 Open Access Article
Open Access ArticleMulti-directional electrodeposited gold nanospikes for antibacterial surface applications†
Aaron
Elbourne
 a,
Victoria E.
Coyle
b,
Vi Khanh
Truong
cd,
Ylias M.
Sabri
b,
Ahmad E.
Kandjani
b,
Suresh K.
Bhargava
a,
Victoria E.
Coyle
b,
Vi Khanh
Truong
cd,
Ylias M.
Sabri
b,
Ahmad E.
Kandjani
b,
Suresh K.
Bhargava
 b,
Elena P.
Ivanova
b,
Elena P.
Ivanova
 a and
Russell J.
Crawford
*a
a and
Russell J.
Crawford
*a
aSchool of Science, College of Science, Engineering and Health, RMIT University, Melbourne, VIC 3001, Australia. E-mail: russell.crawford@rmit.edu.au
bCentre for Advanced Materials and Industrial Chemistry (CAMIC), School of Science, College of Science, Engineering and Health, RMIT University, Melbourne, VIC 3001, Australia
cSchool of Science, Faculty of Science, Engineering and Technology, Swinburne University of Technology, Haw-thorn, VIC 3122, Australia
dARC Research Hub for Australian Steel Manufacturing, Wollongong, New South Wales, Australia
First published on 21st August 2018
Abstract
The incorporation of high-aspect-ratio nanostructures across surfaces has been widely reported to impart antibacterial characteristics to a substratum. This occurs because the presence of such nanostructures can induce the mechanical rupture of attaching bacteria, causing cell death. As such, the development of high-efficacy antibacterial nano-architectures fabricated on a variety of biologically relevant materials is critical to the wider acceptance of this technology. In this study, we report the antibacterial behavior of a series of substrata containing multi-directional electrodeposited gold (Au) nanospikes, as both a function of deposition time and precursor concentration. Firstly, the bactericidal efficacy of substrata containing Au nanospikes was assessed as a function of deposition time to elucidate the nanopattern that exhibited the greatest degree of biocidal activity. Here, it was established that multi-directional nanospikes with an average height of ∼302 nm ± 57 nm (formed after a deposition time of 540 s) exhibited the greatest level of biocidal activity, with ∼88% ± 8% of the bacterial cells being inactivated. The deposition time was then kept constant, while the concentration of the HAuCl4 and Pb(CH3COO)2 precursor materials (used for the formation of the Au nanospikes) was varied, resulting in differing nanospike architectures. Altering the Pb(CH3COO)2 precursor concentration produced multi-directional nanostructures with a wider distribution of heights, which increased the average antibacterial efficacy against both Gram-negative Pseudomonas aeruginosa and Gram-positive Staphylococcus aureus bacteria. Importantly, the in situ electrochemical fabrication method used in this work is robust and straightforward, and is able to produce highly reproducible antibacterial surfaces. The results of this research will assist in the wider utilization of mechano-responsive nano-architectures for antimicrobial surface technologies.
Introduction
Mechano-responsive, nanostructured antimicrobial surfaces have been the focus of intensive research in recent years.1,2 This increase in research activity has been largely a response to the increasing prevalence of multi-resistant bacteria and their associated infections.3–8 The seriousness of this epidemic has been highlighted by the Infectious Diseases Society of America (IDSA), which has labeled antibacterial resistance as a “public health crisis”.9 Medical experts have predicted that this crisis will worsen rapidly in the coming years.3,6,8–10 The adaptation of bacteria to survive in the presence of conventional antibiotics, together with their ability to establish biofilms on currently available antibacterial surfaces, is a major medical challenge. This evolutionary bacterial resistance has led to an increase in the number of persistent infections caused by resistant strains of bacteria, often resulting in patient death. Indeed, species such as carbapenem-resistant Enterobacteriaceae spp. (CRE) and methicillin-resistant Staphylococcus aureus (MRSA) have already evolved to exhibit resistance to many known antimicrobial agents,11–13 rendering conventional therapies largely ineffective.To this end, research has pivoted towards the development of new antimicrobial technologies, with an emphasis being placed on the development of methods to which bacteria cannot develop resistance. One such technology is the formation of surface nanostructures that can physically resist bacterial attachment or deactivate bacteria upon attachment to the surface.1,14 This approach can be largely divided into two categories, namely (1) antifouling surfaces, whereby the extent of bacterial attachment is limited by an unfavourable surface nano-architecture,15–19 or (2) antibacterial surfaces, where the surface induces cell lysis during bacterial attachment.1,2,20–22 In the latter case, the interaction of bacteria with high-aspect-ratio nanostructures prevents biofilm formation via a physico-mechanical mechanism.1,21–27 Upon bacterial adsorption, the cell wall experiences a self-induced mechanical stress via the action of the substratum surface, which results in cell lysis, and in-turn, cell death.22,25 Importantly, since the bacteria are ruptured while attaching onto the surfaces, the bacteria are unable to establish a biofilm. Previous studies have shown that both Gram-negative and Gram-positive bacterial cells are susceptible to this mechanistic-rupturing, as long as the interaction taking place between the bacterial cell and the surface nanostructure results in the bacterial membrane deforming beyond its elastic limit.22,25 More importantly, this process appears to be independent of surface chemistry, meaning that bacteria are unable to develop a protective response.1,2,15–17,22–38 Such surface nanostructuring provides a viable replacement for conventional antibacterial surface technologies that involve the incorporation of antibiotics or additive chemicals. A comparative analysis of biocidal nanostructures has suggested that short, sharp nanofeatures tend to exhibit greater biocidal behavior;1 however, this supposition is not definitive, with several outliers having been noted throughout the literature.24,39
Cross-referencing the data reported in the current literature on biocidal substrata1,2,15–17,22–38 highlights a common theme: the studies have primarily focused on the fabrication of novel nanostructured surfaces almost exclusively for implant materials, which utilizes the aforementioned mechano-responsive mechanism to mitigate the formation of a biofilm at the implant–tissue interface and hence the prevention of any subsequent infections. Moreover, studies have mainly focused on substrata such as black silicon (bSi), titanium, titania, and polymeric surfaces, each fabricated via laboratory scale methods such as reactive ion etching, electron beam lithography, plasma etching, and femtosecond laser irradiation.1,14 These methods are often not scalable, which somewhat limits their applicability to broader applications. To enable the wide-spread application of this technology would require the consideration of a broader range of manufacturing methods for the production of, for example, sterile surface coatings and larger biomedical devices. The identification of a large number of high-aspect-ratio nano-architectures that kill a range of bacteria would be greatly beneficial in order to unlock the full potential of mechano-responsive, antimicrobial nanostructures.
In this study, we employed a simple in situ, low-cost, electrodeposition method for the purposeful production of multi-directional gold (Au) nanospikes on a substratum surface. The resultant nanostructured surfaces were found to exhibit high levels of antibacterial activity. The nanospike geometry (length, density, cap-radii, base-radii and surface roughness) was controlled via both the deposition time and the concentration of the precursor materials (HAuCl4 and the inorganic directional growth agent, Pb(CH3COO)2). Firstly, a range of deposition times were employed in order to fabricate a series of Au nanospikes with average heights of ∼103, 302, 384, and 555 nm. The antibacterial activity of the substrata possessing these nanospikes was then screened against live Pseudomonas aeruginosa (P. aeruginosa) bacterium. This series of tests established that nanospikes resulting from a deposition time of 540 s led to the formation of a substratum with the highest antibacterial efficacy. Following this, a fixed deposition time of 540 s was used to assess the effects of changes in the concentration of the HAuCl4 and Pb(CH3COO)2 precursors on antibacterial efficacy. The aim of this work was to fabricate highly-bactericidal substrata containing multi-directional Au nanospikes in order to provide an insight into the relationship between nanospike geometry and their antibacterial behavior.
Materials and methods
Nanospike fabrication and characterisation
Gold nanospikes were fabricated using protocols previously described.40,41 Briefly, blank silicon substrata were deposited with a 10 nm Ti adhesion layer, followed by a 100 nm Au layer using e-beam evaporation. This substratum was used as the working electrode. Gold nanospikes were then electrodeposited onto the smooth gold layer from a solution containing HAuCl4 and Pb(CH3COO)2. The electrodeposition process was performed using a CH Instruments (CHI 760C) electrochemical analyzer with an electrochemical cell that allowed reproducible positioning of the working electrode (the base substratum), with a Ag/AgCl (3 M KCl) reference electrode, and a graphite counter electrode. Deposition was performed at a constant potential of 0.05 V for varying deposition times (360, 540, 720 and 900 s). For the time dependant deposition series, a solution of 6.8 mM HAuCl4 and 1 mM Pb(CH3COO)2 was used. The surface structure was then altered by varying the HAuCl4 and Pb(CH3COO)2 concentration; for HAuCl4 concentrations of 3.4 mM and 13.6 mM were used, while Pb(CH3COO)2 concentrations of 0.5 mM and 2 mM were investigated. Following electrodeposition, the substrata were washed several times with deionized water, air-dried, and characterized using scanning electron microscopy (SEM), X-ray diffraction (XRD), and energy-dispersive X-ray spectroscopy (EDX). High-resolution electron micrographs of the native surfaces were recorded using a FE-SEM, FEI NOVA nanoSEM (FEI company, Oregon, United States), at 10 kV. EDX measurements were also performed on this instrument. X-ray photoelectron spectroscopy (XPS) characterization of the materials was performed using a Thermo K-Alpha instrument at a pressure better than 1 × 10−9 Torr. Core level binding energies (BEs) were aligned with the adventitious C 1s binding energy of 285 eV. XRD measurements were carried out on a Bruker D8 Discover micro diffraction system with general area detector diffraction system (GADDS) instrument operating at a voltage of 40 kV and a current of 40 mA with Cu Kα radiation.AFM characterization
The substrata were imaged using a combination of MFP-3D and Cypher ES Atomic Force Microscopes (Oxford Instrument, Asylum Research, Santa Barbara, CA, USA) at room temperature (25 °C). All images were obtained using amplitude modulated-AFM (AM-AFM) with OMCL-AC240TS cantilevers (Olympus Corporation, Japan, nominal spring constant kc = 2 N m−1). To minimize the imaging force, a set-point ratio (imaging amplitude (A)/free amplitude (A0)) of >0.7–0.8 was maintained during imaging. Each cantilever was calibrated using the thermal spectrum method, which produced a well-defined resonance peak, and the lever sensitivity was determined using force spectroscopy; the spring constant is resolved via the inverse optical lever sensitivity (InVOLS) using force curve measurements on the hard gold surface in air. The features of all images presented rotated as the scan angle was changed and scaled correctly with scan size, confirming they are not a consequence of imaging artefacts.Bacteria-surface SEM characterisation
High-resolution electron micrographs of the nanospiked surfaces incubated with bacteria for 18 hours were recorded using a field-emission scanning electron microscope (FE-SEM; ZEISS SUPRA 40 VP, Oberkochen, BW, Germany) at 3 kV using methods described previously.37,42AFM and SEM image analysis
AFM data were processed using a combination of the native asylum research software, custom MATLAB codes, and Gwyddion, a free SPM data analysis tool.43 No imaging filtering (noise deconvolution, scar removal, or FFT filtering) was applied, meaning that the data was not artificially improved for publication. SEM images were analyzed using both Gwyddion and ImageJ software.Cell viability analysis
A confocal laser scanning microscopy (CLSM) (Fluoview FV10 inverted microscope, Olympus, Tokyo, Japan) was used to evaluate the proportions of live and dead cells attached to each surface. Surface attached cells were dyed using a LIVE/DEAD® BacLight™ Bacterial Viability Kit, L7012. Specifically, SYTO® 9 and propidium iodide (PI) fluorescent dyes (Molecular Probes™, Invitrogen, Grand Island, NY, USA) were used to stain the cells. SYTO® 9 permeated both intact and damaged cell membranes, binding to nucleic acids and fluorescing green when excited by a 485 nm wavelength laser. PI dominantly enters cells that have undergone membrane damage, which are considered to be non-viable, and binds with higher affinity to nucleic acids than SYTO® 9. Bacterial suspensions were stained according to the manufacturer's protocol and as described in our previously published articles.25,37,44 Importantly, discrepancies in viability assessment were avoided by ensuring that no green (485 nm) and red (543 nm) fluorescence overlap was observed during image assessment. This means that live and dead cell differentiation was adequately achieved, providing a meaningful assessment of the antibacterial activity of the surface. Furthermore, photobleaching of the SYTO® 9 dye was avoided by limiting each surface location to a single confocal scan.Bacterial strains, growth conditions, and sample preparation
The pathogenic bacteria, Pseudomonas aeruginosa ATCC 9721 and Staphylococcus aureus (S. aureus) CIP 65.8T, were obtained from the American Type Culture Collection (ATCC, Manassas, VA, U.S.A.) and the Culture Collection of the Institute Pasteur (CIP, Paris, France), respectively. The selected bacteria represent two large prokaryotic lineages, namely Gram-negative and Gram-positive bacteria, whose responses on nanostructured surfaces will be typical for taxonomically related bacteria. Prior to each experiment, bacterial cultures were refreshed on nutrient agar from stocks (Oxoid, U.K.). Fresh bacterial suspensions were grown overnight at 37 °C in 5 mL of nutrient broth (Oxoid, U.K.). The density of bacterial suspensions was adjusted to OD600 = 0.1, after collection on at the logarithmic stage of cell growth (data not shown). Experimental protocols were kept in accordance with our previously published articles.25,37,44Results and discussion
Surface characterisation as a function of electrodeposition time
A series of nanostructured surfaces were prepared using fixed precursor concentrations of 6.8 mM HAuCl4 and, 1 mM Pb(CH3COO)2, an inorganic directional growth agent, over a range of deposition times (360, 540, 720, and 900 s) using previously established protocols.40,41 These initial electrodeposition conditions were selected in order to promote the growth of multi-directional nanospikes, formed at an orientation that was approximately normal to the substratum surface. Increasing the deposition time alters the nanospike geometry, increasing the average nanospike height, spacing, cap-radius, base-radius, aspect-ratio, and surface roughness, while simultaneously decreasing the pillar density. This decrease in density occurs as a result of an increase in the nanospike lateral growth (widening) as a function of deposition time. Importantly, substrata containing this series of nanospikes were used to determine the most biocidal surface architecture. 5 μm × 3.5 μm tilted (45°) SEM micrographs and 5 μm × 5 μm AFM images of the resulting nanospikes are presented in Fig. 1. Both SEM and AFM measurements were employed to thoroughly determine the nano-architectures present on the substratum surfaces in order to elucidate the relationship that exists between the antibacterial behaviour of a surface and its nanostructure. The deposition time corresponding to the surfaces is given to the left of the images.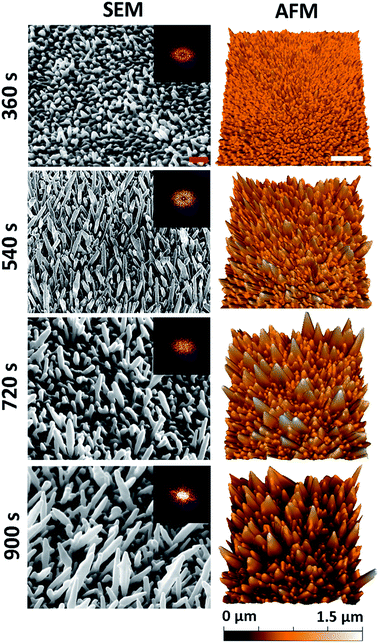 | ||
| Fig. 1 Native surface data for the nanospike deposition series: column (1) SEM (45° tilted) and column (2) AFM images of the gold nanospike surfaces as a function of electrodeposition time. The red and white scale bars are 200 nm and 1 μm, respectively. 2D-FFT data (insets) are shown for the top-down SEM images (see Fig. S2†). | ||
For all deposition times, well-defined Au nanospikes with tapering prismatic ends were observed in both the SEM and AFM data; however, the surface parameters of the nanospikes changed as a function of the deposition time. To quantify this observation, the geometric surface parameters of the nanospikes were assessed using image processing software (Gwyddion43 and Image J), and two-dimensional fast Fourier transform analysis, for both the SEM and AFM data. This analysis allowed the average inter-spike spacing (nm), nanospike height (nm), cap radius (nm), base radius (nm), spike-density (/μm2), and the surface roughness (Ra) (nm) to be determined. The corresponding parameters for each surface type are presented as a function of electrodeposition time in Table 1. These values are consistent with those of the Au nanospikes grown via a similar electrodeposition protocol previously reported in the literature.40,41 In general, the nanospikes exhibited a multimodal, multi-directional surface distribution, forming at angles ranging between 45–90° to the underlying substratum. Additionally, the geometric distribution of the nanospikes tended to increase with deposition time, which is exemplified by the increasing standard deviation associated with the measurements as a function of time. This indicated that a multi-directional surface structure emerged with an increasing deposition time. An AFM image of the initial substratum containing the Ti and Au layers is included in Fig. S1† for comparison.
| Electrodeposition time (s) | Spacing (nm) | Height (nm) | Cap radius (nm) | Base radius (nm) | Density (/μm2) | R a (nm) |
|---|---|---|---|---|---|---|
| a R a: roughness analysis. | ||||||
| 360 | 120 ± 84 | 103 ± 36 | 45 ± 32 | 47 ± 38 | 26.6 | 34.0 |
| 540 | 211 ± 120 | 302 ± 57 | 60 ± 13 | 89 ± 29 | 16.0 | 93.0 |
| 720 | 312 ± 145 | 416 ± 112 | 94 ± 71 | 112 ± 82 | 11.8 | 109.6 |
| 900 | 355 ± 136 | 555 ± 214 | 103 ± 82 | 115 ± 86 | 9.1 | 180.0 |
X-ray diffraction (XRD) data of similarly prepared nanospike surfaces has been previously reported over this deposition time range.40,41,45 The data revealed that the face centered cubic (fcc) Au nanospikes grew preferentially along the Au(200) crystallographic plane, due to the affinity of the Pb2+ ion towards the Au (110) and (100) crystallographic plane, while the AuCl4− ions favoured adsorption onto the Au(111) facet. The differing affinities of these ions to the different planes during the electrodeposition process resulted in the formation of pure gold nanospikes. As both precursor agents are potentially toxic to bacterial species, it was important to establish the surface chemistry of the resultant Au nanospikes. Representative examples of the XRD, energy-dispersive X-ray (EDX), and X-ray photoelectron spectroscopy (XPS) data are shown in Fig. S3.† This data confirms that no Pb2+ or Cl− ions were present on the electrodeposited surfaces following fabrication. A more detailed explanation of the data is also provided in the ESI.†
Antibacterial behaviour of the substrata containing the gold nanospikes
The antibacterial efficacy of the different nanospike-containing surfaces was initially assessed using P. aeruginosa ATCC 9721 cells. This approach was taken since P. aeruginosa bacteria are model cells that have been used in previous studies assessing the efficacy of mechano-responsive antibacterial surfaces.21,25,26 The rationale for this approach was that it would allow us to determine the optimal anti-bacterial substratum prior to undertaking further testing against other bacteria such as the Gram-positive Staphylococcus aureus CIP 65.8T (S. aureus) cells.The substrata were immersed in a suspension of the bacterial cells for 18 h, after which time the antibacterial activity of the substrata were assessed via confocal laser scanning microscopy (CLSM). Representative CLSM images for each of the surface type incubated with P. aeruginosa are presented in Fig. 2a–d. The relative proportion of live-to-dead cells was assessed using a fluorescent staining technique, where intact and damaged cells (or live and dead cells) would fluorescence green or red, respectively. The relative numbers of live and dead cells was then determined and displayed as a percentage in the bar chart presented in Fig. 2e. The associated standard deviation of bactericidal activity is indicated by the error bars. For the 360, 540, 720, and 900 s electrodeposition surfaces, the average proportion of non-viable (dead) P. aeruginosa cells was found to be 52.7, 88.0, 76.0 and 77.5%, respectively. The proportion of dead cells was significantly greater than that obtained on the unmodified gold coated control surface, where only 18.3% of cells were found to be inactivated (cf.Fig. 2 and S4A†). Interestingly, the highest level of antibacterial efficacy was found to be on the surfaces prepared over a 540 s electrodeposition period, which contained nanospikes with an average height and inter-spike spacing of 302 ± 57 nm and 211 ± 120 nm, respectively. This effective nanospike height is consistent with that reported in a number of previous studies,22,23,27,38,46–48 which suggests that a nanofeature height of ∼300 nm tends to display a greater level of biocidal activity towards both Gram-negative and Gram-positive bacteria than substrata possessing nanospikes with greater or less height.22,23,27,38,46–48 The reason for the effectiveness of this ‘optimal’ peak height in killing bacteria is still largely unknown. The inter-pillar spacing on the substrata used in this study was slightly larger than that of a number of previously reported effective antibacterial surfaces, which would initially suggest that nanofeature height plays an important role in the overall bactericidal behaviour of the substrates. This assertion has been often speculated and reported in the relevant literature for both experimental23,32–34,46,49 and theoretical studies22,38 of naturally occurring and biomimetic antibacterial nanostructures. These studies have also shown that the modification of various surface parameters can significantly alter the bactericidal activity of the surface. Given these observation, it appears that single surface parameters alone do not control the biocidal behavior of the surface, but rather the behavior arises as a result of the interplay between several surface parameters.1
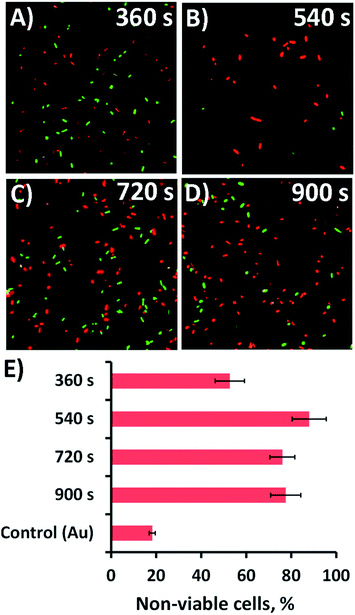 | ||
| Fig. 2 Bactericidal activity of the nanospikes against P. aeruginosa. (A–D) Cell viability of P. aeruginosa (ATCC 9721) cells incubated in the presence of the substrata shown in Fig. 1. Fluorescent staining of the samples with LIVE/DEAD backlight highlights the live (green) and dead (red) cells in the CLSM images. (E) The corresponding average non-viable cell percentage (red cells) for the different substrata are shown in the bar graph. The CLSM image size is 70 μm × 70 μm. | ||
Developing multi-directional nanostructures: the effect of precursor concentration on nanospike morphology
The substratum prepared using a 540 s deposition time exhibited the highest level of bactericidal activity (see Fig. 1 and 2). This deposition time was then used to study the antibacterial behavior of the substrata as a function of precursor concentration and the resultant nanospike morphology. Altering the initial Pb(CH3COO)2 concentration has been shown to promote uneven nanospike growth, resulting in the formation of more bimodal nanostructures.45 The surface geometries resulting from an initial Pb(CH3COO)2 concentration of 0.5 mM and 2 mM are shown in Fig. 3. The resulting surfaces can be compared to the substrata prepared using a 540 s deposition time, as presented in Fig. 1. The surface parameters of these substrata together with those obtained using an initial 1 mM Pb(CH3COO)2 concentration are presented in Table 2.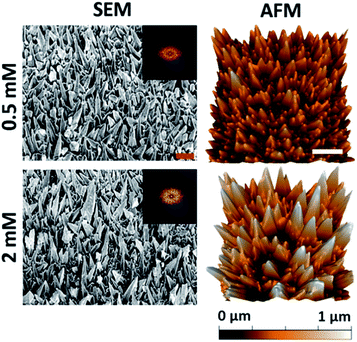 | ||
| Fig. 3 Native surface data for the Pb+ concentration series: column (1) SEM (45° tilted) and column (2) AFM images of the gold nanospike surfaces for 0.5 and 2 mM Pb(CH3COO)2. The 2D-FFT data is shown for the top-down SEM images (see Fig. S5†). The red and white scale bars are 200 nm and 1 μm, respectively. | ||
| Pb(CH3COO)2 conc. (mM) | Spacing (nm) | Height (nm) | Cap radius (nm) | Base radius (nm) | Density (/μm2) | R a (nm) |
|---|---|---|---|---|---|---|
| a R a: roughness analysis. | ||||||
| 0.5 | 270 ± 111 | 298 ± 98 | 70 ± 15 | 98 ± 31 | 14.8 | 100.3 |
| 1 | 211 ± 120 | 302 ± 57 | 60 ± 13 | 89 ± 29 | 16.0 | 93.0 |
| 2 | 389 ± 182 | 412 ± 213 | 87 ± 57 | 140 ± 69 | 12.3 | 152.9 |
It can be seen that reducing the initial Pb2+ concentration results in the formation of a surface with a wider distribution of nanospike heights with increased inter-spike spacing, a slightly lower nanospike density, and an approximately unchanged cap radius compared to the substratum prepared using 1 mM Pb(CH3COO)2 (Fig. 1). The substratum prepared using 2 mM Pb(CH3COO)2 possessed an increased average nanospike height (and distribution) of 480 ± 182 nm.
The effect of the initial HAuCl4 concentration on the resultant surface architecture was also investigated. Increasing and decreasing the AuCl4− ion concentrations to 3.4 mM and 13.6 mM, respectively, produced surfaces with lower average height nanospikes over a deposition time of 540 s. The resulting antibacterial activity of the substrata was commensurately decreased. The corresponding AFM, SEM and CLSM data are given in Fig. S6† and the respective surface parameters are presented in Table S1.† As the antimicrobial efficacy was decreased by changing the initial HAuCl4 concentration, this data was not considered further.
Antibacterial activity of the substrata prepared using different Pb(CH3COO)2 concentrations
The antibacterial activity of the substrata prepared using different Pb(CH3COO)2 concentrations (see Fig. 4) over a 540 s deposition period was determined using both P. aeruginosa and S. aureus bacteria. These were chosen as they represent the two major prokaryotic taxonomic groups: Gram-negative and Gram-positive bacteria, respectively. Duplicate substratum surfaces were immersed with each bacterial suspension for 18 hours and the antibacterial response was then quantified used CLSM imaging. Representative CLSM images and SEM micrographs for each of the substrata after immersion with P. aeruginosa (left column) and S. aureus (right column) are presented in Fig. 4. For comparison, the CLSM and SEM images obtained after incubating the bacterial samples in the presence of the unmodified gold coated control surface are shown in Fig. S4† for both P. aeruginosa (left column) and S. aureus (right column) bacteria. The relative live-to-dead cell population is shown as the proportion (%) of dead cells in the bar chart in Fig. 5. The associated standard deviation of bactericidal activity is indicated by the error bars. The average proportion of inactivated P. aeruginosa cells was found to be 92.5, 88.0 and 89.7%, and average proportion of inactivated S. aureus cells was found to be 64.6, 63.1 and 73.5%, for the substrata formed using the 0.5, 1 and 2 mM Pb(CH3COO)2 precursor concentration, respectively. Again, these values represent a significant increase in antibacterial activity compared to the respective control surfaces for both bacterial species. The average biocidal activity of these substrata towards P. aeruginosa cells was observed to slightly increase for the 0.5 mM and 2 mM Pb(CH3COO)2 system; however, the variance was not statistically significant. As such, the results obtained suggested that all of the substratum surfaces exhibited an approximately equivalent degree of biocidal activity, with the observed fluctuation in average bacterial kill proportions being a consequence of natural variations within a biological system. Importantly, for implant-based applications, nanostructured antibacterial technologies need only to aid osteointegration by reducing the probability of infection.50,51 Here, the so-called “race to the surface” becomes important,52 where host cells and pathogenic species compete for surface colonisation at the implant–tissue interface. Competitive proliferation studies50,51 have highlighted the viability of similar surface architectures, demonstrating that mammalian cells were capable of growing over, and successfully colonizing, pre-infected substrata possessing antibacterial surface nanostructures.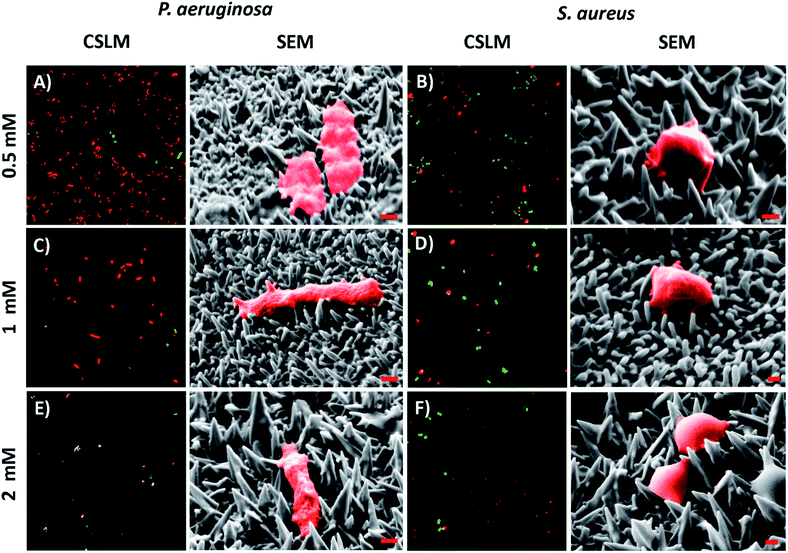 | ||
| Fig. 4 Representative CSLM images and SEM micrographs of (A, C, & E) P. aeruginosa and (B, D, & F) S. aureus cells atop the variant nanospiked surfaces (indicated to the left). For the CSLM images, cell staining was conducted and interpreted as described in Fig. 2. The CSLM image size is 70 μm × 70 μm. For the SEM micrographs, both bacterial species have been false colored red to highlight the cell attachment morphology. The red scale bar is 200 nm in each image. | ||
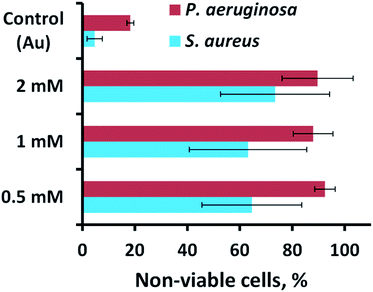 | ||
| Fig. 5 Cell viability of P. aeruginosa ATCC 9721 and S. aureus CIP 65.8T cells incubated in the presence of the substrata shown in Fig. 4. | ||
A number of studies have investigated the mechano-responsive antibacterial mechanism of various nanostructured surfaces.20,22,24–26,31,32,34–36,38,53–57 While the precise mechanism responsible for the antibacterial activity is still the subject of debate,32,58 the consensus between the results obtained from both experimental22–26,31,32,38,59,60 and theoretical32,33 studies suggest that a torsional force is induced across the bacterial membrane upon surface adsorption.1,2 During this event, the adhesion force of the bacteria results in the surface nanofeatures indenting into the bacterial membrane, causing it to stretch between the nanofeatures to the point of rupture, which causes cell death.22,38 Both biophysical and theoretical models have been proposed that describe this interaction, asserting that Gram-positive bacteria are less susceptible to this phenomenon than Gram-negative bacteria as a result of their more rigid cell wall. This phenomenon was also observed in the current study, with the Gram-negative P. aeruginosa cells experiencing greater degrees of cell lysis than the Gram-positive S. aureus cells for each substratum surface formed using different Pb(CH3COO)2 precursor concentrations (see Fig. 5).
Previous studies have established that both Gram-negative and Gram-positive bacteria are capable of modulating their surface attachment patterns in response to a nanostructured substratum.16,17,61 Importantly, when the nanostructural feature spacing is larger than the bacterial cell, the bacteria will orientate themselves so that they sit between the nanofeatures.62 When the nanofeature spacing is smaller than the bacterium, rod-shaped bacteria have been found to orient themselves perpendicularly to the nanofeatures in order to increase the extent of cell-surface contact.31,60,62–64 This behaviour is commensurate with the observed reduction in bactericidal activity towards the Gram-positive S. aureus cells, meaning that the smaller spherical bacterium may be avoiding rupture by adsorbing onto the substratum at a position located between the nanofeatures. The larger P. aeruginosa cells are less able to avoid the surface nanofeatures due to their larger, rod-shaped morphology, which when combined with their thinner cell-membrane, results in a greater degree of cell death (see Fig. 4 and 5).
Recently, Kelleher23et al. and Dickson46et al. investigated a series of nanostructurally varient natural (cicada wing) and biomimetic (PMMA) biocidal surfaces, respectively. Here, the studies investigated the relationship between the geometry of the respective nanotopographies and their corresponding mechano-bactericidal efficacies. The studies concluded that a reduction in the nanofeatural spacing correlated with an increase in the surface killing efficiency. Furthermore, several studies have highlighted that nanofeatures of ∼300 nm in height are capable of exerting a greater amount of localized stress on the membranes of bacterial cells during surface attachment;1,25,27,36,37 however, in all cases it must be stressed that it is the combined influence of all nanostructural surface parameters that results in the overall biocidal behaviour of the substratum.1 Together, these observations are commensurate with the results of this study, whereby there is a correlation between biocidal activity (see Fig. 5) and the nanospike geometry (see Table 2). Indeed, inspection of the nanospikes formed from various initial concentrations of Pb(CH3COO)2 highlighted a balance between nanofeatural height distribution and the nanospike density. Here, multi-directional nanospikes, with an average height of approximately 300 nm, coupled with a nanospike density between ∼12–16 nanospikes per μm2, resulted in a higher antibacterial efficacy towards the bacteria investigated in this study (see Fig. 1, 2, Tables 1 and 2).
Interestingly, for all Pb(CH3COO)2 based variants the average antibacterial activity is invariant, within error (see Fig. 5). However, for the 2 mM Pb(CH3COO)2 system, the error was the largest observed. The larger error can be attributed to the following: for the 2 mM Pb(CH3COO)2 precursor surface, certain regions of the substratum exhibits the greatest bactericidal activity – up to 100% and 94% for P. aeruginosa and S. aureus cells, respectively, but there are also surface locations where the bactericidal response is significantly lower. In comparision, the 1 mM and 0.5 mM Pb(CH3COO)2 precursor surface structures are more uniformly biocidal, which produced a lower associated error (see Fig. 5). This means that the higher variability and distribution of nanospike geometry (see data in Table 2) may lead to the formation of regions of the surface where bacteria can attach between the nanofeatures, avoiding cell rupture. The smaller, more-rigid, spherical S. aureus cells have a higher probability of attaching in these non-lethal regions than the larger P. aeruginosa cells. These multi-directional, bimodal nanostructures may exhibit a greater biocidal activity than that of the more regular nanostructures previously investigated, including both naturally occuring23,25,26,32,34–36,53–55 and synthetic24,27,33,37,48,49,54,57,65 surfaces. This occurs because the bacteria are subjected to forces operating in multiple directions during surface adsorption, which limits their ability to evade critical membrane damage, and hence cell death. By contrast, more conventional, highly-ordered nanostructures21,23,27,46,49 may allow some bacteria to evade the action of the antibacterial nanotopographies, decreasing the resultant biocidal action of these surfaces. This suggestion is supported by the observations of Fisher et al.,47 where multi-level diamond nanocones exhibited higher degrees of bactericidal behaviour than their more regular counterparts.
Lastly, Wu et al.66 recently reported the antibacterial activity of sparsely ordered nanopillars, nanorings, and nanonuggets, each formed via the electrodeposition of gold onto nanoporous templates, towards methicillin-resistant S. aureus (MRSA) cells. Here, the authors reported >99% cell inactivity after 2 hours of incubation for all surface structures investigated. It is likely that the reported antimicrobial activity is an overinterpretation of the actual result, as a proliferation assay was used rather than a staining technique, where the surface was rinsed and the resulting solution was analysed via iterative optical density measurements following incubation. This measurement technique is an indirect method, which overlooks the true extent of cell-surface attachment, which could have been analysed using CLSM and SEM. Analysis of the incubation solution supernatant may not directly correlate to the number of cells attached to the interface. In this study, we used standard methods to determine the bactericidal activity of the nanostructured surfaces.
Conclusions
In this work, we reported the use of a simple, low-cost electrodeposition protocol for the fabrication of substrata that possess highly bactericidal gold nanospikes. Of the fabrication conditions investigated, the greatest bactericidal activity towards both Gram-negative and Gram-positive bacteria was found for substrates with multi-directional nanospikes. This was achieved using different concentrations of the Pb(CH3COO)2 precursor with a deposition time of 540 s. Altering the initial 6.8 mM concentration of the HAuCl4 precursor was found to result in the formation of a substratum with a diminished antibacterial activity.It has been shown that the bactericidal activity of the substrata is physical in nature, and hence the activity can be directly correlated to the geometric parameters of the substrates. Here, nanospikes with heights of ∼300 nm, cap-radii of ∼60 nm, base-radii of ∼90–140 nm, and an inter-spike spacing between 200–400 nm (considering the somewhat random spike orientations) were found to be more effective at killing the attaching bacteria than substrata with smaller or larger features. Every surface investigated in this study was found to be more biocidal than the smooth gold control substrates. The multi-directional structures investigated in this work could be used as a design parameter for incorporation into gold-based dental implants and next-generation intra-cortical electrode materials. Furthermore, the simple electrodeposition method described in this work could be used in the fabrication of large scale, non-planar surfaces, whereas many currently available fabrication techniques are limited to the creation of smaller flat surfaces.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors thank both the Microscopy and Microanalysis Facility (RMMF) and the MicroNano Research Facility (MNRF) at RMIT University for the use of their facilities. AE and RC thank Dr Madeleine Frances Dupont for her graphical abstract design. Funding from the Australian Research Council Industrial Transformation Research Hubs Scheme (Project Number IH130100017) is gratefully acknowledged.References
- A. Elbourne, R. J. Crawford and E. P. Ivanova, J. Colloid Interface Sci., 2017, 508, 603–616 CrossRef CAS PubMed.
- J. Hasan, R. J. Crawford and E. P. Ivanova, Trends Biotechnol., 2013, 31, 295–304 CrossRef CAS PubMed.
- H. W. Boucher, G. H. Talbot, J. S. Bradley, J. E. Edwards, D. Gilbert, L. B. Rice, M. Scheld, B. Spellberg and J. Bartlett, Clin. Infect. Dis., 2009, 48, 1–12 CrossRef PubMed.
- K. Bush, P. Courvalin, G. Dantas, J. Davies, B. Eisenstein, P. Huovinen, G. A. Jacoby, R. Kishony, B. N. Kreiswirth, E. Kutter, S. A. Lerner, S. Levy, K. Lewis, O. Lomovskaya, J. H. Miller, S. Mobashery, L. J. V. Piddock, S. Projan, C. M. Thomas, A. Tomasz, P. M. Tulkens, T. R. Walsh, J. D. Watson, J. Witkowski, W. Witte, G. Wright, P. Yeh and H. I. Zgurskaya, Nat. Rev. Microbiol., 2011, 9, 894–896 CrossRef CAS PubMed.
- D. Campoccia, L. Montanaro and C. R. Arciola, Biomaterials, 2006, 27, 2331–2339 CrossRef CAS PubMed.
- C. f. D. Control and Prevention, Antibiotic resistance threats in the United States, 2013, Centres for Disease Control and Prevention, US Department of Health and Human Services, 2013 Search PubMed.
- U. D. o. Health and H. Services, Antibiotic resistance threats in the United States, CDC, Atlanta, 2013 Search PubMed.
- H. C. Neu, Science, 1992, 257, 1064–1074 CrossRef CAS PubMed.
- B. Spellberg, R. Guidos, D. Gilbert, J. Bradley, H. W. Boucher, W. M. Scheld, J. G. Bartlett and J. J. Edwards, Clin. Infect. Dis., 2008, 46, 155–164 CrossRef PubMed.
- S. B. Levy and B. Marshall, Nat. Med., 2004, 10(12s), S122 CrossRef CAS PubMed.
- J. T. Jacob, E. Klein, R. Laxminarayan, Z. Beldavs, R. Lynfield, A. J. Kallen, P. Ricks, J. Edwards, A. Srinivasan, S. Fridkin, K. J. Rasheed, D. Lonsway, S. Bulens, R. Herrera, C. L. McDonald, J. Patel, B. Limbago, M. Bell and D. Cardo, Morbidity and Mortality Weekly Report, 2013, 62, 165–169 Search PubMed.
- N. Gupta, B. M. Limbago, J. B. Patel and A. J. Kallen, Clin. Infect. Dis., 2011, 53, 60–67 CrossRef PubMed.
- K. A. Rodvold and K. W. McConeghy, Clin. Infect. Dis., 2014, 58, S20–S27 CrossRef CAS PubMed.
- D. P. Linklater, S. Juodkazis and E. P. Ivanova, Nanoscale, 2017, 9, 16564–16585 RSC.
- E. Fadeeva, V. K. Truong, M. Stiesch, B. N. Chichkov, R. J. Crawford, J. Wang and E. P. Ivanova, Langmuir, 2011, 27, 3012–3019 CrossRef CAS PubMed.
- E. P. Ivanova, V. K. Truong, H. K. Webb, V. A. Baulin, J. Y. Wang, N. Mohammodi, F. Wang, C. Fluke and R. J. Crawford, Sci. Rep., 2011, 1, 165 CrossRef PubMed.
- E. P. Ivanova, V. K. Truong, J. Y. Wang, C. C. Berndt, R. T. Jones, I. I. Yusuf, I. Peake, H. W. Schmidt, C. Fluke, D. Barnes and R. J. Crawford, Langmuir, 2010, 26, 1973–1982 CrossRef CAS PubMed.
- S. Gangadoo, S. Chandra, A. Power, C. Hellio, G. S. Watson, J. A. Watson, D. W. Green and J. Chapman, J. Mater. Chem. B, 2016, 4, 5747–5754 RSC.
- C. James and R. Fiona, Adv. Eng. Mater., 2012, 14, B175–B184 CrossRef.
- C. M. Bhadra, V. K. Truong, V. T. Pham, M. Al Kobaisi, G. Seniutinas, J. Y. Wang, S. Juodkazis, R. J. Crawford and E. P. Ivanova, Sci. Rep., 2015, 5, 16817 CrossRef CAS PubMed.
- E. P. Ivanova, J. Hasan, H. K. Webb, G. Gervinskas, S. Juodkazis, V. K. Truong, A. H. F. Wu, R. N. Lamb, V. A. Baulin, G. S. Watson, J. A. Watson, D. E. Mainwaring and R. J. Crawford, Nat. Commun., 2013, 4, 2838 CrossRef PubMed.
- S. Pogodin, J. Hasan, V. A. Baulin, H. K. Webb, Vi K. Truong, T. H. P. Nguyen, V. Boshkovikj, C. J. Fluke, G. S. Watson, J. A. Watson, R. J. Crawford and E. P. Ivanova, Biophys. J., 2013, 104, 835–840 CrossRef CAS PubMed.
- S. M. Kelleher, O. Habimana, J. Lawler, B. O'Reilly, S. Daniels, E. Casey and A. Cowley, ACS Appl. Mater. Interfaces, 2015, 8, 14966–14974 CrossRef PubMed.
- T. Diu, N. Faruqui, T. Sjöström, B. Lamarre, H. F. Jenkinson, B. Su and M. G. Ryadnov, Sci. Rep., 2014, 4, 7122 CrossRef CAS PubMed.
- E. P. Ivanova, J. Hasan, H. K. Webb, V. K. Truong, G. S. Watson, J. A. Watson, V. A. Baulin, S. Pogodin, J. Y. Wang, M. J. Tobin, C. Löbbe and R. J. Crawford, Small, 2012, 8, 2489–2494 CrossRef CAS PubMed.
- J. Hasan, H. K. Webb, V. K. Truong, S. Pogodin, V. A. Baulin, G. S. Watson, J. A. Watson, R. J. Crawford and E. P. Ivanova, Appl. Microbiol. Biotechnol., 2013, 97, 9257–9262 CrossRef CAS PubMed.
- P. L. Denver, N. Huu Khuong Duy, M. B. Chris, J. Saulius and P. I. Elena, Nanotechnology, 2017, 28, 245301 CrossRef PubMed.
- C. M. Magin, S. P. Cooper and A. B. Brennan, Mater. Today, 2010, 13, 36–44 CrossRef CAS.
- C. Serrano, L. García-Fernández, J. P. Fernández-Blázquez, M. Barbeck, S. Ghanaati, R. Unger, J. Kirkpatrick, E. Arzt, L. Funk, P. Turón and A. del Campo, Biomaterials, 2015, 52, 291–300 CrossRef CAS PubMed.
- V. Truong, H. Webb, E. Fadeeva, B. Chichkov, A. Wu, R. Lamb, J. Wang, R. Crawford and E. Ivanova, Biofouling, 2012, 28, 539–550 CrossRef CAS PubMed.
- X. Li, Phys. Chem. Chem. Phys., 2016, 18, 1311–1316 RSC.
- C. D. Bandara, S. Singh, I. O. Afara, A. Wolff, T. Tesfamichael, K. Ostrikov and A. Oloyede, ACS Appl. Mater. Interfaces, 2017, 9(8), 6746–6760 CrossRef CAS PubMed.
- C. M. Bhadra, V. Khanh Truong, V. T. H. Pham, M. Al Kobaisi, G. Seniutinas, J. Y. Wang, S. Juodkazis, R. J. Crawford and E. P. Ivanova, Sci. Rep., 2015, 5, 16817 CrossRef CAS PubMed.
- D. E. Mainwaring, S. H. Nguyen, H. Webb, T. Jakubov, M. Tobin, R. N. Lamb, A. H. F. Wu, R. Marchant, R. J. Crawford and E. P. Ivanova, Nanoscale, 2016, 8, 6527–6534 RSC.
- J. Hasan, H. K. Webb, V. K. Truong, G. S. Watson, J. A. Watson, M. J. Tobin, G. Gervinskas, S. Juodkazis, J. Y. Wang, R. J. Crawford and E. P. Ivanova, Langmuir, 2012, 28, 17404–17409 CrossRef CAS PubMed.
- V. K. Truong, N. M. Geeganagamage, V. A. Baulin, J. Vongsvivut, M. J. Tobin, P. Luque, R. J. Crawford and E. P. Ivanova, Appl. Microbiol. Biotechnol., 2017, 1–8, DOI:10.1007/s00253-017-8205-9.
- E. P. Ivanova, J. Hasan, H. K. Webb, G. Gervinskas, S. Juodkazis, V. K. Truong, A. H. Wu, R. N. Lamb, V. A. Baulin and G. S. Watson, Nat. Commun., 2013, 4, 2838 CrossRef PubMed.
- F. Xue, J. Liu, L. Guo, L. Zhang and Q. Li, J. Theor. Biol., 2015, 385, 1–7 CrossRef CAS PubMed.
- P. M. Tsimbouri, L. Fisher, N. Holloway, T. Sjostrom, A. H. Nobbs, R. M. D. Meek, B. Su and M. J. Dalby, Sci. Rep., 2016, 6, 36857 CrossRef CAS PubMed.
- B. Plowman, S. J. Ippolito, V. Bansal, Y. M. Sabri, A. P. O'Mullane and S. K. Bhargava, Chem. Commun., 2009, 5039–5041, 10.1039/b910830k.
- Y. M. Sabri, S. J. Ippolito, J. Tardio, V. Bansal, A. P. O'Mullane and S. K. Bhargava, Sci. Rep., 2014, 4, 6741 CrossRef CAS PubMed.
- D. E. Mainwaring, S. H. Nguyen, H. Webb, T. Jakubov, M. Tobin, R. N. Lamb, A. H.-F. Wu, R. Marchant, R. J. Crawford and E. P. Ivanova, Nanoscale, 2016, 8, 6527–6534 RSC.
- D. Nečas and P. Klapetek, Gwyddion: an open-source software for SPM data analysis, 2012, vol. 10, p. 181 Search PubMed.
- L. Boulos, M. Prévost, B. Barbeau, J. Coallier and R. Desjardins, J. Microbiol. Methods, 1999, 37, 77–86 CrossRef CAS PubMed.
- E. Coyle Victoria, E. Kandjani Ahmad, M. Sabri Ylias and K. Bhargava Suresh, Electroanalysis, 2017, 29, 294–304 CrossRef.
- M. N. Dickson, E. I. Liang, L. A. Rodriguez, N. Vollereaux and A. F. Yee, Biointerphases, 2015, 10, 021010 CrossRef PubMed.
- L. E. Fisher, Y. Yang, M.-F. Yuen, W. Zhang, A. H. Nobbs and B. Su, Biointerphases, 2016, 11, 011014 CrossRef PubMed.
- P. W. May, M. Clegg, T. A. Silva, H. Zanin, O. Fatibello-Filho, V. Celorrio, D. J. Fermin, C. C. Welch, G. Hazell, L. Fisher, A. Nobbs and B. Su, J. Mater. Chem. B, 2016, 4, 5737–5746 RSC.
- C. M. Bhadra, M. Werner, V. A. Baulin, V. K. Truong, M. A. Kobaisi, S. H. Nguyen, A. Balcytis, S. Juodkazis, J. Y. Wang, D. E. Mainwaring, R. J. Crawford and E. P. Ivanova, Nano-Micro Lett., 2018, 10, 36 CrossRef.
- D. H. Nguyen, V. T. Pham, M. Al Kobaisi, C. Bhadra, A. Orlowska, S. Ghanaati, B. M. Manzi, V. A. Baulin, S. Joudkazis and P. Kingshott, Langmuir, 2016, 32, 10744–10751 CrossRef CAS PubMed.
- V. T. H. Pham, V. K. Truong, A. Orlowska, S. Ghanaati, M. Barbeck, P. Booms, A. J. Fulcher, C. M. Bhadra, R. Buividas, V. Baulin, C. J. Kirkpatrick, P. Doran, D. E. Mainwaring, S. Juodkazis, R. J. Crawford and E. P. Ivanova, ACS Appl. Mater. Interfaces, 2016, 8, 22025–22031 CrossRef CAS PubMed.
- A. Gristina, Science, 1987, 237, 1588–1595 CrossRef CAS PubMed.
- G. S. Watson, D. W. Green, L. Schwarzkopf, X. Li, B. W. Cribb, S. Myhra and J. A. Watson, Acta Biomater., 2015, 21, 109–122 CrossRef CAS PubMed.
- D. W. Green, K. K.-H. Lee, J. A. Watson, H.-Y. Kim, K.-S. Yoon, E.-J. Kim, J.-M. Lee, G. S. Watson and H.-S. Jung, Sci. Rep., 2017, 7, 41023 CrossRef CAS PubMed.
- X. Li, G. S. Cheung, G. S. Watson, J. A. Watson, S. Lin, L. Schwarzkopf and D. W. Green, Nanoscale, 2016, 8, 18860–18869 RSC.
- E. P. Ivanova, S. H. Nguyen, H. K. Webb, J. Hasan, V. K. Truong, R. N. Lamb, X. Duan, M. J. Tobin, P. J. Mahon and R. J. Crawford, PLoS One, 2013, 8, e67893 CrossRef CAS PubMed.
- A. Sakamoto, Y. Terui, C. Horie, T. Fukui, T. Masuzawa, S. Sugawara, K. Shigeta, T. Shigeta, K. Igarashi and K. Kashiwagi, FEMS Microbiol. Lett., 2014, 361, 10–16 CrossRef CAS PubMed.
- D. P. Linklater, S. Juodkazis, S. Rubanov and E. P. Ivanova, ACS Appl. Mater. Interfaces, 2017, 9, 29387–29393 CrossRef CAS PubMed.
- K. Nowlin, A. Boseman, A. Covell and D. LaJeunesse, J. R. Soc., Interface, 2015, 12(102), 20140999 CrossRef.
- X. Li and T. Chen, Phys. Rev. E, 2016, 93, 052419 CrossRef PubMed.
- K. Bazaka, R. J. Crawford and E. P. Ivanova, Biotechnol. J., 2011, 6, 1103–1114 CrossRef CAS PubMed.
- A. I. Hochbaum and J. Aizenberg, Nano Lett., 2010, 10, 3717–3721 CrossRef CAS PubMed.
- A. K. Epstein, A. I. Hochbaum, K. Philseok and J. Aizenberg, Nanotechnology, 2011, 22, 494007 CrossRef CAS PubMed.
- L. C. Hsu, J. Fang, D. A. Borca-Tasciuc, R. W. Worobo and C. I. Moraru, Appl. Environ. Microbiol., 2013, 79, 2703–2712 CrossRef CAS PubMed.
- K. K. Chung, J. F. Schumacher, E. M. Sampson, R. A. Burne, P. J. Antonelli and A. B. Brennan, Biointerphases, 2007, 2, 89–94 CrossRef CAS PubMed.
- S. Wu, F. Zuber, J. Brugger, K. Maniura-Weber and Q. Ren, Nanoscale, 2016, 8, 2620–2625 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8na00124c |
| This journal is © The Royal Society of Chemistry 2019 |
