 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Nickel(II)-promoted specific hydrolysis of zinc finger proteins†
Agnieszka
Belczyk-Ciesielska
a,
Brigitta
Csipak
b,
Bálint
Hajdu
b,
Aleksandra
Sparavier
a,
Masamitsu N.
Asaka
c,
Kyosuke
Nagata
c,
Béla
Gyurcsik
 *b and
Wojciech
Bal
*b and
Wojciech
Bal
 *a
*a
aInstitute of Biochemistry and Biophysics, Polish Academy of Sciences, Pawinskiego 5a; 02-106, Warsaw, Poland. E-mail: wbal@ibb.waw.pl; Fax: +48-226584636
bDepartment of Inorganic and Analytical Chemistry, University of Szeged, Dóm tér 7, Szeged H-6720, Hungary. E-mail: gyurcsik@chem.u-szeged.hu; Fax: +36 62544340
cNagata Special Laboratory, Faculty of Medicine, University of Tsukuba, 1-1-1 Tennodai, Tsukuba 305-8575, Japan
First published on 4th July 2018
Abstract
In this work we demonstrate that the previously described reaction of sequence specific Ni(II)-dependent hydrolytic peptide bond cleavage can be performed in complex metalloprotein molecules, such as the Cys2His2 zinc finger proteins. The cleavage within a zinc finger unit possessing a (Ser/Thr)-X-His sequence is not hindered by the presence of the Zn(II) ions. It results in loss of the Zn(II) ion, oxidation of the SH groups and thus, in a collapse of the functional structure. We show that such natural Ni(II)-cleavage sites in zinc finger domains can be edited out without compromising the DNA binding specificity. Inserting a Ni(II)-susceptible sequence between the edited zinc finger and an affinity tag allows for removal of the latter sequence by Ni(II) ions after the protein purification. We have shown that this reaction can be executed even when a metal ion binding N-terminal His-tag is present. The cleavage product maintains the native zinc finger structure involving Zn(II) ions. Mass spectra revealed that a Ni(II) ion remains coordinated to the hydrolyzed protein product through the N-terminal (Ser/Thr)-X-His tripeptide segment. The fact that the Ni(II)-dependent protein hydrolysis is influenced by the Ni(II) concentration, pH and temperature of the reaction provides a platform for novel regulated DNA effector design.
Significance to metallomicsThe present study shows that Cys2His2 zinc finger proteins including (S/T)XH sequence can be hydrolytically cleaved by Ni(II) ions. The cleavage involving any of the histidines bound to Zn(II)-ions abolishes the functional structure of the affected finger. Designed Ni(II) sensitive sequences however, allow for removal of affinity purification tags (even metal ion-binding oligo-His tag) or other regulatory fusion protein elements. Our study contributes to understanding of the effect of Ni(II) ions on such a complex and sensitive metalloprotein. At the same time it opens a perspective for novel zinc finger fusion protein design which can be activated/regulated by Ni(II)-induced cleavage. |
Introduction
Transcription factors of the Cys2His2 zinc finger (ZF) family control gene functions in living organisms by binding to specific nucleic acid sequences.1,2 The canonical sequence of a finger unit, able to form a ββα secondary structure with a tetrahedral metal ion binding site, was defined on the basis of a library of native sequences as (Tyr,Phe)-X-![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[y with combining low line]](https://www.rsc.org/images/entities/char_0079_0332.gif)
![[s with combining low line]](https://www.rsc.org/images/entities/char_0073_0332.gif) -X2,4-
-X2,4-![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[y with combining low line]](https://www.rsc.org/images/entities/char_0079_0332.gif)
![[s with combining low line]](https://www.rsc.org/images/entities/char_0073_0332.gif) -X3-Phe-X5-Leu-X2-
-X3-Phe-X5-Leu-X2-![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif)
![[i with combining low line]](https://www.rsc.org/images/entities/char_0069_0332.gif)
![[s with combining low line]](https://www.rsc.org/images/entities/char_0073_0332.gif) -X3-5-
-X3-5-![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif)
![[i with combining low line]](https://www.rsc.org/images/entities/char_0069_0332.gif)
![[s with combining low line]](https://www.rsc.org/images/entities/char_0073_0332.gif) (Zn(II) binding residues underlined).3 ZF proteins possess a modular assembly in which each ZF unit recognizes three subsequent nucleotides.1,2 This allows for targeting DNA sequences of a 9–12 base pair length by a cascade of such units in a ZF protein.4–9 Artificial ZF proteins with desired DNA specificities can be obtained by altering the amino acids in the α-helical DNA base recognition region according to the well-known methodologies.10–14 A popular approach is to utilize effector molecules (i.e. ligand or metal-binding regulatory sequences or an endonuclease catalytic domain) linked to ZF proteins, thereby providing an opportunity to generate artificial transcription factors or enzymes for specific DNA actions.2,15–25
(Zn(II) binding residues underlined).3 ZF proteins possess a modular assembly in which each ZF unit recognizes three subsequent nucleotides.1,2 This allows for targeting DNA sequences of a 9–12 base pair length by a cascade of such units in a ZF protein.4–9 Artificial ZF proteins with desired DNA specificities can be obtained by altering the amino acids in the α-helical DNA base recognition region according to the well-known methodologies.10–14 A popular approach is to utilize effector molecules (i.e. ligand or metal-binding regulatory sequences or an endonuclease catalytic domain) linked to ZF proteins, thereby providing an opportunity to generate artificial transcription factors or enzymes for specific DNA actions.2,15–25
Previously we reported on Ni(II)-induced peptide bond hydrolysis that occurs at the N-terminal side of Ser or Thr residues in peptides,26–36 designed recombinant proteins37,38 and flexible regions of native proteins37,39 bearing X-(Ser/Thr)-X-His-X sequences (X being any amino acid except for Cys or Pro before or after Ser/Thr). The method found application in the affinity tag removal from fusion proteins by Ni(II) ions.37,38 The cleavage is site specific and sequence selective making this method unique among other approaches using metal ions.31,37,40 The molecular mechanism of this reaction was proposed earlier31,35 and is depicted in Fig. S1 (ESI†). The formation of a square planar Ni(II) complex is strictly required for the reaction: it contains the Ni(II) ion bound to the imidazole nitrogen of the His residue and three preceding amide nitrogens.31,35
Various human transcription factors contain potential Ni(II) cleavage sites within their Cys2His2 ZF units. The 27 amino acid long peptide, representing the 3rd ZF unit of the Sp1 human transcription factor was shown to be cleaved by Ni(II) ions even though the His residue crucial for the reaction was initially complexed to the structural Zn(II) ion.32 The Zn(II) binding site and the structure of the ZF unit was abolished in the reaction. Naturally occurring Ni(II)-cleavage sites, which are present in nearly 25% of human Cys2His2 ZF sequences32 can thus be considered potentially susceptible to hydrolysis upon the exposure to Ni(II) ions naturally present in human tissues.41 Nickel is absorbed through the diet, skin, lungs and then taken up by cells.42,43 By analogy with the hydrolytic cleavage of histone H2A observed in Ni(II)-exposed cell cultures, endogenous nickel present in human cells may damage ZFs as part of its toxicity.
The knowledge, summarized above, inspired us to challenge sequence specific Ni(II)-induced peptide bond cleavage within a complex metalloprotein such as a Cys2His2-type classical ZF array consisting of three ZF units and also possessing a Ni(II)-cleavable N-terminal deka-His fusion tag. Our aim was to verify that the binding of Zn(II) ions is unaffected by the presence of the Ni(II) ions in the uncleaved finger units, i.e. they are not replaced by Ni(II) ions and the oxidation of cysteines in the binding sites is not catalysed by Ni(II) ions. We analysed and optimized the cleavage of two ZF proteins by Ni(II) ions at a single peptide bond between the protein and the affinity tag, and investigated the structural integrity, as well as the Zn(II) and DNA binding properties of the products by spectrometric and electrophoretic methods.
Materials and methods
Gene construction
Two ZF proteins, denoted by P-1MEY and P-1MEY# were investigated, both consisting of three ZF units. The P-1MEY protein is similar to that included in the ZF protein/DNA crystal structure, with 1MEY PDB code,44 and the P-1MEY# ZF protein is its modified version as described below. The gene of the P-1MEY protein was ordered as a full length DNA with a stop codon (IDT Japan Ltd) and ligated into a pET-16b vector between the NdeI and BamHI sites. The gene of the P-1MEY# protein was constructed starting from the above DNA sequence by the modified QuikChange™ site-directed mutagenesis method (QCM, Stratagene, La Jolla, CA) using the following primers: 1MEY#-F (5′-tcgccatcagcgtacccatac-3′) and 1MEY#-R (5′-accaggttatcgctgcgggaaaatgacttc-3′) according to Scheme S1 (ESI†). Consequently, a His was replaced with Asn (H99N) and a Ser with Val (S101V). After the amplification of the DNA with KOD-FX DNA polymerase (TOYOBO, Japan), the resulted DNA fragments were treated by T4 polynucleotide kinase (TOYOBO, Japan) and T4 ligase in the ligation high premix (TOYOBO, Japan). DH5α cells were transformed with the resulted plasmid, and after the DNA purification the success of the mutagenesis was verified by a standard sequencing procedure.Protein expression and purification
The P-1MEY and P-1MEY# proteins were expressed in BL21 (DE3) E. coli cells, which were cultured to OD of ca. 0.3, then the protein expression was induced by 0.2 mM IPTG at 20 °C for 18 hours. Cells were collected and resuspended in a buffer containing 500 mM NaCl, 100 mM HEPES (pH 8.2), and 10 mM imidazole, and then sonicated. The pET-16b-P-1MEY and pET-16b-P-1MEY# plasmids permitted introduction of the N-terminal deka-His tag and thus, the expressed proteins were subjected to Ni(II)-affinity chromatography as the main purification step. Applying an imidazole concentration gradient in the eluent, most of the target proteins were obtained at 300 mM imidazole as shown in Fig. S2a (ESI†). The Zn(II) load of the ZF proteins was verified by EDTA titration of the ZF proteins monitored by circular dichroism spectroscopy (Fig. S2b, ESI†). The Zn(II) contents of the purified ZF proteins were 3.0 ± 0.2 equivalents/protein molecule.Protein hydrolysis by Ni(II) ions
30–40 μM P-1MEY or P-1MEY# proteins were incubated with 400 μM Ni(II) (∼10-fold molar excess) in a 100 mM HEPES buffer with such an addition of Zn(II) that a 10–100 μM final concentration of Zn(II) ions in excess of the protein remained in solution. The conditions of the hydrolytic experiments varied between 50 °C, pH 8.2 and 37 °C, pH 7.4 (i.e. close to physiological conditions). Control reaction mixtures without Ni(II) were incubated for 72 hours under the same conditions. All samples were prepared and then incubated in low adsorption 1.5 ml tubes (LowBind, Eppendorf), using a thermoblock (TB-941U, JW Electronic).20 μl samples were collected from reaction mixtures at given time points and the reactions were terminated by acidification (2% TFA), as performed and justified before.35 Samples were analysed by polyacrylamide gel-electrophoresis (Tricine-SDS-PAGE45), and in parallel by HPLC (Breeze, Waters) using an ACE C18 analytical column. The eluting solvent A was 0.1% TFA/water and solvent B was 0.1% TFA/90% acetonitrile.
Electrospray ionization mass spectrometry (ESI-MS)
The samples investigated by ESI-MS (Premier, Q-Tof1, Waters) were either obtained from the HPLC separation of the cleavage products as described above (P-1MEY), or taken directly from the reaction mixture (pH 8.2) with or without the sample acidification to pH <4.0 (P-1MEY#). The samples were analysed following a buffer exchange (to 100 mM NH4HCO3, pH 8.1) or desalting on a RP-C18 pre-column (Waters).Circular dichroism (CD) measurements
CD measurements were performed on a JASCO J-815 spectropolarimeter in quartz cuvettes (d = 2 mm or 0.1 mm). The spectra were recorded in the 180–300 nm range, using 0.5 nm resolution, 2 s response time, and scanning speed of 20 nm min−1. The samples were taken directly from the hydrolytic reaction mixture at the beginning and after the termination of the reaction. The solvent was exchanged to 5 mM phosphate buffer (pH 7.4) prior to the CD experiment using an Amicon Ultra-0.5 Centrifugal Filter Unit with Ultracel-3 membrane having a nominal molecular weight limit of 3 kDa.Electrophoretic mobility shift assay (EMSA)
The 1MEY+2bp oligonucleotide probe of the sequence 5′-CG![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ACTTTTGT
ACTTTTGT![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) CG-3′, which included the 1MEY target sequence (underlined) and the ZFP#2+2bp probe of the sequence 5′-CGGGGATAGGCACTTTTGTGCCTATCCCCG-3′ were used for EMSA experiments. These oligonucleotides were labeled with [γ-32P]ATP by T4 polynucleotide kinase (TOYOBO). The labeled oligonucleotides were self-annealed, to yield the double strand sequence stabilized by the TTTT loop, and then purified from native PAGE, and used as probes. EMSA experiments were performed in 10 μl of a 60 mM HEPES buffer, pH 8.2, containing 10% (V/V) glycerol, 45 mM NaCl, 32P-end-labeled probe (15 fmol) and P-1MEY or P-1MEY# protein at a concentration giving rise to a protein excess between 0 and 100 fold, as indicated in Fig. 1. The reaction mixtures were separated on a 6% (m/V) polyacrylamide gel containing 12.5 mM Tris buffer and 96 mM glycine. Radiolabeled probes were visualized by a BAS2000 imager. Further EMSA experiments were carried out with a DNA probe including four tandem copies (underlined) of the favourable 1MEY binding sequence. It was constructed by the annealing and subsequent prolongation of the following deoxy-oligonucletide primers to yield a 72 bp DNA:
CG-3′, which included the 1MEY target sequence (underlined) and the ZFP#2+2bp probe of the sequence 5′-CGGGGATAGGCACTTTTGTGCCTATCCCCG-3′ were used for EMSA experiments. These oligonucleotides were labeled with [γ-32P]ATP by T4 polynucleotide kinase (TOYOBO). The labeled oligonucleotides were self-annealed, to yield the double strand sequence stabilized by the TTTT loop, and then purified from native PAGE, and used as probes. EMSA experiments were performed in 10 μl of a 60 mM HEPES buffer, pH 8.2, containing 10% (V/V) glycerol, 45 mM NaCl, 32P-end-labeled probe (15 fmol) and P-1MEY or P-1MEY# protein at a concentration giving rise to a protein excess between 0 and 100 fold, as indicated in Fig. 1. The reaction mixtures were separated on a 6% (m/V) polyacrylamide gel containing 12.5 mM Tris buffer and 96 mM glycine. Radiolabeled probes were visualized by a BAS2000 imager. Further EMSA experiments were carried out with a DNA probe including four tandem copies (underlined) of the favourable 1MEY binding sequence. It was constructed by the annealing and subsequent prolongation of the following deoxy-oligonucletide primers to yield a 72 bp DNA:
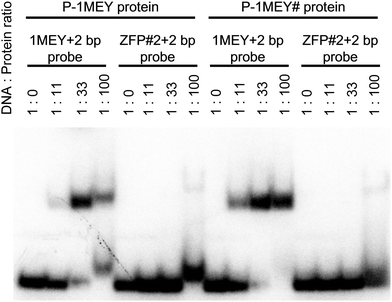 | ||
| Fig. 1 Electrophoretic mobility shift assay of the P-1MEY and P-1MEY# proteins with the specific (1MEY+2 bp) and non-specific (ZFP#2+2 bp) DNA probes. | ||
5′-GGCGAATTC![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) TGCTTA
TGCTTA![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) GAGCTCGAG-3′
GAGCTCGAG-3′
5′-GGCGAATTC![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) TAAGCA
TAAGCA![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) GAGCTCGAG-3′. The DNA samples (cfinal = 4.0 μM) were incubated with increasing amounts of the Ni(II)-cleaved P-1MEY# ZF protein from 1
GAGCTCGAG-3′. The DNA samples (cfinal = 4.0 μM) were incubated with increasing amounts of the Ni(II)-cleaved P-1MEY# ZF protein from 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 1
1 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 molar ratio at pH 7.4 in a 100 mM HEPES buffer. The reaction mixtures were separated on a 2% (m/V) agarose gel and the DNA was visualized by the intercalated ethidium ion fluorescence using an UviDoc gel documentation system.
4 molar ratio at pH 7.4 in a 100 mM HEPES buffer. The reaction mixtures were separated on a 2% (m/V) agarose gel and the DNA was visualized by the intercalated ethidium ion fluorescence using an UviDoc gel documentation system.
Results and discussion
Choice and redesign of the ZF protein
The P-1MEY ZF protein sequence is based on the consensus sequence CP-1 derived by Berg et al.,46 and confirmed by a recent study,47 with the exception of the amino acids responsible for DNA recognition. It is a well studied protein suitable for our model cleavage experiments. The sequence was derived from a ZF protein (denoted as P-1MEY-PDB) previously structurally characterized in the complex with its cognate DNA (PDB code 1MEY).44 The amino acid sequences of the P-1MEY and mutant P-1MEY# proteins investigated by us are aligned to the P-1MEY-PDB protein in Scheme 1. The core sequence of P-1MEY responsible for the functional structure and DNA recognition was kept intact. The sequence was modified here by the deka-His tag at the N-terminus for the purpose of affinity purification. Further amino acids, following the affinity tag sequence were encoded by the pET-16b plasmid: S![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) contains the underlined Ni(II)-sensitive sequence and IEGR is the Factor Xa protease (NEB) recognition sequence. Short extensions (additional amino acids compared to the P-1MEY-PDB) at both termini of the P-1MEY protein sequence (see Scheme 1) can be derived from the consensus sequence as they are found in the Zinc Finger Tools program used for protein redesign (vide infra).13
contains the underlined Ni(II)-sensitive sequence and IEGR is the Factor Xa protease (NEB) recognition sequence. Short extensions (additional amino acids compared to the P-1MEY-PDB) at both termini of the P-1MEY protein sequence (see Scheme 1) can be derived from the consensus sequence as they are found in the Zinc Finger Tools program used for protein redesign (vide infra).13
 | ||
| Scheme 1 Amino acid sequences of ZF proteins applied in this study in aligned with related ZF sequences. P-1MEY-PDB: the protein used in the crystallization study;44 P-1MEY: the original protein extended with a deka-His tag sequence for the purification purpose; P-1MEY#: the redesigned protein with the ATCUN motif masked by the single Ni(II)-hydrolysable N-terminal sequence; ZF Tools: the protein predicted by the Zinc Finger Tools program13 to recognize the specific 5′-GAGGCAGAA-3′ DNA sequence; ZF Tools (FS): the fixed sequences applied by the Zinc Finger Tools program without the variable regions responsible for DNA recognition (denoted by XXXXXXX in each finger unit); Sp1: a ZF protein sequence having two Ni(II) sensitive sites in the 3rd finger unit (similarly to the P-1MEY protein); CP-1 (2015): the 26 amino acids long consensus ZF peptide sequence found by Goldberg et al.;47 CP-1: the 26 amino acids long consensus ZF peptide sequence established and investigated by Berg et al.46 Ni(II)-hydrolytic sites are underlined and bolded, the conserved Zn(II)-binding sites are marked yellow, the modifications suggested by Zinc Finger Tools are marked light blue, and the experimentally performed mutations are marked magenta. | ||
P-1MEY contains three X(S/T)XHX sequence motifs that can undergo hydrolytic cleavage upon Ni(II)-binding. Two of them are located close to each other within the DNA recognition region of the 3rd ZF unit (similarly to the 3rd ZF unit of Sp1 – Scheme 1), while the third sequence is located between the N-terminal deka-His tag and the ZF protein sequence. Based on the experiments with the Sp1 C-terminal peptide32 the possible Ni(II)-dependent hydrolysis within the 3rd ZF unit of P-1MEY may be expected to affect the properties of this finger. Therefore, we redesigned P-1MEY ZF protein into P-1MEY# with a single Ni(II)-sensitive sequence at the N-terminus, by editing out the hydrolysable sequences in the 3rd finger.
Since the necessary substitutions fall within the DNA recognition sequence, the Zinc Finger Tools program13 was utilized to maintain the DNA binding specificity. For the target DNA sequence derived from the 1MEY/DNA structure,44 the program yielded a ZF protein sequence differing at various sites from the original one (Zinc Finger Tools sequence in Scheme 1). To obtain the P-1MEY# protein we introduced only the minimal changes limited to the 3rd ZF unit involving the His to Asn (H99N) and Ser to Val (S101V) substitutions (Scheme 1; see the Materials and methods section for details).
The DNA-binding specificity was maintained in the redesigned ZF protein
The DNA binding properties of the P-1MEY-PDB ZF protein were previously studied and the 5′-GAGGCAGAA-3′ DNA sequence included in the 1MEY crystal structure was considered as a favourable binding site of the P-1MEY protein.44 After the expression and purification procedures the DNA binding specificity of both P-1MEY and the modified P-1MEY# ZF proteins was verified by electrophoretic mobility shift assays (EMSA) using various short DNA molecules. Fig. 1 shows that the modifications at the two termini did not affect the DNA binding specificity of the P-1MEY protein, and that the P-1MEY# mutant has recognition properties comparable to the P-1MEY protein. P-1MEY# preferred to bind the target DNA similarly to the original protein (see also Fig. S3, ESI†). Besides, P-1MEY# was less prone to form non-specific complexes at a 100-fold protein excess with the ZFP#2+2 bp non-specific DNA probe, while under the same conditions the addition of P-1MEY resulted in a shifted DNA band characteristic for this non-specific DNA interaction. A small amount of such non-specific complex between the P-1MEY protein and the 1MEY+2 bp specific DNA probe was observed only at a high protein excess.Monitoring the Ni(II)-induced hydrolysis of the ZF proteins
We verified the Ni(II)-cleavage for all susceptible sequences: three in P-1MEY (SGH, SDH, SRH) and one in P-1MEY# protein (SGH). The hydrolysis reactions were monitored by Tricine-SDS-PAGE and products were analysed by HPLC.The imidazole groups of the N-terminal deka-His tag have the ability to complex Ni(II) ions and compete in this respect with the His residue from the hydrolytic sequence X-(Ser/Thr)-X-His-X. To overcome this effect we used at least a 10-fold molar excess of Ni(II), as demonstrated before to be effective for protein cleavage.37–39 The hydrolysis removes the His-tag, while the catalytic Ni(II) ion remains coordinated to the (Ser/Thr)-X-His motif of the protein (C-terminal product).
The CP-1 ZF peptide showed a high affinity towards Zn(II) ions (Kd ∼ 2 pM),46 and a recent re-investigation of ZF affinities for Zn(II) ions indicated even lower Kd values.48 Accordingly, P-1MEY and P-1MEY# proteins were purified in their zinc-loaded forms. Nevertheless, we used an additional amount of Zn(II) ions (final excess: 10–100 μM) in each reaction mixture to decrease the possibility of the Zn(II)/Ni(II) substitution in ZF domains during the reaction. Metal ion substitution was suggested to change the specificity of DNA recognition due to alteration of the ZF structure, resulting from different coordination geometries of these two metal ions.49 Change of the protein structure was also supported in the presence of other metal ions, such as Cd(II) and Hg(II),50 and the ZF cleavage was initiated by a dinuclear Pt(II) complex initially replacing the Zn(II) ions from the thiolate sites.51
The structural integrity of P-1MEY and P-1MEY# proteins in the presence and absence of Ni(II) ions was verified by CD spectroscopy upon iterative heating and cooling (from 10 °C to 50 °C and back) of solutions adequate for the hydrolytic experiments. The collapse of the structure of ZF proteins at high temperatures would result in a disordered structure and thus, in an altered spectral pattern, similar to that obtained for the apo-protein (Fig. S2b, ESI†). It has to be noted here that this phenomenon was observed upon addition of Cu(II) ions (unpublished results), but it was excluded for Ni(II), as no significant changes were detected in the presence of the latter metal ion in the intensity of the CD signal, exemplified by the P-1MEY protein in Fig. S4 (ESI†).
Typically, three bands were observed in SDS PAGE after the hydrolysis of the P-1MEY protein in the 10–15 kDa range, corresponding to the substrate and the protein cleavage products, and a further one appeared in the low molecular weight range, corresponding to the short terminal peptide fragments (Fig. S5, ESI†). The presence of distinct bands suggested that specific cleavages occurred at least at two of three potential Ni(II) cleavage sites. The sequence redesign process allowed P-1MEY# to be converted into a single high molecular weight hydrolysis product (Fig. 2).
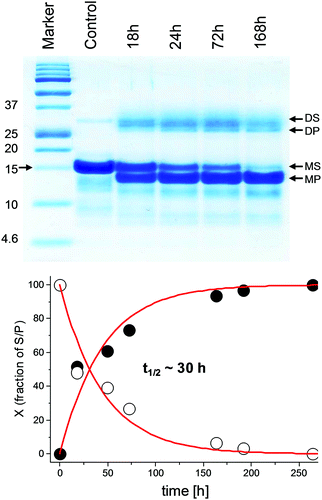 | ||
| Fig. 2 Top: The electrophoretic analysis of the Ni(II)-dependent hydrolysis of P-1MEY# ZF protein at 37 °C and pH 8.2; incubation times in hours (h), S – substrate, P – product; D and M stand for the dimer and monomer, respectively. Bottom: Kinetic plots of substrate formation (filled circles) and product depletion (empty circles) in the hydrolysis reaction based on the densitometric analysis of the Tricine-SDS-PAGE image considering only the bands of the monomeric protein fractions. The red line represents the fitting to the 1st order rate law. | ||
According to the kinetic analysis, the hydrolysis of the P-1MEY# mutant protein occurred more slowly than that of the P-1MEY (t1/2 = 30 h vs. 14 h at 37 °C and pH 8.2). The control experiment without the Ni(II) ions revealed only a marginal degradation of the protein after 72 h of incubation at the elevated temperature and pH.
The Ni(II)-induced hydrolysis of P-1MEY and P-1MEY# ZFs was investigated at 50 or 37 °C and pH 8.2 or 7.4 to demonstrate the significant effect of the temperature and pH on the reaction rate. The hydrolysis experiments repeated under physiological-like conditions at 37 °C and pH 7.4 indicated that the reactivity pattern was maintained, although the reaction rate decreased significantly for both P-1MEY and P-1MEY#. The densitometric results obtained from SDS PAGE gel analysis are shown in Fig. S6 (ESI†). These data were used to calculate reaction rate constants. The comparison of t1/2 values in the 1st order kinetic approximation revealed their strong pH dependence. We observed a 9-fold increase of the reaction rate between pH 7.4 and 8.2 at 37 °C (t1/2 of ∼11 d, and ∼ 30 h, respectively). The increase of the temperature from 37 °C to 50 °C resulted in a further acceleration of the reaction rate (t1/2 ∼ 7.5 h). The pH dependence of the 1st order rate constant for the Ni(II)-assisted hydrolysis reaction has a sigmoidal shape. The reaction rate is maximal at pH above pH 8.2, where the square planar (4N) complex predominates. At lower pH (pH 7.4) the extent of the formation of the active 4N complex is much lower: 20–80 times less, compared to pH 8.2.31,52,53
Verification of the cleavage specificity and binding of the metal ions by MS
The resolution of the gel electrophoresis is not sufficient to precisely identify the hydrolytic cleavage sites. Therefore, we analysed the reaction mixtures by ESI-MS. The experimentally determined masses of the substrate proteins and their Ni(II)-hydrolysis products agreed well with the theoretical predictions. Among the P-1MEY products we identified short GHHHHHHHHHHS and![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif)
![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif)
![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) QRTHTGKKTS N- and C-terminal peptides, respectively. The peptide resulting from the cleavage at the
QRTHTGKKTS N- and C-terminal peptides, respectively. The peptide resulting from the cleavage at the ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif)
![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif)
![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) site,
site, ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif)
![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif)
![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) L
L![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif)
![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif)
![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) QRTHTGKKTS, was not detected as the cleavage within the
QRTHTGKKTS, was not detected as the cleavage within the ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif)
![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif)
![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) sequence was much faster than that at SDH, as predicted.32 However, the
sequence was much faster than that at SDH, as predicted.32 However, the ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif)
![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif)
![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) L tetrapeptide was detected, confirming that both SDH and the SRH cleavage sites were hydrolytically active. The protein product after cleavage at both SGH and SDH sites was also found (Table 1 and Table S1, ESI†). These data revealed that the three Ni(II)-susceptible sequences exerted different activities during the reaction with the Ni(II) ion. The hydrolysis at the SDH site was the least favorable one, consistently with previous studies on model peptides.30–32 The mass of the final P-1MEY product of 9720 Da reflects the coordination of 2 × Zn(II) and 1 × Ni(II) ions and the oxidation of cysteines within the destroyed 3rd ZF unit, which lost the Zn(II) ion binding ability after hydrolysis.
L tetrapeptide was detected, confirming that both SDH and the SRH cleavage sites were hydrolytically active. The protein product after cleavage at both SGH and SDH sites was also found (Table 1 and Table S1, ESI†). These data revealed that the three Ni(II)-susceptible sequences exerted different activities during the reaction with the Ni(II) ion. The hydrolysis at the SDH site was the least favorable one, consistently with previous studies on model peptides.30–32 The mass of the final P-1MEY product of 9720 Da reflects the coordination of 2 × Zn(II) and 1 × Ni(II) ions and the oxidation of cysteines within the destroyed 3rd ZF unit, which lost the Zn(II) ion binding ability after hydrolysis.
| Protein | Cleavage site | M W experimental (Da) | M W calculated (Da) all-SS/all-SH |
|---|---|---|---|
| a Separation of the P-1MEY cleavage products by HPLC was performed under acidic conditions thus, the products did not contain complexed metal ions. b (S) and (L) denote the small and large protein products of the hydrolysis at each site. n.d., not detected. For the sequences see Table S1 (ESI). c HPLC peak broadened due to the presence of various oxidation products. The comparison of the calculated and measured masses of the central large (L) protein portions yielded a 6 Da difference, corresponding to oxidation of Cys residues to disulfides. d No oxidation products were detected when the sample was directly taken from the reaction mixture and immediately acidified. e The protein sample was not acidified. The calculated mass reflected the coordination of 3 × Zn(II) and 1 × Ni(II) ions. f Oxidation of Cys residues within the destroyed finger unit was detected when the sample was directly taken from the reaction mixture and immediately acidified. g The protein sample was not acidified. The calculated mass reflected the coordination of 2 × Zn(II) and 1 × Ni(II) ions. Oxidation of cysteines within the destroyed finger unit was detected. | |||
| P-1MEYa | — | 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 012 012 |
13006.3/13012.3 |
| SRH (L)b | 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500c 500c |
11500.6/11506.6 | |
| SGH (L) | 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 496d 496d |
11490.7/11496.7 | |
11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 743e 743e |
11743.5 | ||
| SGH + SRH (L) | 9986 | 9985.1/9991.1 | |
| SGH + SDH (L) | 9532 | 9532.6/9538.6 | |
| 9536f | 9532.6/9538.6 | ||
| 9720g | 9722,1 | ||
| SRH (S) | 1524 | 1523.6 | |
| SGH (S) | 1534 | 1533.5 | |
| SDH (S) | n.d. | 1976.1 | |
| P-1MEY# | — | 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 001 001 |
13001.3 |
| SGH (L) | 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 487d 487d |
11485.8 | |
| SGH (L) | 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 733e 733e |
11732.6 | |
The ESI MS analysis of the P-1MEY# reaction products at various pH confirmed the identity of the cleaved protein and its metal binding status. The mass obtained under acidic conditions corresponded well to that of the cleaved apoprotein. The mass of the hydrolyzed P-1MEY# protein in a buffered solution was 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 733 Da (Table S1, ESI†). As the calculated mass of the apoprotein is 11485.8 Da, it proves the coordination of three Zn(II) ions within the ZF units and a Ni(II) ion at the N-terminal ATCUN motif formed by the cleavage reaction (the calculated mass of the holoprotein is 11733.6 Da). The thiolate oxidation detected within the 3rd finger of the original P-1MEY was not observed here due to the fact that the Zn(II) binding sites of the redesigned P-1MEY# protein remained intact during the hydrolytic process.
733 Da (Table S1, ESI†). As the calculated mass of the apoprotein is 11485.8 Da, it proves the coordination of three Zn(II) ions within the ZF units and a Ni(II) ion at the N-terminal ATCUN motif formed by the cleavage reaction (the calculated mass of the holoprotein is 11733.6 Da). The thiolate oxidation detected within the 3rd finger of the original P-1MEY was not observed here due to the fact that the Zn(II) binding sites of the redesigned P-1MEY# protein remained intact during the hydrolytic process.
The functional structure is maintained in the Ni(II)-complexed P-1MEY# cleavage product
Circular dichroism spectra of P-1MEY# were recorded before and after the Ni(II)-induced hydrolysis, which resulted in the N-terminal deka-His tag removal. The comparison of these confirmed the preservation of the spectral pattern characteristic for the folded solution structure of a Cys2His2 ZF unit18,54 in the cleavage product (Fig. 3), even though the reaction was carried out at harsh conditions (50 °C and pH 8.2), and the buffer was exchanged before the CD measurement.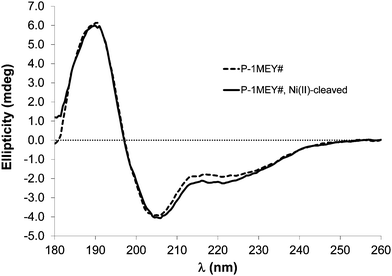 | ||
| Fig. 3 Circular dichroism spectra of the P-1MEY# protein and its Ni(II)-cleaved product in 5 mM phosphate buffer (pH 7.4) at 50 μM protein concentration in a 0.1 mm quartz cell. | ||
The CD results further confirmed that unwanted modifications such as the cleavage at non-specific sites, removal of Zn(II) ions or oxidative damage of the protein did not occur to a detectable extent in the modified P-1MEY# protein. The slight increase of the CD intensity should be the consequence of the removal of the short disordered N-terminal His-tag (GHHHHHHHHHHS). The CD spectrum of the cleaved P-1MEY protein (Fig. S7, ESI†) is practically identical to that of the intact P-1MEY. This suggests that the ordering effect of the His-tag removal and the disordering effect of the remnant peptide sequence after the cleavage of the 3rd finger unit partially compensate for each other, and that the structure of the two uncleaved ZF units was maintained during the hydrolytic reaction in good agreement with the MS results. The consequences of editing out the cleavage sites from the DNA recognition region of P-1MEY in the Ni(II)-induced hydrolytic peptide bond cleavage reaction are manifested in Fig. 4.
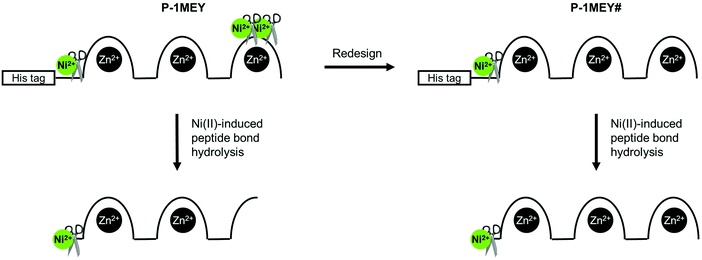 | ||
| Fig. 4 Schematic representation of the Ni(II)-induced cleavage of P-1MEY and the redesigned P-1MEY# ZF proteins. | ||
The question whether the Ni(II)-cleaved P-1MEY# is able to recognize its cognate DNA was investigated by EMSA. The band of the DNA molecule including four copies of the ZF recognition site was already shifted in agarose gel upon addition of one molar equivalent of the protein (Fig. 5). By increasing the protein excess a second distinct shifted band appeared due to the binding of one or more further protein molecules. At a fourfold excess of the protein only the second band was observed. A smear was present to a small extent attributed to minor non-specific binding – natural for all ZF protein as a consequence of the electrostatic component of their interactions with DNA. Similar phenomena were observed with intact P-1MEY#, indicating that both proteins can interact with the favourable DNA sequences equally effectively.
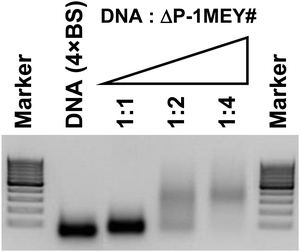 | ||
| Fig. 5 DNA binding of the Ni(II)-cleaved P-1MEY# ZF protein (denoted by ΔP-1MEY#). A short double strand DNA including four tandem segments of the favourable 5′-GAGGCAGAA-3′ sequence was applied in the EMSA experiments. The intensity decrease at higher protein to DNA ratios is presumably due to the impaired ability of the ethidium ion to bind the protein-loaded DNA. | ||
Conclusion
We have tested for the first time the possibility of the specific peptide bond hydrolysis induced by Ni(II) ions within two complex His-tagged ZF metalloproteins without compromising the original metal ion binding sites. The hydrolytic cleavage of the two potential Ni(II)-cleavage sites within the 3rd finger was not hindered by the presence of Zn(II) ions, but concomitantly it abolished the Zn(II) binding site, resulting in loss of the functional structure of this finger. Such partial cleavage of a ZF type protein may lead to altered DNA binding causing unwanted side-effects in cells. Under the applied conditions the deka-His affinity tag did not interfere with the hydrolytic peptide bond cleavage.We redesigned the P-1MEY protein to edit out the naturally occurring Ni(II) cleavage sites in the 3rd ZF unit and to obtain a single N-terminal Ni(II)-catalytic site between the N-terminal His-tag and the ZF protein sequence. The Zinc Finger Tools program was applied to design sequence changes without affecting the DNA binding specificity. We confirmed experimentally that the P-1MEY# protein maintained the Zn(II) and Ni(II)-binding ability and a functional solution structure after the Ni(II)-induced protein hydrolysis. This is crucial for any subsequent applications.
Importantly, the Ni(II) ion remains coordinated to the (Ser/Thr)-X-His motif in the C-terminal cleavage product. Such peptide sequence, characterized by the presence of a His residue in position 3 (referred to as ATCUN motif) provides strong Cu(II) or Ni(II) binding and was shown to cleave DNA in the presence of peroxides.55,56 Functionalization with DNA-binding agents can enhance the efficiency or the selectivity of this reaction,23,55,57–64 and may provide a chance for future treatment of Ni(II)-dependent cancer utilizing the abundant Ni(II) ions present there already.65–71
The advantage of our approach compared to previous reports is that the ATCUN site of the ZF protein is initially masked by the Ni(II) sensitive sequence, and can be activated and regulated by the addition of Ni(II) ions, temperature jump or pH jump (as the rate of the peptide bond cleavage itself is strictly dependent on these three factors).30–32,37–39 Furthermore, the Ni(II) induced peptide-bond hydrolysis can be applied for the removal of any fusion proteins (such as affinity tags, inhibitory or activating domains) from sensitive ZF metalloproteins. Thereby it provides a chance for simple regulation and replacement of the expensive specific protease enzymes. The N-terminal metal-binding site may also promote the application of zinc fingers and eventually their fusion variants as specific carriers of drugs or sensors in their ternary complexes. Recently it has been reported that the ATCUN motif can improve bioactivity of antimicrobial peptides.72
Zinc finger proteins are potential biotechnological tools for specific gene recognition and manipulation. Although the recent competitive TALE and especially the CRISPR technologies prevail this field,73–75 ZF protein based factors may still be attractive choice for eukaryotic applications because of their complex protein–DNA interface (the complexity of intra- and inter-finger interactions allows one for discrimination between similar targets), small size, human origin, as well as their low antigenicity. This is reflected in a number of recent publications.65,76–78 In addition the method described above can be generally applied for any engineered protein, and by optimizing the Ni(II)-induced protein hydrolysis to be significantly accelerated it may significantly contribute to this field.
Abbreviations
| [γ-32P]ATP | Adenosine triphosphate labelled with radioactive phosphorus. |
| 1MEY#-F | Forward primer used during QCM to create the gene of P-1MEY# protein. |
| 1MEY#-R | Reverse primer used during QCM to create the gene of P-1MEY# protein. |
| 1MEY | The PDB code of a ZF protein in complex with its specific DNA. |
| ATCUN | Amino terminal copper and nickel binding motif contains a His in the third position from the N-terminal end of the peptide/protein. |
| BamHI | Type II restriction endonuclease isolated from Bacillus amyloliquefaciens. |
| BL21 (DE3) | Chemically competent E. coli cells suitable for protein expression. |
| CD | Circular dichroism. |
| CP-1 | Consensus peptide 1. |
| CRISPR | Clustered regularly interspaced short palindromic repeats. |
| Cys2His2 | Most commonly occurring Zn(II) binding site donor set in ZF units. |
| DH5α | Competent E. coli cells for cloning applications. |
| EMSA | Electrophoretic mobility shift assay. |
| ESI-MS | Electron spray ionisation mass spectrometry. |
| Factor Xa | Protease, which recognises IEGR amino acid sequence and cleaves after the arginine. |
| FokI | Restriction endonuclease derived from Flavobacterium okeanokoites. |
| H2A | Histone protein involved in the structure of chromatin in eukaryotic cells. |
| HEPES | 4-(2-Hydroxyethyl)-1-piperazineethanesulfonic acid commonly used buffer in biochemistry and molecular biology. |
| His-tag | A dekahistidine-tag is an amino acid motif in proteins that consists of ten histidine (His) residues. The His-tag makes it possible to purify the protein by Ni(II)-affinity chromatography. |
| HPLC | High-performance liquid chromatography. |
| IDT | Integrated DNA technologies®. |
| IEGR | Ile-Glu-Gly-Arg amino acid sequence. |
| IPTG | Isopropyl β-D-1-thiogalactopyranoside |
| KOD-FX | DNA polymerase from Toyobo Life Science Department. |
| native PAGE | Native polyacrylamide gel electrophoresis. |
| NdeI | Type II restriction endonuclease isolated from Neisseria denitrificans. |
| NEB | New England Biolabs®. |
| OD | Optical density. |
| P-1MEY# | Modified P-1MEY protein. |
| P-1MEY | ZF protein similar to the P-1MEY-PDB protein. |
| P-1MEY-PDB | The ZF protein part of the 1MEY complex. |
| PDB | (Protein Data Bank) library of crystal structure data of proteins and protein–DNA complexes. |
| pET-16b | Cloning/expression vector carrying an N-terminal His·Tag® sequence followed by a Factor Xa site. |
| pET-16b-P-1MEY# | pET-16b plasmid containing the gene of P-1MEY# protein. |
| pET-16b-P-1MEY | pET-16b plasmid containing the gene of P-1MEY protein. |
| QuikChange™ (QCM) | Site-directed in vitro mutagenesis method. |
| Sp1 | A ZF-based transcription factor known as Specificity Protein 1. |
| TALE | Transcription activator-like effectors proteins are secreted by Xanthomonas bacteria infecting various plant species. |
| TFA | 2,2,2-Trifluoroacetic acid. |
| Tricine-SDS-PAGE | A tricine modified sodium-dodecyl-sulfate polyacrylamide gel electrophoresis method for protein separation. |
| Tris | Tris(hydroxymethyl)aminomethane commonly used buffer in biochemistry and molecular biology. |
| ZF unit | A peptide of ∼22–24 amino acids, having a tetrahedral metal binding site within a ββα structure. Each unit recognises a three nucleobase sequence. |
| ZF | Zinc finger protein consisting of ZF units. |
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by Foundation for Polish Science TEAM 2009-4/1 program, co-financed by European Regional Development Fund resources within the framework of Operational Program Innovative Economy, by the Hungarian National Research, Development and Innovation Office (GINOP-2.3.2-15-2016-00038 and K_16/120130), and by collaboration of Hungarian and Polish Academies of Sciences (grants NKM-102//2016 and 2/2014-2016, respectively). The equipment used was sponsored in part by the Centre for Preclinical Research and Technology (CePT), a project co-sponsored by European Regional Development Fund and Innovative Economy, The National Cohesion Strategy of Poland. We thank Mr Jacek Olędzki for assistance with ESI-MS experiments.References
- J. Miller, A. D. McLachlan and A. Klug, EMBO J., 1985, 4, 1609–1614 Search PubMed.
- A. Klug, Annu. Rev. Biochem., 2010, 79, 213–231 CrossRef PubMed.
- D. Jantz, B. T. Amann, G. J. Gatto and J. M. Berg, Chem. Rev., 2004, 104, 789–799 CrossRef PubMed.
- Q. Liu, Z.-Q. Xia and C. C. Case, J. Biol. Chem., 2002, 277, 3850–3856 CrossRef PubMed.
- B. Dreier, R. R. Beerli, D. J. Segal, J. D. Flippin and C. F. Barbas, III, J. Biol. Chem., 2001, 276, 29466–29478 CrossRef PubMed.
- B. Dreier, R. P. Fuller, D. J. Segal, C. V. Lund, P. Blancafort, A. Huber, B. Koksch and C. F. Barbas III, J. Biol. Chem., 2005, 280, 35588–35597 CrossRef PubMed.
- B. Dreier, D. J. Segal and C. F. Barbas III, J. Mol. Biol., 2000, 303, 489–502 CrossRef PubMed.
- D. J. Segal, B. Dreier, R. R. Beerli and C. F. Barbas III, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 2758–2763 CrossRef.
- K.-H. Bae, Y. D. Kwon, H.-C. Shin, M.-S. Hwang, E.-H. Ryu, K.-S. Park, H. Y. Yang, D.-K. Lee, Y. Lee, J. Park, H. S. Kwon, H.-W. Kim, B.-I. Yeh, H. W. Lee, S. H. Sohn, J. Yoon, W. Seol and J.-S. Kim, Nat. Biotechnol., 2003, 21, 275–280 CrossRef PubMed.
- J. Liu and G. D. Stormo, Bioinformatics, 2008, 24, 1850–1857 CrossRef PubMed.
- M. S. Bhakta and D. J. Segal, Methods Mol. Biol., 2010, 649, 3–30 CrossRef PubMed.
- F. Fu and D. F. Voytas, Nucleic Acids Res., 2013, 41, D452–D455 CrossRef PubMed.
- J. G. Mandell and C. F. Barbas III, Nucleic Acids Res., 2006, 34, W516–W523 CrossRef PubMed.
- J. D. Sander, M. L. Maeder, D. Reyon, D. F. Voytas, J. K. Joung and D. Dobbs, Nucleic Acids Res., 2010, 38, W462–W468 CrossRef PubMed.
- T. Mino, T. Mori, Y. Aoyama and T. Sera, Arch. Virol., 2008, 153, 1291–1298 CrossRef PubMed.
- M. G. Di Certo, N. Corbi, G. Strimpakos, A. Onori, S. Luvisetto, C. Severini, A. Guglielmotti, E. M. Batassa, C. Pisani, A. Floridi, B. Benassi, M. Fanciulli, A. Magrelli, E. Mattei and C. Passananti, Hum. Mol. Genet., 2010, 19, 752–760 CrossRef PubMed.
- M. Imanishi, T. Nakaya, T. Morisaki, D. Noshiro, S. Futaki and Y. Sugiura, ChemBioChem, 2010, 11, 1653–1655 CrossRef PubMed.
- A. Nomura and A. Okamoto, Chem. Commun., 2009, 1906–1908 RSC.
- M. Papworth, P. Kolasinska and M. Minczuk, Gene, 2006, 366, 27–38 CrossRef PubMed.
- A. J. Wood, T. W. Lo, B. Zeitler, C. S. Pickle, E. J. Ralston, A. H. Lee, R. Amora, J. C. Miller, E. Leung, X. Meng, L. Zhang, E. J. Rebar, P. D. Gregory, F. D. Urnov and B. J. Meyer, Science, 2011, 333, 307 CrossRef PubMed.
- D. Carroll, Annu. Rev. Biochem., 2014, 83, 409–439 CrossRef PubMed.
- M. Dhanasekaran, S. Negi and Y. Sugiura, Acc. Chem. Res., 2006, 39, 45–52 CrossRef PubMed.
- M. Nagaoka, M. Hagihara, J. Kuwahara and Y. Sugiura, J. Am. Chem. Soc., 1994, 116, 4085–4086 CrossRef.
- E. Németh, G. K. Schilli, G. Nagy, C. Hasenhindl, B. Gyurcsik and C. Oostenbrink, J. Comput.-Aided Mol. Des., 2014, 28, 841–850 CrossRef PubMed.
- E. Németh, M. N. Asaka, K. Kato, Z. Fábián, C. Oostenbrink, H. E. M. Christensen, K. Nagata and B. Gyurcsik, ChemBioChem, 2018, 19, 66–75 CrossRef PubMed.
- W. Bal, J. Lukszo, K. Bialkowski and K. S. Kasprzak, Chem. Res. Toxicol., 1998, 11, 1014–1023 Search PubMed.
- W. Bal, R. Liang, J. Lukszo, S.-H. Lee, M. Dizdaroglu and K. S. Kasprzak, Chem. Res. Toxicol., 2000, 13, 616–624 Search PubMed.
- M. Mylonas, A. Krezel, J. C. Plakatouras, N. Hadjiliadis and W. Bal, J. Chem. Soc., Dalton Trans., 2002, 4296–4306 RSC.
- A. Krezel, M. Mylonas, E. Kopera and W. Bal, Acta Biochim. Pol., 2006, 53, 721–727 Search PubMed.
- A. Krezel, E. Kopera, A. M. Protas, J. Poznanski, A. Wysłouch-Cieszynska and W. Bal, J. Am. Chem. Soc., 2010, 132, 3355–3366 CrossRef PubMed.
- E. Kopera, A. Krezel, A. M. Protas, A. Belczyk, A. Bonna, A. Wyslouch-Cieszynska, J. Poznanski and W. Bal, Inorg. Chem., 2010, 49, 6636–6645 CrossRef PubMed.
- E. Kurowska., J. Sasin-Kurowska, A. Bonna, M. Grynberg, J. Poznanski, L. Knizewski, K. Ginalski and W. Bal, Metallomics, 2011, 3, 1227–1231 RSC.
- A. M. Protas, H. H. N. Ariani, A. Bonna, A. Polkowska-Nowakowska, J. Poznanski and W. Bal, J. Inorg. Biochem., 2013, 127, 99–106 CrossRef PubMed.
- H. H. N. Ariani, A. Polkowska-Nowakowska and W. Bal, Inorg. Chem., 2013, 52, 2422–2431 CrossRef PubMed.
- E. I. Podobas, A. Bonna, A. Polkowska-Nowakowska and W. Bal, J. Inorg. Biochem., 2014, 136, 107–114 CrossRef PubMed.
- N. Wezynfeld, K. Bossak, W. Goch, A. Bonna, W. Bal and T. Fraczyk, Chem. Res. Toxicol., 2014, 27, 1996–2009 Search PubMed.
- E. Kopera, A. Belczyk and W. Bal, PLoS One, 2012, 7, e36350 CrossRef PubMed.
- E. Kopera, S. Krzywda, M. Lenarcic Živkovic, A. Dvornyk, B. Kłudkiewicz, K. Grzelak, I. Zhukov, W. Zagórski-Ostoja, M. Jaskólski and W. Bal, PLoS One, 2014, 9, e106936 CrossRef PubMed.
- N. E. Wezynfeld, A. Bonna, W. Bal and T. Fraczyk, Metallomics, 2015, 7, 596–604 RSC.
- N. E. Wezynfeld, T. Fraczyk and W. Bal, Coord. Chem. Rev., 2016, 327–328, 166–187 CrossRef.
- W. Rezuke, J. Knight and F. Sunderman, Jr., Am. J. Ind. Med., 1987, 11, 419–426 CrossRef PubMed.
- Occupational, Industrial, and Environmental Toxicology, ed. M. I. Greenberg, R. J. Hamilton, S. D. Phillips and G. J. McCluskey, Pennsylvania, Mosby, 2nd edn, 2003 Search PubMed.
- J. J. Hostynek and H. I. Maibach, Nickel and the skin: Absorption, Immunology, Epidemiology, and Metallurgy, CRC Press, Taylor and Francis Group, 2002, p. 158 Search PubMed.
- C. A. Kim and J. M. Berg, Nat. Struct. Biol., 1996, 3, 940–945 CrossRef PubMed.
- H. Schägger, Nat. Protoc., 2006, 1, 16–22 CrossRef PubMed.
- B. A. Krizek, B. T. Amann, V. J. Kilfoil, D. L. Merkle and J. M. Berg, J. Am. Chem. Soc., 1991, 113, 4518–4523 CrossRef.
- A. N. Besold, L. R. Widger, F. Namuswe, J. L. Michalek, S. L. J. Michel and D. P. Goldberg, Mol. BioSyst., 2016, 12, 1183–1193 RSC.
- A. Miloch and A. Krezel, Metallomics, 2014, 6, 2015–2024 RSC.
- M. Nagaoka, J. Kuwahara and Y. Sugiura, Biochem. Biophys. Res. Commun., 1993, 194, 1515–1520 CrossRef PubMed.
- U. Heinz, L. Hemmingsen, M. Kiefer and H.-W. Adolph, Chem. – Eur. J., 2009, 15, 7350–7358 CrossRef PubMed.
- J. B. Mangrum, I. Zgani, S. D. Tsotsoros, Y. Qu and N. P. Farrell, Chem. Commun., 2013, 49, 6986–6988 RSC.
- A. Belczyk-Ciesielska, I. A. Zawisza, M. Mital, A. Bonna and W. Bal, Inorg. Chem., 2014, 53, 4639–4646 CrossRef PubMed.
- N. E. Wezynfeld, T. Frączyk and W. Bal, Coord. Chem. Rev., 2016, 327–328, 166–187 CrossRef.
- Y. Berezovskaya, C. T. Armstrong, A. L. Boyle, M. Porrini, D. N. Woolfson and P. E. Barran, Chem. Commun., 2011, 47, 412–414 RSC.
- C. Harford and B. Sarkar, Acc. Chem. Res., 1997, 30, 123–130 CrossRef.
- W. Bal, J. Christodoulou, P. J. Sadler and A. Tucker, J. Inorg. Biochem., 1998, 70, 33–39 CrossRef PubMed.
- D. P. Mack and P. B. Dervan, Biochemistry, 1992, 31, 9399–9405 CrossRef PubMed.
- W. Bal, J. Lukszo and K. S. Kasprzak, Chem. Res. Toxicol., 1997, 10, 915–921 Search PubMed.
- X. Huang, M. E. Pieczko and C. E. Long, Biochemistry, 1999, 38, 2160–2166 CrossRef PubMed.
- Y. Y. Fang, C. A. Claussen, K. B. Lipkowitz and E. C. Long, J. Am. Chem. Soc., 2006, 128, 3198–3207 CrossRef PubMed.
- C. Harford, S. Narindrasorasak and B. Sarkar, Biochemistry, 1996, 35, 4271–4278 CrossRef PubMed.
- Y. Jin and J. A. Cowan, J. Am. Chem. Soc., 2005, 127, 8408–8415 CrossRef PubMed.
- Y. Jin, M. A. Lewis, N. H. Gokhale, E. C. Long and J. A. Cowan, J. Am. Chem. Soc., 2007, 129, 8353–8361 CrossRef PubMed.
- Z. Yu, M. Han and J. A. Cowan, Angew. Chem., Int. Ed., 2014, 53, 1901–1905 CrossRef PubMed.
- E. Canaz, M. Kilinc, H. Sayar, G. Kiran and E. Ozyurek, J. Trace Elem. Med. Biol., 2017, 43, 217–223 CrossRef PubMed.
- J. Jiménez-Lamana and J. Szpunar, Metallomics, 2017, 9, 1014–1027 RSC.
- C. M. Agbale, M. H. Cardoso, I. K. Galyuona and O. L. Franco, Metallomics, 2016, 8, 1159–1169 RSC.
- H. Romanowicz-Makowska, E. Forma, M. Bryś, W. M. Krajewska and B. Smolarz, Pol. J. Pathol., 2011, 62, 257–261 Search PubMed.
- C. Zhao, X. Chen, D. Zang, X. Lan, S. Liao, C. Yang, P. Zhang, J. Wu, X. Li, N. Liu, Y. Liao, H. Huang, X. Shi, L. Jiang, X. Liu, Z. He, Q. P. Dou, X. Wang and J. Liu, Oncogene, 2016, 35, 5916–5927 CrossRef PubMed.
- L. Wang, J. Fan, J. A. Hitron, Y.-O. Son, J. T. F. Wise, R. V. Roy, D. Kim, J. Dai, P. Pratheeshkumar, Z. Zhang and X. Shi, Toxicol. Sci, 2016, 151, 376–387 CrossRef PubMed.
- A. Arita and M. Costa, Metallomics, 2009, 1, 222–228 RSC.
- M. D. Libardo, J. L. Cervantes, J. C. Salazar and A. M. Angeles-Boza, ChemMedChem, 2014, 9, 1892–1901 Search PubMed.
- B. L. Oakes, D. F. Xia, E. F. Rowland, D. J. Xu, I. a. Ankoudinova, J. S. Borchardt, L. Zhang, P. Li, J. C. Miller, E. J. Rebar and M. B. Noyes, Nat. Commun., 2016, 7, 10194 CrossRef PubMed.
- L. S. Qi, M. H. Larson, J. A. Doudna, J. S. Weissmann, A. P. Arkin and W. A. Lim, Cell, 2013, 152, 1173–1183 CrossRef PubMed.
- J. C. Miller, S. Tan, G. Qiao, K. A. Barlow, J. Wang, D. F. Xia, X. Meng, D. E. Paschon, E. Leung, S. J. Hinkley, G. P. Dulay, K. L. Hua, I. Ankoudinova, G. J. Cost, F. D. Urnov, H. S. Zhang, M. C. Holmes, L. Zhang, P. D. Gregory and E. J. Rebar, Nat. Biotechnol., 2011, 29, 143–148 CrossRef PubMed.
- P. Yang, Y. Wang, D. Hoang, M. Tinkham, A. Patel, M.-A. Sun, G. Wolf, M. Baker, H.-C. Chien, K.-Y. N. Lai, X. Cheng, C.-K. J. Shen and T. S. Macfarlan, Science, 2017, 356, 757–759 CrossRef PubMed.
- M. Imbeault, P.-Y. Helleboid and D. Trono, Nature, 2017, 543, 550–554 CrossRef PubMed.
- A. Gutierrez-Guerrero, S. Sanchez-Hernandez, G. Galvani, J. Pinedo-Gomez, R. Martin-Guerra, A. Sanchez-Gilabert, A. Aguilar-González, M. Cobo, P. Gregory, M. Holmes, K. Benabdellah and F. Martin, Hum. Gene Ther., 2018, 29, 366–380 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8mt00098k |
| This journal is © The Royal Society of Chemistry 2018 |
