospital of Stomatology, Jilin University, 1500 Qinghua Road, Changchun 130021, China. E-mail: linct@jlu.edu.cn; Tel: +86-431-88796039
bDepartment of Orthodontics, School and Hospital of Stomatology, Jilin University, 1500 Qinghua Road, Changchun 130021, China
First published on 20th February 2018
Abstract
Oral bone defects are difficult to treat. Recently, endogenous miR-34a was shown to be involved in bone anabolism. Clinical application of such microRNAs requires the inherent instability of microRNAs to be overcome by an efficient delivery system. In this study, we employed N-acetyl-L-leucine-modified polyethylenimine (N-Ac-L-Leu-PEI) as an miR-34a carrier and evaluated its delivery ability, transfection efficiency, cytotoxicity and whether it enhanced osteogenic differentiation and bone formation in vitro and in vivo. Stable N-Ac-L-Leu-PEI/miR-34a nanocomplexes were synthesized at a mass ratio of 4 and had a small size (190.34 nm), a low zeta potential (21.1 mV), a high transfection efficiency (69.39%) and no cytotoxicity in MG63 cells. N-Ac-L-Leu-PEI-mediated miR-34a delivery in vitro promoted ALP activity and expression of osteogenic differentiation markers, Runx2, SP7 and ColI to higher levels than those produced by Lipofectamine 2000-mediated delivery. N-Ac-L-Leu-PEI also achieved delivery of miR-34a in vivo to a local cranial bone defect area with miR-34a retaining the ability to initiate significant new bone formation 12 weeks post-implantation. This demonstrates the potential for N-Ac-L-Leu-PEI as a gene therapy vehicle for the regeneration of bone defects.
1. Introduction
Defects in oral bones are serious sequelae of tumors, periodontitis, fractures and surgery, and the ultimate goal of complete bone tissue regeneration after injury is still a great challenge. Various therapeutic methods have been applied, such as bone transplantation, guided bone regeneration, bone replacement technology, and tissue engineering; however, all these strategies have limitations.1 To improve these methods, many studies have recently applied gene therapy to bone regeneration.miR-34a is regarded as an efficient regulator of bone metabolism.2–6 miR-34a is an evolutionarily conserved miRNA and is an endogenous inhibitor of osteoclasts to block osteoporosis and bone metastasis by suppressing osteoclastogenesis and Tgif2.4 miR-34a is also implicated in tooth development, being expressed during various stages of tooth growth, and regulates the differentiation and proliferation of human dental papilla cells.7,8 The effects of miR-34a on osteogenic differentiation are multiple. miR-34s inhibit osteogenic proliferation and differentiation in mice,5 but miR-34a plays a positive role in osteogenesis and bone formation in human adipose-derived stromal cells6 via the RBP2/NOTCH1/CYCLIN D1 coregulatory network. From these findings it was suggested that gene therapy based on miR-34a could be a valuable approach to achieve bone regeneration. However, microRNA instability and low intracellular penetration limited its application in bone tissue regeneration.9 It is, therefore, necessary to apply an effective delivery system for miR-34a-mediated gene therapy.
Non-viral carriers, such as cationic polymers polyethylenimine (PEI) and cyclodextrin, are regarded as “gold standard” delivery systems due to their high gene-loading capacity, low production cost and low immunogenicity.10–12 Branched PEI, with an average molecular weight of 25 kDa (PEI25K), exhibits high gene transfection efficiency owing to its high cationic density. This characteristic also promotes condensation of nucleic acids into polyplexes and facilitates their transfer into cells and endosome release.13,14 However, the excessive positive charge of PEI25K can also cause cytotoxicity and hemolysis as well as serum instability.15,16 To solve these problems, PEI25K has been chemically modified by grafting hydrophobic groups or polymers to enhance transfection efficiency and to decrease cytotoxicity.17–19 In a previous study, the derivative N-acetyl-L-leucine-modified polyethylenimine (N-Ac-L-Leu-PEI) was constructed by grafting hydrophobic N-acetyl-L-leucine onto PEI25K, and the carrier improved transfection efficiency and biocompatibility. This carrier has been successfully applied in delivery of the p53 gene20 and a DNAzyme21 to inhibit the proliferation and migration of tumor cells.
In this study, N-Ac-L-Leu-PEI was employed as a carrier for miR-34a to investigate whether N-Ac-L-Leu-PEI could effectively deliver miR-34a without toxicity so as to regulate osteogenesis in vitro and in vivo. The characteristics of N-Ac-L-Leu-PEI/miR-34a were determined by gel retardation, particle size and zeta potential assays. Transfection efficiency and cytotoxicity were detected by flow cytometry and MTT assays. Finally, the miR-34a regulation of osteogenic differentiation of MG63 cells in vitro and of bone regeneration in a cranial bone defect model in vivo was evaluated.
2. Materials and methods
2.1 Materials
The miR-34a mimics and FAM-labeled miR-34a were synthesized by Gene Pharma (Suzhou, China): miR-34a, sense: 5′-AGCCGCTGGCAGTGTCTTA-3′; antisense: 5′-CAGAGCAGGGTCCGAGGTA-3′. The derivative N-Ac-L-Leu-PEI was constructed according to a previous report20 and branched PEI25K (Sigma-Aldrich, St. Louis, USA) was provided by the School of Life Sciences, Jilin University. Lipofectamine 2000 was purchased from Invitrogen. Dulbecco's Modified Eagle's Medium (DMEM), fetal bovine serum (FBS) and trypsin were purchased from Hyclone (Hyclone, USA). 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) was sourced from Sigma (USA). The annexin V-FITC/propidium iodide (PI) apoptosis detection kit was purchased from Shenggong (Shanghai, China). The PrimeScript™ RT reagent kit and SYBR-Green premix ExTaq™ were purchased from Takara (Dalian, China). All other chemicals used were of the highest reagent grade commercially available and used as received. The following instruments were used: Olympus IX71 fluorescence microscope (Olympus, Japan), microplate reader (Rayto, USA), FACSCalibur (BD Bioscience, Mountain View, USA), NanoDrop 1000 spectrophotometer (Thermo Scientific, USA), and Mx3005P fluorescence quantitative PCR instrument (Agilent Technologies, USA).2.2 Gel retardation assay
N-Ac-L-Leu-PEI/miR-34a nanocomplexes were prepared at different mass ratios by incubation at room temperature for 30 minutes. The ability of N-Ac-L-Leu-PEI to bind to miR-34a was assessed by 1.5% agarose gel electrophoresis in a Tris-acetate-ethylenediaminetetraacetic acid buffer (80 V, 25 minutes).2.3 Size distribution and zeta potential
The particle sizes and zeta potentials of N-Ac-L-Leu-PEI/miR-34a nanocomplexes were examined using a Zetasizer Nano ZS90 (Malvern Instruments, Malvern, UK). The morphologies of N-Ac-L-Leu-PEI/miR-34a nanocomplexes were determined by transmission electron microscopy (TEM) (FEI Tecnai G2 STWINF20). These tests were repeated three times.2.4 Flow cytometry (FCM) and fluorescence observation
The in vitro gene transfection efficacy of Lipofectamine 2000 and N-Ac-L-Leu-PEI was evaluated in MG63 cells. MG63 cells were seeded in 6-well plates at 3 × 105 cells per well22 and cultured for 24 h prior to transfection (when they had reached 70–80% confluency). Next, the medium was replaced with serum-free medium containing polymer/FAM-miR-34a polyplexes at an miR-34a concentration of 0.1 μg mL−1. Six hours after transfection, cells were washed with PBS and analyzed on an Olympus IX71 fluorescence microscope (Osaka, Japan) at 480 nm. Transfection efficiency was evaluated by scoring the percentage of FAM-miR-34a+ cells by flow cytometry using a FACSCalibur (BD Bioscience Mountain View, USA).2.5 Cell viability assay
The cytotoxicity of Lipofectamine 2000-, PEI25K- and N-Ac-L-Leu-PEI-mediated miR-34a delivery was evaluated using an MTT assay. MG63 cells were seeded into a 96-well plate at a density of 5 × 103 cells per well and pre-incubated at 37 °C in 5% CO2 for 24 h. Cells were then cultured with polymer/miR-34a nanocomplex at a mass ratio of 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. After co-culture for 24, 48 or 72 h, 20 μL MTT solution (1 mg mL−1 in PBS) was added to each well and the plate was incubated for an additional 4 h. After removing the MTT solution, 150 μL per well DMSO was added to dissolve the formazan crystals. Following an additional incubation for 10 min, the absorbance at 492 nm was recorded using a microplate reader (Rayto, USA). The cell viability (%) was calculated as Asample/Acontrol × 100.
1. After co-culture for 24, 48 or 72 h, 20 μL MTT solution (1 mg mL−1 in PBS) was added to each well and the plate was incubated for an additional 4 h. After removing the MTT solution, 150 μL per well DMSO was added to dissolve the formazan crystals. Following an additional incubation for 10 min, the absorbance at 492 nm was recorded using a microplate reader (Rayto, USA). The cell viability (%) was calculated as Asample/Acontrol × 100.
2.6 Apoptosis and live/dead cell assays
miR-34a-induced apoptosis was analyzed using an Annexin V-FITC/PI apoptosis detection kit (Shenggong, Shanghai, China). MG63 cells were seeded in a 6-well plate at a density of 3 × 105 cells per well22 and incubated for 24 h before transfection. Transfection was performed using the PEI25K or N-Ac-L-Leu-PEI/miR-34a nanocomplex (4 wt/wt ratio, 2 μg miR-34a) in serum-free DMEM for 6 h. After incubation in DMEM containing 10% FBS for 72 h, cells were harvested, washed twice with PBS and resuspended in binding buffer according to the manufacturer's protocol. Cells were then incubated with Annexin V-FITC/PI at room temperature for 20 min in the dark. Apoptosis was assessed by analyzing 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells gated through a FACSCalibur (BD Biosciences) with Cell Quest software (BD Biosciences).
000 cells gated through a FACSCalibur (BD Biosciences) with Cell Quest software (BD Biosciences).
Transfected MG63 cells for live/dead cell assays were washed with PBS and then incubated with Calcein-AM/PI at room temperature for 30 min in the dark. Cell images were collected under blue-ray and green-ray to calculate the percentage of cells.
2.7 Real-time PCR assay
MG63 cells were harvested after incubation with N-Ac-L-Leu-PEI/miR-34a or Lipofectamine 2000/miR-34a for 72 h. RNAs were isolated using a HiPure Total RNA kit (Magen, China) according to the manufacturer's instructions. miRNA was subjected to reverse transcription and PCR using a SYBR Green Hairpin-it miRNAs qRT-PCR kit. mRNAs were also reverse transcribed and amplified using a PrimerScript® RT reagent kit and SYBR Green Premix Ex Taq. The PCR products were evaluated with an MxPro Mx3005P real-time PCR detection system (Agilent Technologies, Santa Clara, CA, USA). The internal controls of mRNAs and miR-34a were GAPDH and U6, respectively. The cycling conditions of mRNAs were: 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s, 55 °C for 30 s, and 72 °C for 1 min. The cycling conditions for miRNAs were: 95 °C for 3 min, followed by 40 cycles of 95 °C for 12 s, and 62 °C for 40 s. The 2−ΔΔCt method was used to calculate relative expression levels, and the obtained values were the average of triplicate measurements. Primers used for qPCR were:U6, F:5′-ATTGGAACGATACAGAGAAGATT-3′/R:5′-GGAACGCTTCACGAATTTG-3′; miR-34a, F:5′-AGCCGCTGGCAGTGTCTTA-3′/R:5′-CAGAGCAGGGTCCGAGGTA-3′.
GAPDH, F:5′-ACCACAGTCCATGCCATCAC-3′/R: 5′-TCCACCACCCTGTTGCTGTA-3′; Runx2, F:5′-TTGCAGTCTTCCTGGAGAAAGTT-3′/R:5′-CCTCTCGACCCGACTGCAGATC-3′;
SP7, F:5′-CACAGCTCTTCTGACTGTCTG-3′/R:5′-CTGGTGAAATGCCTGCATGGAT-3′;
COL-1, F:5′-AGGGCCAAGACGAAGACATC-3′/R:5′-AGATCACGTCATCGCACAACA-3′.
2.8 Western blot analysis
The effect of miR-34a transfection on the expression of proteins associated with osteoblast-related factors was evaluated by western blotting, in which protein levels were normalized against those of GAPDH. Seventy-two hours after transfection with the N-Ac-L-Leu-PEI/miR-34a nanocomplex (4 wt/wt ratio, 2 μg miR-34a), MG63 cells were harvested, washed twice with ice-cold PBS and lysed with RIPA lysis buffer on ice for 5 min. The lysates were centrifuged at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 15 min and supernatants collected. Protein concentrations were determined using a BCA protein assay kit. Equal amounts of protein were subjected to SDS-PAGE and subsequently transferred to PVDF membranes by electroblotting. The membranes were blocked in PBS containing 5% skimmed milk and 0.1% Tween-20 for 1 h, and then incubated with desired antibodies at 4 °C overnight. Membranes were subsequently incubated with an HRP-goat anti-rabbit IgG (H + L) antibody for 1 h and detected using enhanced chemical luminescence (Amersham, UK).
000 × g for 15 min and supernatants collected. Protein concentrations were determined using a BCA protein assay kit. Equal amounts of protein were subjected to SDS-PAGE and subsequently transferred to PVDF membranes by electroblotting. The membranes were blocked in PBS containing 5% skimmed milk and 0.1% Tween-20 for 1 h, and then incubated with desired antibodies at 4 °C overnight. Membranes were subsequently incubated with an HRP-goat anti-rabbit IgG (H + L) antibody for 1 h and detected using enhanced chemical luminescence (Amersham, UK).
2.9 Rat cranial bone defect model
All animal procedures were approved by the International Animal Care and Use Committee of Jilin University, Changchun, People's Republic of China. The animal experiments were performed in accordance with the requirements of the Experimental Animal Ethics and Welfare guidelines (Permit Number: 20160205). Eight-week-old male Wistar rats with an average weight of 180 g were obtained from the Animal Experiment Center of Jilin University. The bone defect model was prepared according to Yin23 and Zhao.24 Rats were anesthetized using ketamine (60 mg kg−1) and xylazine (8 mg kg−1). The experimental area was shaved and sterilized and the scalp tissue separated along the midline to expose the calvarias area. Unilateral critical bone defects (6 mm in diameter) were created using a bone trephine (3i Implant Innovation, Palm Beach Gardens, FL, USA). The N-Ac-L-Leu-PEI/miR-34a and Lipofectamine 2000/miR-34a nanoparticles were synthesized with 2 μg miR-34a at a W/W ratio of 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, and were then absorbed into a 5 mm diameter, 1.5 mm thick gelatin sponge. The gelatin sponge covered the defect area, and stratified suture wound. The experimental rats were sacrificed at 12 weeks after implantation and exsanguinated via the auricula dextra and perfused via the ventriculus dexter using 4% paraformaldehyde solution. Cone Beam Computerized Tomography (CBCT) and Microcomputed tomography (micro-CT) (micro-CT 35; Scanco Medical AG, Bassersdorf, Switzerland) was used to evaluate new bone formation. Quantification of new bone formation was calculated using Image-Pro Plus software.
1, and were then absorbed into a 5 mm diameter, 1.5 mm thick gelatin sponge. The gelatin sponge covered the defect area, and stratified suture wound. The experimental rats were sacrificed at 12 weeks after implantation and exsanguinated via the auricula dextra and perfused via the ventriculus dexter using 4% paraformaldehyde solution. Cone Beam Computerized Tomography (CBCT) and Microcomputed tomography (micro-CT) (micro-CT 35; Scanco Medical AG, Bassersdorf, Switzerland) was used to evaluate new bone formation. Quantification of new bone formation was calculated using Image-Pro Plus software.
2.10 Statistical analysis
Experiments were performed on at least three individuals. Results are presented as the mean ± standard deviation and were statistically analyzed with Student's t-test unless noted otherwise. A two-tailed P-value of <0.05 was considered statistically significant. P values were *P < 0.05, **P < 0.01, and ***P < 0.001, with n.s. (non-significant) P > 0.05. Statistical analysis was conducted in SPSS version 20.0 (IBM).3. Results
3.1 Characterization of the N-Ac-L-Leu-PEI/miR-34a nanocomplex
The binding of N-Ac-L-Leu-PEI and miR-34a was assessed using an agarose gel retardation assay. Partial to complete binding was observed between N-Ac-L-Leu-PEI and miR-34a at a mass rate of 2 (Fig. 1A), indicating the formation of a stable N-Ac-L-Leu-PEI/miR-34a complex. Compared with plasmid DNA (critical mass rate of 0.6–0.8),20 N-Ac-L-Leu-PEI exhibited lower binding ability for microRNAs and single-stranded oligoDNAs (critical mass rate of 2.0).21 The ability of PEI to condense DNA constructs into nanocomplexes and to decrease their negative charge prevents them escaping from lysosomes, promoting their delivery into cells.13,14 We examined the particle size and zeta potential of N-Ac-L-Leu-PEI/miR-34a nanocomplexes at different mass ratios (Fig. 1B and C). The size of N-Ac-L-Leu-PEI/miR-34a complexes rapidly declined with an increasing w/w ratio (Fig. 1B). The smallest N-Ac-L-Leu-PEI/miR-34a complex was 190.34 ± 6.76 nm at a mass ratio of 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The zeta potential also decreased with increasing w/w ratio, with the lowest being 12.93 ± 1.67 mV at a 6 w/w ratio (Fig. 1C). TEM images of N-Ac-L-Leu-PEI/miR-34a complexes showed a globular morphology with an average size of approximately 200 nm (Fig. 1D). This indicated that N-Ac-L-Leu-PEI was able to form stable nanoparticles with miR-34a.
1. The zeta potential also decreased with increasing w/w ratio, with the lowest being 12.93 ± 1.67 mV at a 6 w/w ratio (Fig. 1C). TEM images of N-Ac-L-Leu-PEI/miR-34a complexes showed a globular morphology with an average size of approximately 200 nm (Fig. 1D). This indicated that N-Ac-L-Leu-PEI was able to form stable nanoparticles with miR-34a.
3.2 Transfection efficiency of N-Ac-L-Leu-PEI/miR-34a
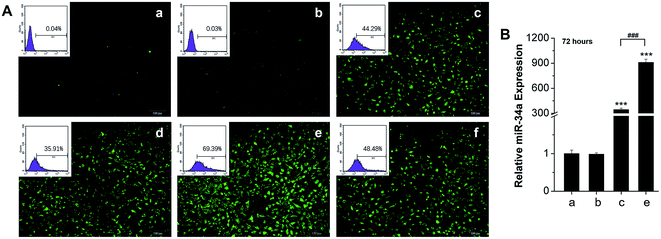
The derivative N-Ac-L-Leu-PEI has achieved successful delivery of the p53 gene and a DNAzyme with higher transfection efficiency compared with PEI25K and Lipofectamine 2000.20,21 To determine whether N-Ac-L-Leu-PEI can achieve acceptable transfection efficiency with microRNAs, we employed fluorescence microscopy, flow cytometry (FCM) and qRT-PCR. Inverted fluorescence microscopy identified FAM-miR-34a+ cells in both Lipofectamine 2000- and N-Ac-L-Leu-PEI-mediated transfection of FAM-miR-34a groups with 30–40% and 60–70% fluorescence intensity, respectively. FCM quantitative analysis of FAM-miR-34a+ cells revealed the highest positive rates in the N-Ac-L-Leu-PEI/miR-34a-FAM group to be at a w/w ratio of 4 (Fig. 2A). Moreover, qRT-PCR showed that 72 h after transfection of MG63 cells, N-Ac-L-Leu-PEI-mediated delivery of miR-34a produced a higher expression level of miR-34a (910 fold) compared with that for Lipofectamine 2000 (344 fold) (Fig. 2B). These data demonstrate that N-Ac-L-Leu-PEI can achieve high transfection efficiency and gene expression of miR-34a in cells, with an optimal mass ratio of 4.3.3 Cytotoxicity of N-Ac-L-Leu-PEI in vitro
PEI has high cytotoxicity; therefore, Li20 synthesized N-Ac-L-Leu-PEI by a 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide/N-hydroxysuccinimide (EDC/NHS)-mediated coupling reaction of N-acetyl-L-leucine on the primary amino group of PEI25K. This derivative has high resistance to protein adsorption and low hemolytic activity. In our MTT assay, the cell viability of three groups was more than 90%, and there was no cytotoxicity in either the Lipofectamine 2000/miR-34a group or the N-Ac-L-Leu-PEI/miR-34a group at 24, 48 and 72 h after transfection (Fig. 3A). Similarly, further apoptosis assays (Fig. 3B) and live/dead cell viability assays (Fig. 4) revealed no significant influence of N-Ac-L-Leu-PEI on MG63 cells. N-Ac-L-Leu-PEI produced a slightly reduced population of early apoptotic cells and more live cells than Lipofectamine 2000. However, in group of PEI25K/miR-34a, the cell viability on 24, 48, 72 h was approximately 85%, 72%, 70%, respectively (Fig. 3), as well as elevated early and late cell apoptosis (23.54%) (Fig. 3) and red cells population in live/dead cell detection (Fig. 4). These results indicate that N-Ac-L-Leu-PEI owned lower toxicity of MG63 cells than PEI25K. We therefore considered N-Ac-L-Leu-PEI to be suitable for further transfection experiments and applications in vitro and in vivo.3.4 Osteogenic differentiation of MG63 cells in vitro
Several recent reports have suggested that miR-34a is involved in the regulation of bone homeostasis. However, the reported roles of miR-34a in osteogenesis are not consistent, with some in vitro studies indicating a positive effect4,6 and some a negative effect.3,4 Most in vivo studies, showed up-regulation of miR-34a during osteogenesis. To determine whether miR-34a induces osteogenic differentiation of MG63 cells in vitro, we evaluated the early secretion function, and gene expression and protein levels during osteogenic differentiation. The early function assay showed that miR-34a markedly improved ALP activity of MG63 cells at 24, 48 and 72 h after transfection (Fig. 5A) and that N-Ac-L-Leu-PEI-mediated miR-34a delivery led to significantly superior ALP activity augmentation compared with Lipofectamine 2000. The expression of Runx2, SP7, and ColI is related to osteoblastogenesis and is required for osteoblast differentiation.25–29 Gene expression and protein levels for Runx2, SP7 and ColI were clearly elevated in miR-34a groups. The effect of miR-34a on genes and proteins of osteogenic differentiation (except for ColI mRNA levels) was greater with N-Ac-L-Leu-PEI delivery compared with Lipofectamine 2000 delivery (Fig. 5B–D).Our data clearly indicate that miR-34a elevates osteogenic differentiation of MG63 cells. This is in agreement with Grilli et al., who confirmed osteogenic differentiation of osteosarcoma by miR-34a.30 Moreover, compared with the commercial reagent, Lipofectamine 2000, N-Ac-L-Leu-PEI promoted higher levels of miR-34a-mediated osteogenic differentiation at the gene and protein expression levels and in early function.
3.5 Bone formation in vivo
We employed a rat cranial bone defect model to evaluate the in vivo regulation of N-Ac-L-Leu-PEI-mediated miR-34a delivery on bone formation. CBCT showed new bone formation to be rare, limited, and significantly increased in blank, Lipofectamine 2000/miR-34a and N-Ac-L-Leu-PEI/miR-34a groups, respectively (Fig. 6A). This was in accordance with the results of micro-CT. N-Ac-L-Leu-PEI-mediated miR-34a delivery clearly produced more new bone formation (61.79% new bone formation in the bone defect area) at 12 weeks post-implantation than Lipofectamine 2000-mediated miR-34a delivery (13.6% new bone formation in the bone defect area) (Fig. 6A and B). Moreover, significant augmentation of Runx2, SP7 and especially ColI expression was observed in the N-Ac-L-Leu-PEI/miR-34a-treated bone defect area (Fig. 6C). Different from osteogenic differentiation in vitro, Lipofectamine 2000 failed to produce a new bone formation effect of miR-34a in vivo. These results indicate that N-Ac-L-Leu-PEI can deliver miR-34a to local tissues and that miR-34a retains the ability to promote new bone formation in an in vivo local bone defect model.4. Discussion
Bone repair requires bone proliferation and differentiation to be continuously stimulated31 and the involvement of microRNAs in bone regeneration has been established. miR-34a has a prominent role in bone metabolism and has been employed in phase I clinical trials.32 In this article, we applied N-Ac-L-Leu-PEI to transport miR-34a with the aim to elevate its stability, cellular penetration and capability of promoting bone formation.N-Ac-L-Leu-PEI possesses high resistance to protein adsorption, low hemolytic activity, low zeta potential and small particle size, as well as high transfection efficiency both in the presence and absence of FBS.20 These characteristics make N-Ac-L-Leu-PEI suitable for forming stable nanocomplexes and as a biocompatible gene delivery vehicle in vitro and in vivo. In this study, we describe using N-Ac-L-Leu-PEI as an miR-34a carrier for stimulating MG63 cells in vitro and for local bone regeneration in vivo. N-Ac-L-Leu-PEI formed a stable nanocomplex with miR-34a and had high cellular uptake rates, superior to those of Lipofectamine 2000. Of note, N-Ac-L-Leu-PEI showed lower cytotoxicity than PEI25K, this can be explained as: the derivative N-Ac-L-Leu-PEI neutralized the excess positive charge of nanocomplex surface to induce less cell membrane disruption; N-Ac-L-Leu had lower non-specific protein adsorption capacity to improve the cell viability.20 In addition, N-Ac-L-Leu-PEI not only delivered miR-34a into cells, but also released it to enable it to play a positive role in osteogenic differentiation in vitro. This was observed in the upregulated expression of Runx2, SP7 and ColI by qRT-PCR and western blotting and by ALP activity. Runx2 is a bone differentiation factor,25,26 and Runx2 activation in turn activates SP7 to launch osteogenic differentiation.27,28 ColI mainly provides a fibrous skeleton for the deposition of bone matrix.28 ALP activity indicates the early secretion of bone tissue. Therefore, all the above are essential for bone formation. In addition, it was interesting to find that the in vivo effect of Lipofectamine 2000-mediated miR-34a delivery on osteogenesis was different to its in vitro effect. Because of complex microenvironments in vivo, carriers may easily be eliminated by immunological rejection or other reactions. Krzeszinski et al. obtained an approximately 5-fold increase in pre-miR-34a levels in bone marrow using chitosan nanoparticles after a single injection. Unfortunately, the osteogenesis promoted by miR-34a was insufficient to resist systemic bone loss in OVX mice and cancer bone metastases.4 Lipofectamine 2000 was unable to effectively deliver miR-34a to the bone defect area in vivo, while, miR-34a carried by N-Ac-L-Leu-PEI led to greater local bone formation. This was similar to the results of Fan et al. who demonstrated dramatic heterotopic bone formation in Bio-Oss collagen scaffolds.6 Our results indicate that N-Ac-L-Leu-PEI can protect miR-34a from degradation in complex in vivo environments.
5. Conclusions
The derivative N-Ac-L-Leu-PEI formed stable nanocomplexes with miR-34a and achieved high transfection efficiency without toxicity. Moreover, the carrier-mediated miR-34a delivery could trigger an effective promotion of osteogenic differentiation in vitro and new bone formation in vivo. miR-34 delivery by this PEI derivative signals a new gene therapy approach for bone regeneration.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
We thank Pro. Quanshun Li in Jilin University School of Life Sciences for providing the N-Ac-L-Leu-PEI. This research was supported by grants from the National Natural Science Foundation of China (No. 81371153), Jilin Natural Science Foundation (No. 20170311032YY, No. 20160101335JC), the Medical Support Program of the Jilin university (No. 470110000423) and Jilin provincial education department project (Ji jiao ke he zi [2016] No. 485).References
- U. Ripamonti and J. C. Petit, Cytokine Growth Factor Rev, 2009, 20, 489–499 CrossRef CAS PubMed.
- H. Kang, H. Chen, P. Huang, J. Qi, N. Qian, L. Deng and L. Guo, Osteoporosis Int., 2016, 27, 1493–1505 CrossRef CAS PubMed.
- L. Chen, K. Holmstrom, W. Qiu, N. Ditzel, K. Shi, L. Hokland and M. Kassem, Stem Cells, 2014, 32, 902–912 CrossRef CAS PubMed.
- J. Y. Krzeszinski, W. Wei, H. Huynh, Z. Jin, X. Wang, T. C. Chang, X. J. Xie, L. He, L. S. Manqala, G. Lopez-Berestein, A. K. Sood, J. T. Mendell and Y. Wan, Nature, 2014, 512, 431–435 CrossRef CAS PubMed.
- J. Wei, Y. Shi, L. Zheng, B. Zhou, H. Inose, J. Wang, X. E. Guo, R. Grosschedl and G. Karsenty, J. Cell Biol., 2012, 197, 509–521 CrossRef CAS PubMed.
- C. Fan, L. Jia, Y. Zheng, C. Jin, Y. Liu, H. Liu and Y. Zhou, Stem Cell Rep., 2016, 7, 236–248 CrossRef CAS PubMed.
- M. Wan, B. Gao, F. Sun, Y. Tang, L. Ye, Y. Fan, O. D. Klein, X. Zhou and L. Zheng, PLoS One, 2012, 7, e50090 CAS.
- F. Sun, M. Wan, X. Xu, B. Gao, Y. Zhou, J. Sun, L. Cheng, O. D. Klein, X. Zhou and L. Zheng, J. Dent. Res., 2014, 93, 589–595 CrossRef CAS PubMed.
- G. Lambert, E. Fattal and P. Couvreur, Adv. Drug Delivery Rev., 2001, 47, 99–112 CrossRef CAS PubMed.
- D. W. Pack, A. S. Hoffman, S. Pun and P. S. Stayton, Nat. Rev. Drug Discovery, 2005, 4, 581–593 CrossRef CAS PubMed.
- M. A. Mintzer and E. E. Simanek, Chem. Rev., 2009, 109, 259–302 CrossRef CAS PubMed.
- K. Xiu, J. J. Yang, N. N. Zhao, J. S. Li and F. J. Xu, Acta Biomater., 2013, 9, 4726–4733 CrossRef CAS PubMed.
- A. E. Nel, L. Madler, D. Velegol, T. Xia, E. M. Hoek, P. Somasundaran, F. Klaessiq, V. Castranova and M. Thompson, Nat. Mater., 2009, 8, 543–557 CrossRef CAS PubMed.
- Z. ur Rehman, D. Hoekstra and I. S. Zuhorn, ACS Nano, 2013, 7, 3767–3777 CrossRef CAS PubMed.
- D. Fischer and Y. Li, Biomaterials, 2003, 24, 1121–1131 CrossRef CAS PubMed.
- A. M. Funhoff, C. F. van Nostrum, M. C. Lok, M. M. Fretz, D. J. Crommelin and W. E. Hennink, Bioconjugate Chem., 2004, 15, 1212–1220 CrossRef CAS PubMed.
- H. Tian, W. Xiong, J. Wei, Y. Wang, X. Chen, X. Jing and Q. Zhu, Biomaterials, 2007, 28, 2899–2907 CrossRef CAS PubMed.
- H. Tian, F. Li, J. Chen, Y. Huang and X. Chen, Macromol. Biosci., 2012, 12, 1680–1688 CrossRef CAS PubMed.
- Z. Guo, H. Tian, L. Lin, J. Chen, C. He, Z. Tang and X. Chen, Macromol. Biosci., 2014, 14, 1406–1414 CrossRef CAS PubMed.
- Z. Li, L. Zhang and Q. Li, Colloids Surf., B, 2015, 135, 630–638 CrossRef CAS PubMed.
- Z. Xing, S. Gao, Y. Duan, H. Hao, L. Li, Y. Yang and Q. Li, Int. J. Nanomed., 2015, 10, 5715–5727 CAS.
- Y. Zheng, C. Lin and X. Hou, et al., RSC Adv., 2017, 7, 27121–27127 RSC.
- H. Yin, L. Cui and Y. Cao, Chin. J. Aesthetic Med., 2009, 18, 727–729 CAS.
- L. Zhao, K. Zhang, W. H. Bu, X. W. Xu, H. Jin, B. Chang, B. C. Wang, Y. J. Sun, B. Yang, C. Y. Zheng and H. C. Sun, RSC Adv., 2016, 41, 34081–34089 RSC.
- K. Redlich and J. S. Smolen, Nat. Rev. Drug Discovery, 2012, 11, 234–250 CrossRef CAS PubMed.
- Y. Zhang, R. Xie, C. M. Croce, J. L. Stein, J. B. Lian, A. J. van Wignen and G. S. Stein, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 9863–9868 CrossRef CAS PubMed.
- X. Zhou, Z. Zhang, J. Q. Feng, V. M. Dusevich, K. Sinha, H. Zhang, B. G. Darnay and B. de Crombrugghe, Proc. Natl. Acad. Sci. U. S. A., 2010, 10, 12919–12924 CrossRef PubMed.
- K. Nakashima, X. Zhou, G. Kunkel, Z. Zhang, J. M. Deng, R. R. Behringer and B. de Crombrugghe, Cell, 2002, 108, 17–29 CrossRef CAS PubMed.
- E. Hesse, T. E. Hefferan, J. E. Tarara, C. Haasper, R. Krettek, L. Lu and M. J. Yaszemski, J. Biomed. Mater. Res., Part A, 2010, 94, 442–449 Search PubMed.
- A. Grilli, M. Sciandra, M. Terracciano, P. Picci and K. Scotlandi, BMC Med. Genomics, 2015, 8, 34 CrossRef CAS PubMed.
- K. Andreas, R. Zehbe, M. Kazubek, K. Grzeschik, N. Sternberg, H. Baumler, H. Schubert, M. Sittinger and J. Ringe, Acta Biomater., 2011, 7, 1485–1495 CrossRef CAS PubMed.
- M. Agostini and R. A. Knight, OncoTargets Ther., 2014, 5, 872 Search PubMed.
| This journal is © The Royal Society of Chemistry 2018 |