Sulfonate-functionalized tetraphenylethylenes for selective detection and wash-free imaging of Gram-positive bacteria (Staphylococcus aureus)†
Viraj G.
Naik
 a,
Sharanabasava D.
Hiremath
a,
Sharanabasava D.
Hiremath
 a,
Avijit
Das
a,
Avijit
Das
 b,
Dishant
Banwari
b,
Dishant
Banwari
 a,
Ram U.
Gawas
a,
Ram U.
Gawas
 a,
Malabika
Biswas
a,
Malabika
Biswas
 b,
Mainak
Banerjee
b,
Mainak
Banerjee
 *a and
Amrita
Chatterjee
*a and
Amrita
Chatterjee
 *a
*a
aDepartment of Chemistry, BITS Pilani, Goa Campus, NH 17 B Bypass Road, Zuarinagar, Goa 403726, India. E-mail: amrita@goa.bits-pilani.ac.in; mainak@goa.bits-pilani.ac.in
bDepartment of Biological Sciences, BITS Pilani, Goa Campus, NH 17B Bypass Road, Zuarinagar, Goa 403726, India
First published on 17th September 2018
Abstract
Two sulfonate functionalized tetraphenylethylene (TPE) derivatives were synthesized and used as probes for the detection and imaging of Gram-positive bacteria (e.g. Staphylococcus aureus). The strategy is based on AIE-based turn-on fluorescence resulting from simple electrostatic interactions between the negatively charged probe and positively charged functionalities of lipoteichoic acid (LTA) which is a major component of the outer cell wall of Gram-positive bacteria. TPE-mono-sulfonate (1) and TPE-di-sulfonate (2) both exhibited fast detection ability but the latter, owing to its high-water solubility and higher sensitivity, is the preferred probe for fluorimetric studies and wash-free bacterial imaging. Probe 2 efficiently interacts with the cell-wall of Gram-positive bacteria (S. aureus or B. subtilis) and the bacteria fluoresce blue, whereas, it showed negligible response to both Gram-negative and acid-fast bacteria. Up to 43 fold increase in fluorescence intensity was observed for probe 2 within the bacterial concentration of 0–12 × 108 CFU mL−1. Interestingly, probe 2 showed significant antibacterial activity against S. aureus with an MIC of 23.4 μg mL−1. This simple strategy is advantageous in many ways such as cost-effectiveness and easy synthesis, high water solubility of the probe, fast response, etc. and can be applicable for real sample analysis which is demonstrated by artificially contaminating apple juice with S. aureus.
Introduction
Staphylococcus aureus is a widely known Gram-positive pathogen closely related to human health.1,2 It is marked as one of the most common carriers of hospital-acquired infections, causing minor skin infections to life-threatening diseases such as pneumonia,3 meningitis,4 endocarditis,5 and septicaemia.5 Additionally, most of the food poisoning cases caused due to unhygienic cooking conditions result from contamination by S. aureus.6 Among various Staphylococcus species, S. aureus poses an enormous threat to human health because it shows strong resistance to antibiotics. Other Gram-positive bacteria (such as Bacillus species) sometimes cause serious infections to humans.7 Hence, rapid detection and quantification of these bacteria are essential, making it an active field of research in environmental hygiene, clinical diagnosis and food safety.8 The conventional bacterial culture and metabolic test is the standard method for bacterial detection due to its high sensitivity and reliability.9–11 However, it is a labor-intensive and time-consuming process and requires sophisticated laboratory equipment.12 Meanwhile, other techniques including polymerase-chain-reaction (PCR),13 quartz-crystal-microbalance (QCM),14 surface plasmon resonance (SPR),15 electrochemiluminescence,16 and flow cytometry (FCM)17 have also been exploited for the detection of bacteria but suffer from low sensitivity, and involve complicated and expensive procedures.In recent years, fluorimetry has become the most common technique in sensory, imaging, and screening applications because of its high sensitivity and selectivity, non-invasiveness, easy operation, rapid response, etc.18 In this context, a new class of fluorogenic molecules with aggregation-induced emission (AIE) properties has drawn the attention of researchers worldwide.19–22 These molecules can overcome aggregation-caused quenching (ACQ) in traditional fluorophores through emission owing to the restriction of intermolecular rotations (RIR) and proscription of energy dissipation upon aggregation.21 AIE-active molecules exhibit high quantum efficiency, good biocompatibility and photo-stability, and therefore, find a multitude of applications in diverse fields such as chemo-/bio-sensors, electroluminescent materials, cell imaging, optical devices, etc.20 Among the reported AIE-gens, tetraphenylethylene (TPE) is one of the most studied luminophores, extensively used by our group23 and others,24 due to its easy synthesis and simple functionalization strategies for the construction of novel sensing systems.
Bacterial imaging has been reported using various fluorescent molecular probes.25–29 For example, Lai et al. used aptamer modified gold nanoparticles for the selective detection of S. aureus.29 In this context, the AIE-gens have also been used for bacterial imaging in the recent past.30–35 Notably, Feng et al. reported a light-up probe based on vancomycin tagged TPE, named AIE-2Van, for the selective recognition, naked-eye detection and image-guided photodynamic killing of Gram-positive bacteria.30 Zhao et al. used a TPE derived cationic amphiphilic molecule with ammonium groups for imaging and killing of both Gram-positive and Gram-negative bacteria by electrostatic interactions.31 However, these methods are based on the use of bio-conjugates to achieve selectivity or involve intricate multi-step synthesis of the probe. These facts encouraged us to devise a simple and cost-effective strategy for bacterial detection and wash-free imaging. Encouraged by the successful application of sulfonate functionalized TPE in the electrostatic interaction based detection of various analytes by others36 and our group,23b we envisaged to capitalize on this simple method in developing detection tools for Gram-positive bacteria. As is well-known, lipoteichoic acid (LTA), an amphiphilic molecule with NH3+ groups on its backbone, is a major constituent of the outer cell wall of Gram-positive bacteria.37 We presumed that a negatively charged probe could interact with the NH3+ groups of LTA to form a definite array leading to an AIE-based turn-on fluorescence response. As a proof-of-concept, S. aureus was selected as the primary species of Gram-positive bacteria for detection, imaging and antimicrobial studies as it is one of the most harmful pathogenic bacteria.
Results and discussion
We successfully synthesized TPE with one and two sulfonate functionalities and labelled them as probe 1 and 2, respectively, by adopting a two-step procedure in good overall yields (see ESI† for details). First, TPE-mono-OH (3) and TPE-di-OH (4) were synthesized by McMurray coupling between benzophenone and 4-hydroxy benzophenone, and 4-hydroxy benzophenone, respectively23d and were then converted to probes 1 and 2, respectively, by treating with 1,4-butane sultone in the presence of a suitable base. The probes were characterized by 1H NMR, 13C NMR, ESI-MS and HRMS analysis. The spectra for probe 1 matched the values reported by us.23b The spectra of probe 2 were also in good agreement with the expected values. Probes 1 and 2 were applied for the detection of bacteria in bulk solutions.Probe 1 was found to be sparingly soluble in water and its AIE characteristic was determined by carrying out fluorescence studies in variable ratio water–THF solvent systems (Fig. S1, ESI†). Accordingly, 94% water–THF was considered for bacterial sensing studies in which probe 1 showed negligible fluorescence. On increasing the concentration of S. aureus, the fluorescence intensities increased significantly (Fig. S2, ESI†). This increase in fluorescence is associated with the AIE effect presumably caused by electrostatic interactions between the negatively charged probe and the positively charged moieties present on the cell wall of S. aureus. For the fluorimetric study involving probe 1 use of some amount of organic solvent is inevitable for solubility and upon considering their toxic effect on biological systems, incorporation of one more sulfonate functionality was envisaged to achieve a probe that is more effective and completely soluble in water.36
The solubility profile of probe 2 was studied and it was observed that up to 20 mg of the probe was completely soluble in 1 mL of water, while a 1 mM solution showed minimum background fluorescence (Fig. S3, ESI†). The ideal probe concentration for bacterial sensing studies was considered as 30 μM in water and the fluorescence output of this aqueous solution of probe 2 was negligible.
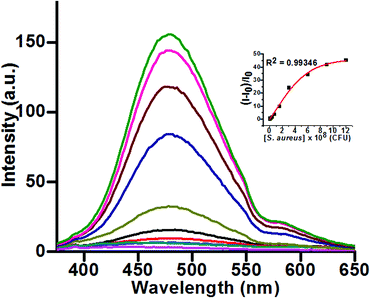
However, incubation of probe 2 with increasing bacterial concentrations led to a gradual increment in fluorescence intensity (Fig. 1). A plot of fluorescence intensity vs. concentration of S. aureus led to a sigmoidal curve with regression coefficient R2 = 0.9934 within the range of 1.19 × 106 to 12 × 108 CFU mL−1 (inset, Fig. 1), and the maximum enhancement in fluorescence intensity was found to be 43 times at the bacterial concentration of 12 × 108 CFU mL−1 (Fig. 1). The detection limit was estimated to be approx. 1.19 × 106 CFU mL−1 (Fig. S4, ESI†). As compared to probe 1 the water-soluble probe 2 showed high sensitivity towards S. aureus and based on this merit further studies were carried out using probe 2 only in an aqueous phase devoid of organic solvents.
The fluorescence output against increasing number of bacterial colonies (viz. S. aureus) showed the same trend of increment as compared to OD@600 values obtained using the standard UV-vis method (Fig. S6, ESI†). This indicates that the present fluorimetric technique can also be used in quantifying bacterial colonies (for Gram-positive bacteria) with equal efficacy. Notably, the more sensitive fluorimetric technique allows the detection and quantification of bacteria at a much lower concentration whereas the UV method fails to produce any stable data because of its poor sensitivity.
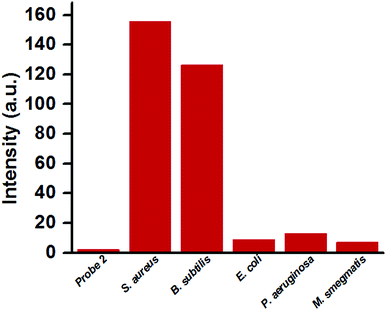
To examine whether the probe could selectively detect Gram-positive bacteria (S. aureus), similar fluorimetric studies were carried out on other bacterial species viz. Gram-positive bacteria, B. subtilis, Gram-negative bacteria E. coli and P. aeruginosa, and acid-fast M. smegmatis keeping the sensing condition the same. As expected, an aqueous solution of B. subtilis showed similar fluorimetric response like S. aureus in the presence of probe 2 (Fig. S7, ESI†) and the LOD was estimated as 2.34 × 106 CFU mL−1 (Fig. S8, ESI†). On the other hand, the fluorescence intensities of the aqueous solutions containing P. aeruginosa, E. coli and M. smegmatis remained negligible (Fig. 2). These experiments unambiguously established the utility of probe 2 for the detection of Gram-positive bacteria.
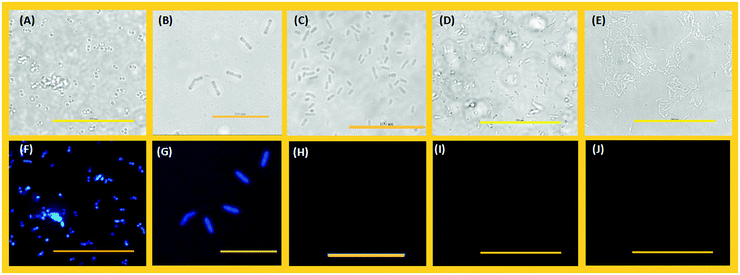
The high water solubility and negligible fluorescence of probe 2 in aqueous solution make it an efficient candidate for wash-free imaging31 of Gram-positive bacteria (viz. S. aureus). The imaging studies were carried out using an inverted microscope with fluorescence attachment. Both phase contrast and fluorescence images of S. aureus, B. subtilis, E. coli, P. aeruginosa and M. smegmatis were obtained by incubating probe 2 with the bacterial cultures for 15 min before capturing the images. As shown in Fig. 3, probe 2 can efficiently interact with the cell-walls of both S. aureus and B. subtilis, and the bacteria fluoresce blue with minimum to negligible background fluorescence. Therefore, there is no requirement to wash off the media before imaging, which makes the imaging process simple with minimum loss of bacteria and this can increase the accuracy of quantification. On the contrary, other bacteria, namely E. coli, P. aeruginosa and M. smegmatis, do not show any fluorescence on the cell surface. This study once again indicates that probe 2 can be used for selective detection and imaging of Gram-positive bacteria over other classes of bacteria.
Next, efforts were made to establish the pre-conceived mechanism for bacterial detection. It was expected that the negatively charged sulfonate groups of the probe would interact with the accessible NH3+ groups of LTA situated on the cell-wall of any Gram-positive bacteria to form a definite array of TPE molecules resulting in a turn-on type fluorescence response by the AIE effect. To prove the proposed phenomenon, a sensing study was carried out with LTA (1 mg mL−1) in water. The sensing conditions were kept the same as before. Fig. S10A (ESI†) depicts the fluorescence emission spectra of probe 2 after incubation with increasing concentrations of LTA. A steady rise in fluorescence intensity was observed upon incremental addition of LTA to the probe solution to obtain a sigmoidal curve with an R2 value of 0.9938 (Fig. S10B, ESI†). However, the maximum fluorescence output was relatively lower than that of the solution of Gram-positive bacteria. This may be due to more randomness of the small and flexible LTA molecules as compared to the rigid cell-wall of bacteria, which does not allow the TPE molecules to interact with them with the same efficiency to form a highly ordered array; a necessary condition for the AIE-effect. However, this study proves that the probe molecules can successfully interact with LTA to emit fluorescence from the solution. Based on the above observations the following mechanism may be proposed: (1) the positively charged ammonium groups of the LTA backbone protrude outward from the thick peptidoglycan outer layer of the cell wall of a Gram-positive bacterium (viz. S. aureus) rendering them accessible for interaction with the negatively charged species, whereas, the phosphate groups are deep seated and prevented from any repulsive interactions; (2) at least one of the negatively charged sulfonate groups of probe 2 could undergo strong electrostatic interaction with the ammonium groups of LTA leading to the stacking of several TPE moieties in a definite array; (3) the TPE moieties on the surface of the bacteria start emitting strong blue fluorescence attributed to their AIE-characteristics (Scheme 1). The proposed mechanism is in concordance with the high selectivity shown by the probe towards Gram-positive bacteria. Acid-fast M. smegmatis has mycolic acid, a long chain aliphatic acid with no other functionality on the cell wall and the cell surface of Gram-negative bacteria (E. coli or P. aeruginosa) is constituted by lipopolysaccharide (LPS) which has negatively charged phosphate groups in the backbone. The absence of suitable positively charged moieties on the cell surface of these classes of bacteria fails to attract negatively charged probes resulting in no fluorescence output.
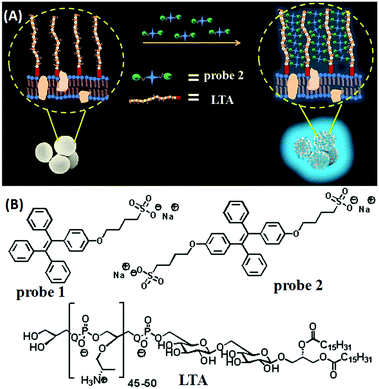 | ||
| Scheme 1 (A) A schematic representation of the sensing mechanism for the detection of Gram-positive bacteria (e.g. S. aureus); (B) structures of probe 1, probe 2 and LTA. | ||
The applicability of this sensing system was tested by real sample analysis. For this, commercially available apple juice was contaminated with S. aureus and probe 2 was applied to obtain the required fluorescence response to match it with the standard plot. Fig. 4 shows the fluorescence response of probe 2 (30 μM) in apple juice contaminated with a measured amount of S. aureus (1.5 × 108 CFU mL−1) upon incubation for 16 h at 37 °C. The fluorescence output was plotted on the standard curve and the data matched closely with the concentration of the contaminant demonstrating the potential applicability of the method in the detection of Gram-positive bacteria in real samples.
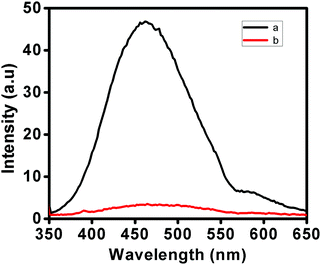 | ||
| Fig. 4 Fluorimetric response of probe 2 (30 μM) upon addition of apple juice spiked with a known concentration of S. aureus, a: probe with apple juice and b: only probe [λex 345 nm; λem 474 nm]. | ||
Antimicrobial study
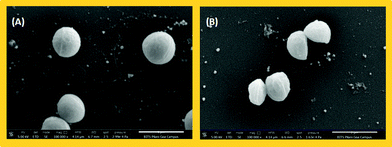
As a common phenomenon, bacteria develop resistance to existing antibiotics over a period of time, so the development of new antibacterial agents is essential.38 The large turn-on fluorescence enhancement upon interaction with Gram-positive bacteria and the simple imaging procedure encouraged us to carry out initial screening of the antibacterial activities of the probes against S. aureus. The spot plate technique was applied for probes 1 and 2 for the initial screening of inhibition zones against S. aureus (Fig. S11, ESI†). Although no significant inhibition zone developed for probe 1, the other probe 2 showed strong inhibition zones on addition of 5 μL and 10 μL of both probes (1 mM) to the bacterial colonies and incubating them for 16 h at 37 °C. Based on the early results, the MIC (minimum inhibitory concentration) was determined for probe 2 by the broth microdilution method.39 The MIC of probe 2 was found to be 23.4 μg mL−1, which is sufficiently high making it a suitable agent for killing Gram-positive bacteria. The antibacterial activity of probe 2 was captured in SEM images (Fig. 5). The images were taken before and after the treatment of probe 2 with bacterial culture of S. aureus. As seen in Fig. 5, cell division of healthy bacteria (Fig. 5A) was inhibited in the presence of probe 2 as the bacteria crumpled during cell division (Fig. 5B). Furthermore, a bacterial growth inhibition study of probe 2 was carried out on S. aureus using kanamycin as a control. 10 fold of the MIC was used for the study for both probe 2 and kanamycin and the optical density (OD@600) over time was measured (Fig. S13, ESI†). The optical density showed a gradual inhibition of growth and the growth kinetics matched well with well-known antibacterial agent kanamycin, making probe 2 an efficient antibacterial agent for S. aureus. It requires further study to get a clear insight about the antibacterial ability of probe 2.Experimental
Materials and methods
4-Hydroxybenzophenone and benzophenone were purchased from Sigma Aldrich (India). TiCl4 was purchased from Spectrochem Pvt. Ltd Mumbai (India). All other common chemicals and solvents of AR grade were obtained from different commercial suppliers and were used without further purification. THF was dried over sodium and freshly distilled before use. The reactions were monitored using thin layer chromatography (TLC) carried out on 0.25 mm silica gel plates (60F-254) under UV light (254 or 365 nm) for visualization. Stock solutions of probe 1 and probe 2 (1 mM) were prepared in THF and water, respectively. Then, the stock solutions of probe 1 and 2 were diluted to different concentrations with deionized water as per the requirements of the experiments. Deionized water (Milli-Q, 18 MΩ) was purged with N2 for 15 min prior to use and as per requirements for dilution purposes.Synthesis of probe 1 and 2
Probe 1 and 2 were synthesized following previously reported procedures (for details see ESI†).23b,36Culture conditions for bacteria
Five cultures of different classes of bacteria, namely S. aureus, B. subtilis, E. coli, P. aeruginosa and M. smegmatis were selected for the study. These cultures were routinely grown on Tryptic Soy Broth (Becton-Dickinson and Company, USA) for S. aureus and B. subtilis, Luria Bertani broth (Hi-Media, India) for E. coli and P. aeruginosa, and Middlebrook 7H9 broth base (Hi-Media, India) for M. smegmatis. Agar was added as per requirement for routine culture maintenance. At OD600 ∼0.5, the grown cells were harvested. The OD of the bacterial cultures was measured using a UV-visible spectrophotometer and the colony-forming units (CFU) were obtained by McFarland standards (for details see ESI†).40Procedure for fluorescence measurement
1 mM stock solutions of probe 1 and probe 2 were prepared using THF and water, respectively. The grown bacterial cells were harvested by centrifugation at 4 °C, washed twice with sterilized phosphate-buffered saline (1× PBS, pH 7.4) solution and used for fluorescence measurements. The solutions prepared were incubated for 15 min before recording the fluorescence spectrum. The excitation wavelength was set at 345 nm and emission spectra were recorded in the range of 346 to 650 nm. All the experiments were performed at room temperature.Method for growth kinetics study for S. aureus
S. aureus RN4220 strain was allowed to grow in Tryptic Soy Broth (TSB) overnight at 37 °C with shaking at 120 rpm. This culture was used for inoculating (1% inoculum) three conical flasks having 100 mL TSB (for control, kanamycin and probe 2) each to an OD600 of ∼0.05. Each of the three cultures was allowed to grow at 37 °C with shaking (at 120 rpm) and the OD600 of the cultures was measured after every 30 min. At 2 h, one set of cultures was treated with kanamycin (10× concentration of MIC i.e. 19.5 μg mL−1) and another set was treated with probe 2 (10× concentration of MIC i.e. 23.4 μg mL−1) and the OD600 of the cultures was measured every 30 min for 8 h. The third set served as a control.Scanning electron microscopy (SEM)
The S. aureus cells were also employed for SEM. Probe 2 treated cells as well as the untreated cells were harvested after 4 h. Cells were then washed twice by centrifugation using 1× PBS (pH 7.4) for 10 min at 7500g. The probe 2 treated and untreated cells were further processed for SEM.41 PBS washed cells were pre-fixed with 3% glutaraldehyde and treated with 1% OsO4 at 4 °C. Next the cells were dehydrated using a series of ethanol solutions (10%, 30%, 50%, 70% and 90%, 5 min each) and finally transferred into 100% ethanol. Complete dehydration was carried out using a CPD chamber (Leica EM CPD300). The specimens were coated with a gold–palladium mixture with 5 nm thickness using Leica EM ACE200. The SEM images of the cells were obtained using a FEI Quanta FEG 250 scanning electron microscope.Conclusions
In this work we have demonstrated that simple electrostatic interactions can be successfully utilized in fast and selective detection and imaging of Gram-positive bacteria (such as S. aureus) using adequately functionalized AIE-gens. Mono- and di-sulfonate derivatives of TPE (1 and 2) were synthesized and successfully used as probes to detect the presence of Gram-positive bacteria by virtue of the AIE effect. The sensing phenomenon is based on electrostatic interactions between the negatively charged probes and NH3+ groups on LTA, which is a major constituent of the outer cell-wall of Gram-positive bacteria. Up to 43 fold increase in fluorescence intensity was observed for probe 2 within a bacterial concentration of 0–12 × 108 CFU mL−1 in the case of S. aureus. TPE-di-sulfonate (2) was preferred over mono-sulfonate (1) owing to its high-water solubility and much higher sensitivity to S. aureus. Probe 2 was successfully utilized in the imaging of Gram-positive bacteria. It was observed that probe 2 efficiently interacts with the cell-wall of S. aureus and B. subtilis, and the bacteria fluoresce blue, whereas, other classes of bacteria such as Gram-negative E. coli and P. aeruginosa, and acid-fast M. smegmatis do not show any fluorescence on the cell surface. The utility of probe 2 in real sample analysis was demonstrated using artificially contaminated apple juice. Interestingly, probe 2 showed significant antibacterial activity against S. aureus with an MIC of 23.4 μg mL−1. The merits of the current probe include simple but effective strategy, cost-effectiveness and easy synthesis, high water solubility to ensure that fluorimetric studies can be carried out in pure water and a fast response and it surely supersedes existing probes in many ways.Conflicts of interest
The authors declare no personal, financial or organisational conflict of interests.Acknowledgements
A. C. thanks DST-SERB (India) (project no. EMR/2016/001416) for financial support. M. B. is also thankful to DST-SERB (India) (project no. EMR/2016/002253) for the research fund. V. G. N. is indebted to DST-SERB (India) for the research fellowship. R. U. G. thanks BITS, Pilani for providing an institute fellowship for research. A. D. gratefully acknowledges CSIR for the SRFship. The authors also acknowledge the Central Research Facility, BITS Pilani, K. K. Birla Goa Campus for ESI-MS and SEM images and the HRMS facility at BITS Pilani, Pilani campus funded by DST-FIST.Notes and references
- A. Ogston, Rev. Infect. Dis., 1984, 6, 122 CrossRef.
- N. Woodford and D. M. Livermore, J. Infect., 2009, 59, S4 CrossRef PubMed.
- G. Lina, Y. Piémont, F. Godail-Gamot, M. Bes, M. O. Peter, V. Gauduchon, F. Vandenesch and J. Etienne, Clin. Infect. Dis., 1999, 29, 1128 CrossRef CAS PubMed.
- J. J. Gordon, D. H. Harter and J. P. Phir, Am. J. Med., 1985, 78, 965 CrossRef CAS PubMed.
- V. G. Fowler Jr., et al. , N. Engl. J. Med., 2006, 355, 653 CrossRef PubMed.
- CDC Staphylococcal Food Poisoning Centers for Disease Control and Prevention, Atlanta, 2010. http://www.cdc.gov/nczved/divisions/dfbmd/diseases/staphylococcal/ Retrieved 4th July 2014.
- (a) N. Woodford and D. M. Livermore, J. Infect., 2009, 59(S1), S4 CrossRef PubMed; (b) R. Silman, S. Rehm and D. M. Shlaes, Medicine, 1987, 66, 218 CrossRef; (c) H. E. Pearson, Am. J. Clin. Pathol., 1970, 53, 506 CrossRef CAS PubMed.
- (a) S. Chen, Q. Li, X. Wang, Y.-W. Yang and H. Gao, J. Mater. Chem. B, 2018, 6, 5198 RSC; (b) S. Yang, H. Ouyang, X. Su, H. Gao, W. Kong, M. Wang, Q. Shu and Z. Fu, Biosens. Bioelectron., 2016, 78, 174 CrossRef CAS PubMed; (c) S. Sandhu, J. A. Schouten, J. Thompson, M. Davis and T. D. H. Bugg, Analyst, 2012, 137, 1130 RSC.
- P. Breeuwer and T. Abee, Int. J. Food Microbiol., 2000, 55, 193 CrossRef CAS PubMed.
- A. S. Kaprelyants and D. B. Kell, J. Appl. Bacteriol., 1992, 72, 410 CrossRef CAS.
- T. A. Camesano, M. J. Natan and B. E. Logan, Langmuir, 2000, 16, 4563 CrossRef CAS.
- O. Lazcka, F. J. D. Campo and F. X. Muñoz, Biosens. Bioelectron., 2007, 22, 1205 CrossRef CAS PubMed.
- H. Sakai, G. Procop, N. Kobayashi, D. Togawa, D. Wilson, L. Borden, V. Krebs and T. Bauer, J. Clin. Microbiol., 2004, 42, 5739 CrossRef CAS PubMed.
- D. Le, F. J. He, T. J. Jiang, L. Nie and S. Yao, J. Microbiol. Methods, 1995, 23, 229 CrossRef.
- A. N. Naimushin, S. D. Soelberg, D. K. Nguyen, L. Dunlap, D. Bartholomew, J. Elkind, J. Melendez and C. E. Furlong, Biosens. Bioelectron., 2002, 17, 573 CrossRef CAS PubMed.
- M. Zhu, W. Liu, H. Liu, Y. Liao, J. Wei, X. Zhou and D. Xing, ACS Appl. Mater. Interfaces, 2015, 7, 12873 CrossRef CAS PubMed.
- N. K. Shrestha, N. M. Scalera, D. A. Wilson, B. Brehm-Stecher and G. W. Procop, J. Clin. Microbiol., 2011, 49, 3383 CrossRef PubMed.
- (a) J. Chan, S. C. Dodani and C. J. Chang, Nat. Chem., 2012, 4, 973 CrossRef CAS PubMed; (b) M. J. Culzoni, A. M. De la Pena, A. Machuca, H. C. Goicoechea and R. Babiano, Anal. Methods, 2013, 5, 30 RSC; (c) H. N. Kim, W. X. Ren, J. S. Kim and J. Yoon, Chem. Soc. Rev., 2012, 41, 3210 RSC; (d) M. E. Jun, B. Roy and K. H. Ahn, Chem. Commun., 2011, 47, 7583 RSC; (e) M. Beija, C. A. M. Afonso and J. M. G. Martinho, Chem. Soc. Rev., 2009, 38, 2410 RSC.
- B. Z. Tang and A. Qin, Aggregation-Induced Emission: Fundamentals, Wiley, New York, 2013 Search PubMed.
- (a) J. Mei, N. L. C. Leung, R. T. K. Kwok, J. W. Y. Lam and B. Z. Tang, Chem. Rev., 2015, 115, 11718 CrossRef CAS PubMed; (b) Z. Yang, Z. Chi, Z. Mao, Y. Zhang, S. Liu, J. Zhao, M. P. Aldred and Z. Chi, Mater. Chem. Front., 2018, 2, 861 RSC; (c) Z. He, C. Ke and B. Z. Tang, ACS Omega, 2018, 3, 3267 CrossRef CAS; (d) D. Ding, K. Li, B. Liu and B. Z. Tang, Acc. Chem. Res., 2013, 46, 2441 CrossRef CAS PubMed; (e) J. Mei, Y. Huang and H. Tian, ACS Appl. Mater. Interfaces, 2018, 10, 12217 CrossRef CAS PubMed; (f) M. Gao and B. Z. Tang, ACS Sens., 2017, 2, 1382 CrossRef CAS PubMed; (g) C. Zhan, X. You, G. Zhang and D. Zhang, Chem. Rec., 2016, 16, 2142 CrossRef CAS PubMed; (h) G. Feng, R. T. K. Kwok, B. Z. Tang and B. Liu, Appl. Phys. Rev., 2017, 4, 021307 CrossRef; (i) R. T. Kwok, C. W. Leung, J. W. Lam and B. Z. Tang, Chem. Soc. Rev., 2015, 44, 4228 RSC; (j) S. Chen, H. Wang, Y. Hong and B. Z. Tang, Mater. Horiz., 2016, 3, 283 RSC; (k) D. D. La, S. V. Bhosale, L. A. Jones and S. V. Bhosale, ACS Appl. Mater. Interfaces, 2018, 10, 12189 CrossRef CAS PubMed.
- N. L. C. Leung, N. Xie, W. Yuan, Y. Liu, Q. Wu, Q. Peng, Q. Miao, J. W. Y. Lam and B. Z. Tang, Chem. – Eur. J., 2014, 20, 15349 CrossRef CAS PubMed.
- (a) K. Ma, F. Zhang, N. Sayyadi, W. Chen, A. G. Anwer, A. Care, B. Xu, W. Tian, E. M. Goldys and G. Liu, ACS Sens., 2018, 3, 320 CrossRef CAS PubMed; (b) M. Fang, S. Xia, J. Bi, T. P. Wigstrom, L. Valenzano, J. Wang, W. Mazi, M. Tanasova, F.-T. Luo and H. Liu, Chem. Commun., 2018, 54, 1133 RSC; (c) M. J. Chang and M. H. Lee, Dyes Pigm., 2018, 149, 915 CrossRef CAS; (d) B. Wu, W. Wang, J. Wang, S. Li and Y. He, Dyes Pigm., 2018, 157, 290 CrossRef CAS; (e) C. Liu, Y. Hang, T. Jiang, J. Yang, X. Zhang and J. Hua, Talanta, 2018, 178, 847 CrossRef CAS PubMed; (f) H.-Q. Peng, X. Zheng, T. Han, R. T. K. Kwok, J. W. Y. Lam, X. Huang and B. Z. Tang, J. Am. Chem. Soc., 2017, 139, 10150 CrossRef CAS PubMed.
- (a) A. Chatterjee, M. Banerjee, D. G. Khandare, R. U. Gawas, S. C. Mascarenhas, A. Ganguly, R. Gupta and H. Joshi, Anal. Chem., 2017, 89, 12698 CrossRef CAS PubMed; (b) D. G. Khandare, H. Joshi, M. Banerjee, M. S. Majik and A. Chatterjee, Anal. Chem., 2015, 87, 10871 CrossRef CAS PubMed; (c) A. Chatterjee, D. G. Khandare, P. Saini, A. Chattopadhyay, M. S. Majik and M. Banerjee, RSC Adv., 2015, 5, 31479 RSC; (d) D. G. Khandare, H. Joshi, M. Banerjee, M. S. Majik and A. Chatterjee, RSC Adv., 2014, 4, 47076 RSC; (e) D. G. Khandare, V. Kumar, A. Chattopadhyay, M. Banerjee and A. Chatterjee, RSC Adv., 2013, 3, 16981 RSC.
- (a) G. Xu, Y. Tang, Y. Ma, A. Xu and W. Lin, Spectrochim. Acta, Part A, 2018, 188, 197 CrossRef CAS PubMed; (b) Y. Li, Z. Zhuang, G. Lin, Z. Wang, P. Shen, Y. Xiong, B. Wang, S. Chen, Z. Zhao and B. Z. Tang, New J. Chem., 2018, 42, 4089 RSC; (c) Y.-H. Zhao, Y. Luo, H. Wang, T. Guo, H. Zhou, H. Tan, Z. Zhou, Y. Long and Z. Tang, ChemistrySelect, 2018, 3, 1521 CrossRef CAS; (d) X.-G. Liu, C.-L. Tao, H.-Q. Yu, B. Chen, Z. Liu, G.-P. Zhu, Z. Zhao, L. Shen and B. Z. Tang, J. Mater. Chem. C, 2018, 6, 2983 RSC; (e) Z. Wang, Y. Gu, J. Liu, X. Cheng, J. Z. Sun, A. Qin and B. Z. Tang, J. Mater. Chem. B, 2018, 6, 1279 RSC; (f) L. Shi, Y. Liu, Q. Wang, T. Wang, Y. Ding, Y. Cao, Z. Li and H. Wei, Analyst, 2018, 143, 741 RSC; (g) M. T. Gabr and F. C. Pigge, Mater. Chem. Front., 2017, 1, 1654 RSC; (h) P. F. Li, Y. Y. Liu, W. J. Zhang and N. Zhao, ChemistrySelect, 2017, 2, 3788 CrossRef CAS; (i) X. Cheng, R. Zhang, X. Cai and B. Liu, J. Mater. Chem. B, 2017, 5, 3565 RSC; (j) G. Jiang, G. Zeng, W. Zhu, Y. Li, X. Dong, G. Zhang, X. Fan, J. Wang, Y. Wu and B. Z. Tang, Chem. Commun., 2017, 53, 4505 RSC; (k) G. Jiang, X. Liu, Q. Chen, G. Zeng, Y. Wu, X. Dong, G. Zhang, Y. Li, X. Fan and J. Wang, Sens. Actuators, B, 2017, 252, 712 CrossRef CAS; (l) J. Shi, Q. Deng, C. Wan, M. Zheng, F. Huang and B. Tang, Chem. Sci., 2017, 8, 6188 RSC.
- V. N. Mehta, S. Jha and S. K. Kailasa, Mater. Sci. Eng. Carbon, 2014, 38, 20 CrossRef CAS PubMed.
- B. Xing, T. Jiang, W. Bi, Y. Yang, L. Li, M. Ma, C. K. Chang, B. Xu and E. K. L. Yeow, Chem. Commun., 2011, 47, 1601 RSC.
- C. Zhu, Q. Yang, L. Liu and S. Wang, Angew. Chem., 2011, 50, 9607 CrossRef CAS PubMed.
- H. Chen, B. Wang, J. Zhang, C. Nie, F. Lv, L. Liu and S. Wang, Chem. Commun., 2015, 51, 4036 RSC.
- H. Z. Lai, S. G. Wang, C. Y. Wu and Y. C. Chen, Anal. Chem., 2015, 87, 2114 CrossRef CAS PubMed.
- G. Feng, Y. Yuan, H. Fang, R. Zhang, B. Xing, G. Zhang, D. Zhang and B. Liu, Chem. Commun., 2015, 51, 12490 RSC.
- E. Zhao, Y. Chen, S. Chen, H. Deng, C. Gui, C. W. Leung, Y. Hong, J. W. Lam and B. Z. Tang, Adv. Mater., 2015, 27, 4931 CrossRef CAS PubMed.
- E. Zhao, Y. Hong, S. Chen, C. W. Leung, C. Y. Chan, R. T. Kwok, J. W. Lam and B. Z. Tang, Adv. Healthcare Mater., 2014, 3, 88 CrossRef CAS PubMed.
- E. Zhao, Y. Chen, H. Wang, S. Chen, J. W. Lam, C. W. Leung, Y. Hong and B. Z. Tang, ACS Appl. Mater. Interfaces, 2015, 7, 7180 CrossRef CAS PubMed.
- Y. Li, X. Hu, S. Tian, Y. Li, G. Zhang, G. Zhang and S. Liu, Biomaterials, 2014, 35, 1618 CrossRef CAS PubMed.
- L. Zhang, L. Jiao, J. Zhong, W. Guan and C. Lu, Mater. Chem. Front., 2017, 1, 1829 RSC.
- (a) M. Wang, X. Gu, G. Zhang, D. Zhang and D. Zhu, Anal. Chem., 2009, 81, 4444 CrossRef CAS PubMed; (b) Y. Hong, C. Feng, Y. Yu, J. Liu, J. W. Y. Lam, K. Q. Luo and B. Z. Tang, Anal. Chem., 2010, 82, 7035 CrossRef CAS PubMed.
- N. W. Schröder, S. Morath, C. Alexander, L. Hamann, T. Hartung, U. Zähringer, U. B. Göbel, J. R. Weber and R. R. Schumann, J. Biol. Chem., 2003, 278, 15587 CrossRef PubMed.
- L. Miyanaga, S. Takano, Y. Morono, K. Hori, H. Unno and Y. Tanji, Biochem. Eng. J., 2007, 37, 56 CrossRef.
- M. B. Coyle, Manual of Antimicrobial Susceptibility Testing, American Society for Microbiology, 2005 Search PubMed.
- J. McFarland, J. Am. Med. Assoc., 1907, 49, 1176 CrossRef.
- A. Das, S. Biswas and M. Biswas, Open Microbiol. J., 2018, 12, 107 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8qm00417j |
| This journal is © the Partner Organisations 2018 |