Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Bacteria killing in ICU associated infections: antibacterial nanosheets as disinfectant†
Li Zhanga,
Jiaming Loub,
Wei Zhangc,
Chaoyang Wud and
Zhaocheng Jin *a
*a
aDepartment of Critical Care Medicine Unit, The Affiliated People's Hospital of Jiangsu University, Zhenjiang, Jiangsu, China. E-mail: JinZhaoC@163.com
bDepartment of Emergency Medicine, The Affiliated People's Hospital of Jiangsu University, Zhenjiang, Jiangsu, China
cDepartment of Pain Medicine, The Affiliated People's Hospital of Jiangsu University, Zhenjiang, Jiangsu, China
dDepartment of Oncology, The Affiliated People's Hospital of Jiangsu University, Zhenjiang, Jiangsu, China
First published on 2nd January 2018
Abstract
Here we report hybrid antibacterial nanosheets (Zn–CuO@GO) where graphene oxide (GO) is decorated with zinc-doped copper dioxide (Zn–CuO) nanoparticles and applied as disinfectant agents to combat multidrug resistant bacterial strains from intensive care units (ICUs). The difference between different Zn–CuO decoration ratios was carefully evaluated and the results indicated when Zn–CuO was deposited onto GO at 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 weight ratio (w/w), the obtained Zn–CuO@GO exhibited the best dispersity, which was suitable as an antibacterial supplement for disinfectant. Importantly, the Zn–CuO@GO (4
1 weight ratio (w/w), the obtained Zn–CuO@GO exhibited the best dispersity, which was suitable as an antibacterial supplement for disinfectant. Importantly, the Zn–CuO@GO (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, w/w) based disinfectant could actively kill 100.0% multidrug bacteria strains within 10 min, including multidrug resistant (MDR) E. coli and methicillin resistant S. aureus (MRSA). Moreover, we measured the antibacterial activities of Zn–CuO@GO at other decoration ratios (i.e., 2
1, w/w) based disinfectant could actively kill 100.0% multidrug bacteria strains within 10 min, including multidrug resistant (MDR) E. coli and methicillin resistant S. aureus (MRSA). Moreover, we measured the antibacterial activities of Zn–CuO@GO at other decoration ratios (i.e., 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1
1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, w/w) to study the effect of Zn–CuO decoration density on the antibiosis activity of Zn–CuO@GO. It was also found that decreased Zn–CuO deposition density not only caused the agglomerateion of Zn–CuO@GO nanosheets (largely due to the relatively increased GO ratio), but also extended their required function time to 30 min for complete bacterial killing. Taking Zn–CuO@GO (4
1, w/w) to study the effect of Zn–CuO decoration density on the antibiosis activity of Zn–CuO@GO. It was also found that decreased Zn–CuO deposition density not only caused the agglomerateion of Zn–CuO@GO nanosheets (largely due to the relatively increased GO ratio), but also extended their required function time to 30 min for complete bacterial killing. Taking Zn–CuO@GO (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, w/w) as the optimized antibacterial nanosheet, further TEM, SEM ad LCSM images revealed that Zn–CuO@GO functions via penetrating and wrapping into the cell wall, inducing bacterial sedimentation and cytoplasma leakage, which make them promising for next-generation disinfectant substitution.
1, w/w) as the optimized antibacterial nanosheet, further TEM, SEM ad LCSM images revealed that Zn–CuO@GO functions via penetrating and wrapping into the cell wall, inducing bacterial sedimentation and cytoplasma leakage, which make them promising for next-generation disinfectant substitution.
Introduction
Disinfectant agents are essential components in the practice of control and prevention of bacterial infections, especially in intensive care units (ICUs), where there have been extensive reports on multidrug resistant bacteria such as carbapenem-resistant acinetobacter baumannii, methicillin resistant S. aureus (MRSA) and multidrug resistant (MDR) E. coli, etc.1a Traditionally, iodophor and hydrogen peroxide (H2O2) were widely used as ICU disinfectant due to their strong oxidizing abilities to cause the denaturing of bacteria and consequently bacterial progressive lysis.1b,c Given the fact that ICU associated bacteria could develop mutations to covert to multi-drug resistance, Kempf et al., demonstrated that iodophor and H2O2 resistant microorganism emerged recently, and these multi-drug resistant bacteria were growing more invulnerable.1d Additionally, Campos et al., have recently found that bacteria (e.g., MRSA) was resistant against a variety of other candidate disinfectants including sodium hypochlorite, glutaraldehyde, formaldehyde, ethanol, chlorhexidine gluconate, and quaternary ammonium.1e A practical method to combat against bacterial resistance is thus urgently to be developed.The application of nanomaterials into bacterial killing is a promising alternative solution, since these nanoparticles have been proven to be highly reactive in antibiosis compared with those chemical compounds.2a,b Several mechanisms, including reactive oxygen species (ROS), metal ion leaking, electrostatic interactions and structural associated physical damage,2c,d are proposed to contribute to their antibacterial behaviours. Among them, physical damage to bacteria is irreversible, and this class of materials emerges as ideal candidates for the development of disinfectants. One of the promising nanomaterials is graphene oxide (GO), a strongly oxygenated and highly hydrophilic two dimensional (2D) layered material with biocompatibility, large specific surface area, high chemical and thermal stability,3a–c and most importantly, bacterial killing abilities. Fan et al., have recently introduced a GO based antibacterial paper which exhibited minimal cytotoxicity to mammalian cells, but effective towards bacterial inhibition.3d Elimelech et al., further demonstrated that GO might cause bacterial lysis by means of their sharpened edges and elevated membrane pressure.3e Similarly, Ivanova et al., proposed that GO inactivated microorganism through the density and orientation of their graphene edges, which led to pore forming in bacterial membrane.3f Despite these recent achievements, GO itself, however, was rarely evaluated against multi-drug resistant bacteria, such as multidrug resistant (MDR) E. coli, methicillin resistant S. aureus (MRSA).4a–c Moreover, GO were typically evaluated on assembled GO integrity, including GO paper3e and GO membrane,3f while antibacterial effect from individually dispersed GO nanosheet was unsatisfying.3c Strategy to decorate GO nanosheets with structure–featured metal oxides that share the physical damage towards bacteria was recently proposed to address this issue, as it not only took advantage of the large specific surface area and morphological features from GO, but also introduced the bacterial activities of metal oxides simultaneously.4d–f,5
Previously, Wu et al., have proposed a Zn–CuO@GO decorated commercial porous nickel (Ni) electrode and demonstrated its ability for efficient capture, rapid killing and ultrasensitive detection of bacteria (normal bacterial stains such as E. coli (ATCC 25922) and S. aureus (ATCC 29213) was evaluated).6a Its behaviour upon contact with multi-drug resistant strains remained elusive, and despite of its effectivity, the electrode was not practical to apply into ICU-associated disinfectant due to its size (6 mm × 24 mm). In contrast, we were able to synthesis Zn–CuO@GO nanosheets to apply as disinfectants and demonstrated their activities to combat against multi-drug resistant bacteria strains. Different from previous works, we obtained Zn–CuO@GO nanosheet solely through in situ deposition and growth, and also, we prepared Zn–CuO@GO with different deposition ratios (i.e., Zn–CuO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) GO = 4
GO = 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 2
1, 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1
1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, w/w) where Zn–CuO that possessed a prickly architecture was allowed to deposit and grow in situ onto GO nanosheets to obtain the hybrid antibacterial nanosheets.6b,c The Zn–CuO@GO suspensions were applied as a disinfectant. Taking multi-drug resistant E. coli (MDR E. coli) and a methicillin resistant S. aureus (MRSA) as examples, these antibacterial nanosheets could effectively inhibit bacterial growth via physical damage, which was confirmed through TEM and SEM. Results indicated that these hybrid nanosheets function as effective antibacterial agents to penetrate into the cellular wall and cause bacterial lysis upon contacting, which was largely due to the fact that they possessed the ‘blades’ from GO and ‘pierces’ from prickly Zn–CuO simultaneously.7a As these antibacterial nanosheets function to cause bacterial lysis through physical damage, possible genetic mutation and development of other drug resistant mechanism might not be applicable.7 To this end, antibacterial Zn–CuO@GO nanosheets is promising as an alternative disinfectant applied in ICU.
1, w/w) where Zn–CuO that possessed a prickly architecture was allowed to deposit and grow in situ onto GO nanosheets to obtain the hybrid antibacterial nanosheets.6b,c The Zn–CuO@GO suspensions were applied as a disinfectant. Taking multi-drug resistant E. coli (MDR E. coli) and a methicillin resistant S. aureus (MRSA) as examples, these antibacterial nanosheets could effectively inhibit bacterial growth via physical damage, which was confirmed through TEM and SEM. Results indicated that these hybrid nanosheets function as effective antibacterial agents to penetrate into the cellular wall and cause bacterial lysis upon contacting, which was largely due to the fact that they possessed the ‘blades’ from GO and ‘pierces’ from prickly Zn–CuO simultaneously.7a As these antibacterial nanosheets function to cause bacterial lysis through physical damage, possible genetic mutation and development of other drug resistant mechanism might not be applicable.7 To this end, antibacterial Zn–CuO@GO nanosheets is promising as an alternative disinfectant applied in ICU.
Results and discussion
By taking advantage of sonochemical method, we fabricated GO nanosheets packaged with prickly Zn–CuO. Briefly, the metal ions (Cu2+ and Zn2+) were firstly allowed to chelate onto GO nanosheets, where ultrasmall nanoclusters were then deposited and readily grew in situ via sonochemical assisted green energies (Fig. 1A).7a As indicated in the transmission electron microscopy image (TEM, Fig. 1B), the prickly Zn–CuO were randomly and successfully deposited onto individual GO nanosheets (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, w/w), which was similar to previous reports (specifically, the deposition weight ratio was calculated from ICP results (Table S1, ESI†)). The dark rugby-like shape and the sublayer sheet indicated for the prickly Zn–CuO and GO, respectively. As indicated in Fig. 1B, the pricks from Zn–CuO were well preserved, arising from their successful in situ nanocrystal deposition and nanoparticle growth. Different from previous works, we obtained Zn–CuO@GO solely from the sonochemical reactions other than scratched from the electrode, and prepared hybrid antibacterial nanosheet in other deposition ratios, i.e., 2
1, w/w), which was similar to previous reports (specifically, the deposition weight ratio was calculated from ICP results (Table S1, ESI†)). The dark rugby-like shape and the sublayer sheet indicated for the prickly Zn–CuO and GO, respectively. As indicated in Fig. 1B, the pricks from Zn–CuO were well preserved, arising from their successful in situ nanocrystal deposition and nanoparticle growth. Different from previous works, we obtained Zn–CuO@GO solely from the sonochemical reactions other than scratched from the electrode, and prepared hybrid antibacterial nanosheet in other deposition ratios, i.e., 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (Fig. 1C) and 1
1 (Fig. 1C) and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (Fig. 1D) (w/w) for comparison. Compared with Zn–CuO@GO obtained at 4
1 (Fig. 1D) (w/w) for comparison. Compared with Zn–CuO@GO obtained at 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (w/w), Zn–CuO@GO prepared at 2
1 (w/w), Zn–CuO@GO prepared at 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1
1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (w/w) were largely in agglomerate (Fig. 1C and D). It is possibly due to the comparably over-abundance of GO nanosheets, as there exists strong π–π stacking between GO nanosheets. Also, the zeta potential of the hybrid antibacterial nanosheet was measured in saline (ESI, Fig. S1†), which indicated that it was positively charged after Zn–CuO (ζ > 0) was deposited into GO nanosheets (ζ < 0).
1 (w/w) were largely in agglomerate (Fig. 1C and D). It is possibly due to the comparably over-abundance of GO nanosheets, as there exists strong π–π stacking between GO nanosheets. Also, the zeta potential of the hybrid antibacterial nanosheet was measured in saline (ESI, Fig. S1†), which indicated that it was positively charged after Zn–CuO (ζ > 0) was deposited into GO nanosheets (ζ < 0).
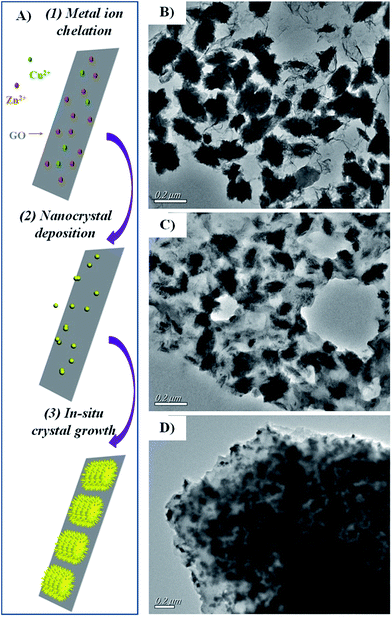 | ||
Fig. 1 (A) Schematic illustration of the formation of Zn–CuO@GO from sonochemical method. TEM image of Zn–CuO@GO (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) (B), Zn–CuO@GO (2 1) (B), Zn–CuO@GO (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) (C), and Zn–CuO@GO (1 1) (C), and Zn–CuO@GO (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) (D). 1) (D). | ||
As indicated in Fig. 2, compared with Zn–CuO (Fig. 2a) and GO (Fig. 2b), the typical XRD peaks of the hybrid antibacterial nanosheet (Fig. 2c–e) exhibited similar pattern with pristine prickly Zn–CuO at 2θ = 32.47°, 35.49°, 38.68°, 48.65°, 58.25°, and 61.45° (Fig. 2a–c), indicating the existence of Zn–CuO in the GO layers.7a The reason why there were no obvious peaks from GO that may bear a typical peak at 12° (Fig. 2b) was possibly due to the coverage of other strong peaks from Zn–CuO NPs, which was also in consistence with previous reports.4g In synergistic with TEM images in Fig. 1B–D, the synthesis of Zn–CuO@GO was demonstrated successful.
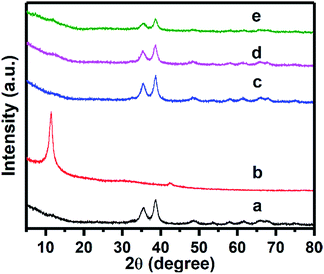 | ||
Fig. 2 XRD pattern of Zn–CuO (a), GO (b), Zn–CuO@GO (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) (c), Zn–CuO@GO (2 1) (c), Zn–CuO@GO (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) (d), and Zn–CuO@GO (1 1) (d), and Zn–CuO@GO (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) (e), respectively. 1) (e), respectively. | ||
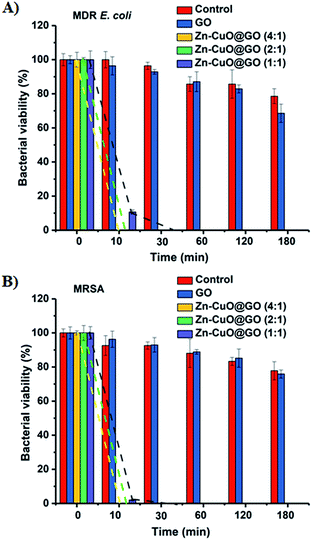
Further, the biocidal activity of Zn–CuO@GO suspension based disinfectant was quantitatively evaluated against a multi-drug resistant E. coli (MDR E. coli) and a methicillin resistant S. aureus (MRSA).7a Briefly, after treatment with Zn–CuO@GO suspensions (at concentrations of 0.1 mg mL−1), the viable bacteria were monitored by counting the number of colony-forming units (CFU) with respect to time.7a As presented in Fig. 3, representative results clearly indicated that Zn–CuO@GO effectively inactivated those multi-drug resistant strains nearly completely after 10 min of treatment (yellow and green dashed lines), except for Zn–CuO@GO (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) (purple dashed lines). Compared with GO which exerted minimal cytotoxicity towards these bacterial strains even after over 3 h of treatment (blue bar), it could be concluded that after integrated with GO sheets, the deposited prickly Zn–CuO could greatly enhance their combinational bacterial killing activity.4f,7a Notably, the antibacterial abilities of Zn–CuO@CuO at different mass ratios were different, as Zn–CuO@GO (4
1) (purple dashed lines). Compared with GO which exerted minimal cytotoxicity towards these bacterial strains even after over 3 h of treatment (blue bar), it could be concluded that after integrated with GO sheets, the deposited prickly Zn–CuO could greatly enhance their combinational bacterial killing activity.4f,7a Notably, the antibacterial abilities of Zn–CuO@CuO at different mass ratios were different, as Zn–CuO@GO (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and Zn–CuO@GO (2
1) and Zn–CuO@GO (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) shared similarity in superiority in combating multi-drug resistant microbes, while Zn–CuO@GO (1
1) shared similarity in superiority in combating multi-drug resistant microbes, while Zn–CuO@GO (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) took more time (∼30 min) for a complete bacterial inhibition. Based on a 10 min time scale after treatment, GO might account for a maximum 3.6% antibacterial activity while Zn–CuO accounted for at least 96.4%. Thus, we concluded that the deposition density of Zn–CuO contributed significantly to the combinational antibacterial activities of Zn–CuO@GO, and Zn–CuO@GO (4
1) took more time (∼30 min) for a complete bacterial inhibition. Based on a 10 min time scale after treatment, GO might account for a maximum 3.6% antibacterial activity while Zn–CuO accounted for at least 96.4%. Thus, we concluded that the deposition density of Zn–CuO contributed significantly to the combinational antibacterial activities of Zn–CuO@GO, and Zn–CuO@GO (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) exhibited the best performance. Additionally, taking consideration of its flexible architecture and good dispersity, which introduced many possibilities for further biomedical application,7b,c the Zn–CuO@GO (4
1) exhibited the best performance. Additionally, taking consideration of its flexible architecture and good dispersity, which introduced many possibilities for further biomedical application,7b,c the Zn–CuO@GO (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) was recognized and chosen as the optimized composite for further evaluation. If not otherwise mentioned, the following Zn–CuO@GO refers to Zn–CuO@GO (4
1) was recognized and chosen as the optimized composite for further evaluation. If not otherwise mentioned, the following Zn–CuO@GO refers to Zn–CuO@GO (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). In comparison with selected works on similar materials (Table S2, ESI†), it was suggested Zn–CuO@GO actively inhibited multi-drug resistant bacteria stains almost completely within 10 min, which rendered themselves promising candidates as a ICU applicable disinfectant.
1). In comparison with selected works on similar materials (Table S2, ESI†), it was suggested Zn–CuO@GO actively inhibited multi-drug resistant bacteria stains almost completely within 10 min, which rendered themselves promising candidates as a ICU applicable disinfectant.
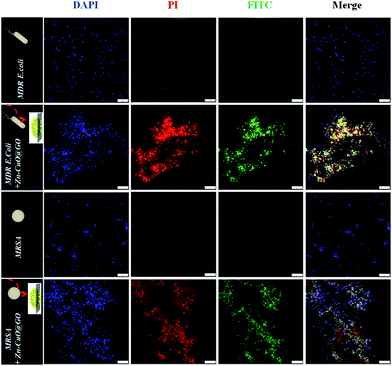
Live/dead dual-staining was conducted to investigate the bacterial viability after exposure to Zn–CuO@GO, and bacterial without any treatment was chosen as control (Fig. 4).4a,7c PI was used specifically to stain membrane perturbed cell with red fluorescence and DAPI was used to label all bacterial nucleus with blue fluorescence. Zn–CuO@GO was also pre-stained with FITC, a green fluorescence utilized widely, to indicate its existence. Notably, bacteria were stained with PI before fixation to indicate the ratio of dead bacteria. It was found that there was scarcely red fluorescence detected for both MDR E. coli and MRSA without treatment, which was in consistence with previous reports. After treated with Zn–CuO@GO at 0.1 mg mL−1, however, it was found that there were severe aggregations observed, and almost all bacteria were dead as they were stained with strong red fluorescent. The merged images where blue, red and green fluorescent almost fitted together further suggested that compared with previous reports,7a the treated bacteria were largely accumulated and adsorbed into the Zn–CuO@GO, where their green fluorescent was in good coincidence with bacteria. This phenomenon demonstrated that after integrated with GO sheets, Zn–CuO could effectively cause the accumulation of bacteria onto their recombinant sheet structure to accelerate bacterial lysis.
To elucidate the bacteria–material interactions after treatment, we first tested whether possible antibacterial mechanisms from zeta potential, metal ions and ROS was dominant. Assuming the fact that if zeta potential was dominant in contributing to the antibacterial activities, Zn–CuO@GO (ζ > 0, Fig. S1, ESI†) should have better activity against Gram-negative MDR E. coli, we careful examined the antibacterial results in Fig. 3 again. The only clue we could detect, however, was that Zn–CuO@GO might be more effective against Gram-positive MRSA (Fig. 3), given that Zn–CuO@GO (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) possessed a better activity towards this strain after 10 min treatment. Such a difference was not detectable for Zn–CuO@GO (4
1) possessed a better activity towards this strain after 10 min treatment. Such a difference was not detectable for Zn–CuO@GO (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 or 2
1 or 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), as they could gave a complete inhibition of both strains within 10 min and exhibited no difference. It was thus indicated that zeta potential might not be a determining factor. Further, we examined the effect from trace amount of metal ion leakage towards bacterial inhibition. As provided in Table S3 (ESI†), however, the time-lapse release of metal ions in Zn–CuO@GO indicated that, there were minimal leakage of metal ions such as Cu2+ and Zn2+ (only 1.2 mmol L−1 of Cu2+ and 0.004 mmol L−1 Zn2+ was detected over 3 h of treatment). The trace amount of Cu2+ and Zn2+ into surrounding environment in turn suggested the leakage of metal ions was hardly a determining factor for Zn–CuO@GO associated bacterial killing. Additionally, the effects of oxidative stress and ROS production were also tested. For ROS production, a ESR spinning technique was applied, which was effective towards the detection of ROS signals. An important part in ROS generation, OH radicals (˙OH), however, was not detectable here under the actual doses of 0.1 mg mL−1 even upon contact with bacteria. From the ESR curves in Fig. S2,† the arrow pointed area, which should be indication of the existence of ROS, gave no significant difference even after treatment. To advance our knowledge of possible oxidative stress introduced by Zn–CuO@GO, we also took an in vitro GSH (glutathione) oxidation assay, of which GSH was an antioxidant in bacteria that could prevent damages to cellular components caused by oxidative stress.7d Through examination of loss of GSH concentration after incubation with Zn–CuO@GO (ESI, Fig. S2†), we found that after 2 h of treatment there were no evident difference in the GSH loss. In fact, loss of GSH was minimal (Zn–CuO@GO (∼23.45%) and GO (∼21.35%)) in comparison with H2O2 (100%), indicating that Zn–CuO@GO was not functioning through oxidative stress mechanism. Taken together, we have concrete reasons to believe that Zn–CuO@GO cause severe bacterial lysis differently.
1), as they could gave a complete inhibition of both strains within 10 min and exhibited no difference. It was thus indicated that zeta potential might not be a determining factor. Further, we examined the effect from trace amount of metal ion leakage towards bacterial inhibition. As provided in Table S3 (ESI†), however, the time-lapse release of metal ions in Zn–CuO@GO indicated that, there were minimal leakage of metal ions such as Cu2+ and Zn2+ (only 1.2 mmol L−1 of Cu2+ and 0.004 mmol L−1 Zn2+ was detected over 3 h of treatment). The trace amount of Cu2+ and Zn2+ into surrounding environment in turn suggested the leakage of metal ions was hardly a determining factor for Zn–CuO@GO associated bacterial killing. Additionally, the effects of oxidative stress and ROS production were also tested. For ROS production, a ESR spinning technique was applied, which was effective towards the detection of ROS signals. An important part in ROS generation, OH radicals (˙OH), however, was not detectable here under the actual doses of 0.1 mg mL−1 even upon contact with bacteria. From the ESR curves in Fig. S2,† the arrow pointed area, which should be indication of the existence of ROS, gave no significant difference even after treatment. To advance our knowledge of possible oxidative stress introduced by Zn–CuO@GO, we also took an in vitro GSH (glutathione) oxidation assay, of which GSH was an antioxidant in bacteria that could prevent damages to cellular components caused by oxidative stress.7d Through examination of loss of GSH concentration after incubation with Zn–CuO@GO (ESI, Fig. S2†), we found that after 2 h of treatment there were no evident difference in the GSH loss. In fact, loss of GSH was minimal (Zn–CuO@GO (∼23.45%) and GO (∼21.35%)) in comparison with H2O2 (100%), indicating that Zn–CuO@GO was not functioning through oxidative stress mechanism. Taken together, we have concrete reasons to believe that Zn–CuO@GO cause severe bacterial lysis differently.
Given its unique nanostructure, we then evaluated physical damage associated antibacterial mechanism, and scanning emission microscope (SEM) was first applied to give detailed elaboration on morphological changes of bacterial after co-incubating with Zn–CuO@GO in saline (Fig. 5). It was found that bacteria without treatment maintained their intact membrane without significant morphological changes (ESI, Fig. S3†). After 30 min extensive contact with Zn–CuO@GO, however, it was observed that Zn–CuO@GO nanosheets were attached tightly onto bacterial membrane, which was similar to Zn–CuO alone. It was shown in Fig. 5A and B, that the bacteria lost their cell membrane integrity, as indicated by their severe shrinkage, which was especially obvious in Fig. 5B. Notably, Zn–CuO@GO may act similarly but in a more effective way taking advantage of Zn–CuO and GO together. Through closer examination of the bacterial surface after treatment, it was found that the surface of Zn–CuO treated bacteria was largely corrugated due to their lost of membrane integrity (ESI, Fig. S4†), while that from Zn–CuO@GO treated bacteria was smoother (Fig. 5a and b), which was attributed to the attachment of Zn–CuO@GO – a layered structure that covered the bacterial membrane. As illustrated in Scheme 1(I and II), in linear with prickly Zn–CuO NPs alone (I), the deposited Zn–CuO NPs on Zn–CuO@GO nanosheets enable them to effectively anchor into the cell membrane (II), and their delicate nano-pricks could then penetrate into the cell walls from diverse positions, causing the leakage of cytoplasm and subsequent bacteria lysis, which was evidenced by the existence of Zn–CuO remaining tightly in the cell wall even after extensive washing (Fig. 5). Synergistically, the layered Zn–CuO@GO could then wrap onto the bacteria surface to deprive of oxygen and nutrient exchange (II), owing to the layered structure of GO nanosheets, which led to a second disastrous inactivation.
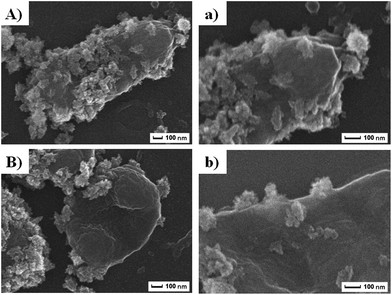 | ||
| Fig. 5 (A and B) were SEM images of MDR E. coli and MRSA after treatment with Zn–CuO@GO, respectively. (a and b) were magnified SEM images revealing detailed cell–material interactions. | ||
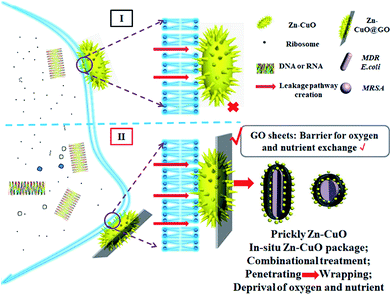 | ||
| Scheme 1 Schematic illustration of the bacterial killing mechanism of Zn–CuO@GO in comparison with prickly Zn–CuO. | ||
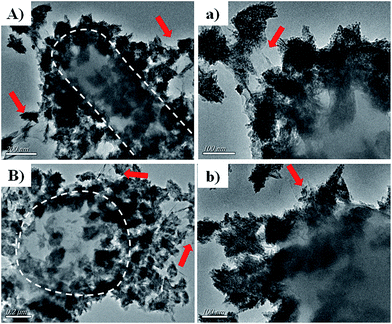
To reveal the vivid mechanism of Zn–CuO@GO towards bacterial-killing as proposed in Scheme 1(II), we investigated their interactions with MDR E. coli and MRSA by TEM. Compared with Zn–CuO (ESI, Fig. S5†), the red arrow in Fig. 6 indicated the existence of GO sheets which was layer structured and well in combination with prickly Zn–CuO. As shown in Fig. 6A and B, the Zn–CuO@GO could bind to bacterial surface extensively due to their strong nonspecific interaction upon contacting, and distributed well throughout the cell wall (corresponding magnified figures were shown in Fig. 6a and b). To make it clearer, we also obtained zoomed figures illustrating the interaction between Zn–CuO@GO and MDR E. coli (Fig. 6), which clearly revealed that Zn–CuO@GO could then anchor and effectively penetrate into cellular membrane, to cause the cytoplasm leakage (cell lysis) subsequently. Especially, the Zn–CuO@GO also form a sublayer wrapping MDR E. coli, as indicated in Fig. 7A. It was also evident in Fig. 7B, that the anchored Zn–CuO could exert their pierce effectively into the cell membrane. To this aspect, we concluded that Zn–CuO@GO could strongly bind to bacterial by wrapping the whole cell wall and dissociate the cell integrity, which also accounted for bacterial accumulation as indicated in Fig. S6 (ESI†), and verified the results obtained in Fig. 4. Thus, it is believable that the Zn–CuO@GO cause bacterial lysis from stepwise penetrating to wrapping upon contacting, namely, through exerting nano-pierce to damage cell walls and subsequently behave to wrap into the cell, which may deprive of oxygen and nutrient exchange and lead to ever-enhanced bacterial inactivation.7e
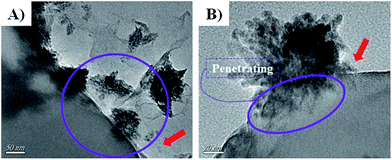 | ||
| Fig. 7 (A and B) were further magnified TEM images uncovering the detailed antibacterial route of Zn–CuO@GO in association with MDR E. coli as an example. | ||
Conclusions
In summary, we described a facial, bottom up and environment-friendly protocol for the scalable fabrication of Zn–CuO@GO nanocomposite for combinational bacterial-killing, especially towards multi-drug resistant strains, such as MDR E. coli and MRSA. Owing to their combinational antibacterial ability, the Zn–CuO@GO could accelerate bacteria lysis and inactivate them nearly completely within 10 min upon contacting. Functioning in a stepwise penetrating and wrapping manner, the deposited prickly Zn–CuO in association with GO layer could penetrate and wrap into bacteria membrane to damage bacteria integrity, induce cytoplasm leakage, deprive of possible oxygen/nutrient exchange, and lead to progressive bacteria lysis. We thus concluded that suspensions containing Zn–CuO@GO nanosheets were promising disinfectant for combating multi-drug resistant bacterial stains from ICU to avoid severe bacterial infections.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The acknowledgements come at the end of an article after the conclusions and before the notes and references.Notes and references
- (a) M. Doidge, A. Allworth, M. Woods, P. Marshall, M. Terry, K. O'brien, H. Goh, N. George, G. Nimmo, M. Schembri and J. Lipman, Infection control and hospital epidemiology, 2010, 31, 418–420 CrossRef PubMed; (b) W. Guo, K. Shan, B. Xu and J. Li, Pathog. Global Health, 2015, 109, 184–192 CrossRef CAS PubMed; (c) W. Rutala, M. Gergen, E. Sickbert-Bennett, D. Williams and D. Weber, Am. J. Infect. Control, 2014, 42, 426–428 CrossRef CAS PubMed; (d) M. Kempf, F. Chen, R. Kern and K. Venkateswaran, Astrobiology, 2005, 5, 391–405 CrossRef CAS PubMed; (e) G. Campos, S. Souza, T. Lobão, D. Da Silva, D. Sousa, P. Oliveira, V. Santos, A. Amorim, S. Farias, M. Cruz and R. Yatsuda, New Microbiol., 2012, 35, 183–190 CAS.
- (a) T. Kalaiyarasan, V. Bharti and O. Chaurasiaa, RSC Adv., 2017, 7, 51130–51141 RSC; (b) A. Chanda, S. Gupta, M. Vasundhara, S. Joshi, G. Mutta and J. Singh, RSC Adv., 2017, 7, 50527–50536 RSC; (c) M. Moritz and M. Geszke-Moritz, Chem. Eng. J., 2013, 228, 596–613 CrossRef CAS; (d) Y. Li, W. Zhang, J. Niu and Y. Chen, ACS Nano, 2012, 6, 5164–5173 CrossRef CAS PubMed.
- (a) X. Xu, Z. Zhang, L. Qiu, J. Zhuang, L. Zhang, H. Wang and Z. Hu, Nat. Nanotechnol., 2016, 11, 930 CrossRef CAS PubMed; (b) Y. Tu, M. Lv, P. Xiu, T. Huynh, M. Zhang, M. Castelli and R. Zhou, Nat. Nanotechnol., 2013, 8, 594 CrossRef CAS PubMed; (c) C. He, C. Cheng, S. Nie, L. Wang, C. Nie, S. Sun and C. Zhao, J. Mater. Chem. B, 2016, 4, 6143 RSC; (d) W. Hu, C. Peng, W. Luo, M. Lv, X. Li, D. Li and C. Fan, ACS Nano, 2010, 4, 4317 CrossRef CAS PubMed; (e) F. Perreault, A. De Faria, S. Nejati and M. Elimelech, ACS Nano, 2015, 9, 7226 CrossRef CAS PubMed; (f) V. Pham, V. Truong, M. Quinn, S. Notley, Y. Guo, V. Baulin and E. Ivanova, ACS Nano, 2015, 9, 8458 CrossRef CAS PubMed.
- (a) C. Courtney, S. Goodman, J. McDaniel, N. Madinger, A. Chatterjee and P. Nagpal, Nat. Mater., 2016, 15, 529 CrossRef CAS PubMed; (b) A. áde Leon, Chem. Commun., 2015, 51, 2886 RSC; (c) K. O'Connell, J. Hodgkinson, H. Sore, M. Welch, G. Salmond and D. Spring, Angew. Chem., Int. Ed., 2013, 52, 10706 CrossRef PubMed; (d) M. Wu, A. Deokar, J. Liao, P. Shih and Y. Ling, ACS Nano, 2013, 7, 1281 CrossRef CAS PubMed; (e) Z. Qi, P. Bharate, C. Lai, B. Ziem, C. Böttcher, A. Schulz and R. Haag, Nano Lett., 2015, 15, 6051 CrossRef CAS PubMed; (f) J. Tang, Q. Chen, L. Xu, S. Zhang, L. Feng, L. Cheng and R. Peng, ACS Appl. Mater. Interfaces, 2013, 5, 3867 CrossRef CAS PubMed; (g) Y. Zhou, J. Yang, T. He, H. Shi, X. Cheng and Y. Lu, Small, 2013, 9, 3445 CrossRef CAS PubMed.
- (a) B. Atiyeh, M. Costagliola, S. Hayek and S. Dibo, Burns, 2017, 33, 139 CrossRef PubMed; (b) W. Jung, H. Koo, K. Kim, S. Shin, S. Kim and Y. Park, Appl. Environ. Microbiol., 2008, 74, 2171 CrossRef CAS PubMed; (c) H. Su, S. Lin, J. Wei, I. Pao, S. H. Chiao, C. Huang and J. Lin, PLoS One, 2011, 6, e21125 CAS; (d) T. Kavitha, A. Gopalan, K. Lee and S. Park, Carbon, 2012, 50, 2994 CrossRef CAS; (e) E. Zanni, E. Bruni, C. Chandraiahgari, G. De Bellis, M. Santangelo, M. Leone, A. Bregnocchi, P. Mancini, M. Sarto and D. Uccelletti, J. Nanobiotechnol., 2017, 15, 57 CrossRef PubMed.
- (a) R. Wu, Y. Ma, J. Pan, S. Lee, J. Liu, H. Zhu, R. Gu, K. Shea and G. Pan, Biosens. Bioelectron., 2017, 101, 52 CrossRef PubMed; (b) E. Malka, I. Perelshtein, A. Lipovsky, Y. Shalom, L. Naparstek, N. Perkas, T. Patick, R. Lubart, Y. Nitzan, E. Banin and A. Gedanken, Small, 2013, 9, 4069 CrossRef CAS PubMed; (c) M. Eshed, J. Lellouche, A. Gedanken and E. Banin, Adv. Funct. Mater., 2014, 24, 1382 CrossRef CAS.
- (a) R. Wu, H. Zhang, J. Pan, H. Zhu, Y. Ma, W. Cui and G. Pan, Adv. Mater. Interfaces, 2016, 3 DOI:10.1002/admi.201600472; (b) M. Hoop, Y. Shen, X. Chen, F. Mushtaq, L. Iuliano, M. Sakar and S. Pané, Adv. Funct. Mater., 2016, 26, 1063 CrossRef CAS; (c) J. Wang, J. Li, S. Qian, G. Guo, Q. Wang, J. Tang and P. K. Chu, ACS Appl. Mater. Interfaces, 2016, 8, 11162 CrossRef CAS PubMed; (d) E. Piskounova, M. Agathocleous, M. Murphy, Z. Hu, S. Huddlestun, Z. Zhao, A. Leitch and T. Johnson, Nature, 2015, 527, 186 CrossRef CAS PubMed; (e) C. Nie, C. Cheng, L. Ma, J. Deng and C. Zhao, Langmuir, 2016, 5955 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra12148b |
| This journal is © The Royal Society of Chemistry 2018 |