Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Interdependence of free zinc changes and protein complex assembly – insights into zinc signal regulation†
Anna
Kocyła
 ,
Justyna
Adamczyk
and
Artur
Krężel
,
Justyna
Adamczyk
and
Artur
Krężel
 *
*
Department of Chemical Biology, Faculty of Biotechnology, University of Wrocław, Joliot-Curie 14a, 50-383 Wrocław, Poland. E-mail: artur.krezel@uwr.edu.pl
First published on 14th December 2017
Abstract
Cellular zinc (Zn(II)) is bound with proteins that are part of the proteomes of all domains of life. It is mostly utilized as a catalytic or structural protein cofactor, which results in a vast number of binding architectures. The Zn(II) ion is also important for the formation of transient protein complexes with a Zn(II)-dependent quaternary structure that is formed upon cellular zinc signals. The mechanisms by which proteins associate with and dissociate from Zn(II) and the connection with cellular Zn(II) changes remain incompletely understood. In this study, we aimed to examine how zinc protein domains with various Zn(II)-binding architectures are formed under free Zn(II) concentration changes and how formation of the Zn(II)-dependent assemblies is related to the protein concentration and reactivity. To accomplish these goals we chose four zinc domains with different Zn(II)-to-protein binding stoichiometries: classical zinc finger (ZnP), LIM domain (Zn2P), zinc hook (ZnP2) and zinc clasp (ZnP1P2) folds. Our research demonstrated a lack of changes in the saturation level of intraprotein zinc binding sites, despite various peptide concentrations, while homo- and heterodimers indicated a concentration-dependent tendency. In other words, at a certain free Zn(II) concentration, the fraction of a formed dimeric complex increases or decreases with subunit concentration changes. Secondly, even small or local changes in free Zn(II) may significantly affect protein saturation depending on its architecture, function and subcellular concentration. In our paper, we indicate the importance of interdependence of free Zn(II) availability and protein subunit concentrations for cellular zinc signal regulation.
Significance to metallomicsThe emerging and growing field of metallomics within biological systems results in the need to explore the factors that drive metal-mediated protein complex formation and functionality to a wider extent. The relevance of Zn(II) ions in proper functioning of the cell is undisputable, but the particular pathways inducing their regulatory role are not completely elucidated. Our investigation considers the influence of free Zn(II) availability along with protein concentration, which impinge on the action of the Zn(II)-dependent interactome. We provide a comprehensive view of the regulation of zinc assemblies relevant to understanding the molecular mechanisms of Zn(II)-involved cellular network driving processes. |
Introduction
Protein complexes are key molecular entities that integrate multiple gene products to perform cellular functions. Being the main effectors of multiple processes driving cell homeostasis, the fundamental challenge is to understand their organization and distribution within the cell. For proper macromolecular assembly, they require a set of factors to be accomplished, e.g. a particular order of interaction, energy-driven conformational changes, post-translational modifications or chaperone assistance.1 To obtain specific macromolecular complexes, a certain availability and abundance of the substrates is strictly required. In the case of metalloproteins, besides the protein assembly components synthesized within the cell, there are metal ions that can neither be produced nor destroyed under physiological conditions. It is worth noting that to form a functional metal–protein assembly within the cell there must be the correct metal ions and proteins in the correct concentrations at the correct place and time. Changes in spatiotemporal cellular conditions impinge on the complex abundance and perturb many processes in consequence.2Zinc (formally Zn(II) in biological systems) is the most widespread d-block metal ion in all domains of life, performing an enormous number of biological functions mostly as protein complexes.3 Zinc-binding macromolecules are considered to represent ∼10% of proteins encoded in the human genome.4 Moreover, transients and interfacial interactions are not included in the estimation.5,6 In metalloproteins, the Zn(II) coordination sphere is generated in a large variety of combinations where the metal ion is bound either solely or in a clustered manner with various geometries and a range of stoichiometries: equimolar (ZnP) or with excess of Zn(II) (ZnnP) or protein (ZnPn or ZnP1P2Pn).5,7 The total Zn(II) concentration in eukaryotic cells is submillimolar,8,9 but the loosely bound Zn(II) (mobile pool), which is also frequently named “available”, “labile” or “free” Zn(II) (Zn(II)free), reflects the affinity of Zn(II) towards cellular protein and varies in mammalian cells from 10−11 to 10−9 M based on the tissue, cell type9–11 and subcellular localization.12 Depending on the protein architecture, coordination sphere and a number of stabilizing and destabilizing effects, Zn(II)-to-protein affinity varies from the pico- to femtomolar range for catalytic and structural binding sites to moderate nano- or subnanomolar affinity for proteins that are regulated by Zn(II) changes such as metal sensors, transcription factors, enzymes or transporting proteins.13,14 Both excessively low and high levels of free Zn(II) render many metalloproteins dysfunctional and likely result in cell death.15 Alterations in the steady state of free Zn(II) are also dictated by the state of the cell, e.g. differentiation, proliferation or rest.16 Therefore intracellular Zn(II) is restrained by a specialized buffering system consisting of metallothionein/thionein to prevent its cytotoxicity.17 Muffling processes are also involved and dampen increases in free Zn(II) through time-dependent effects of binding and flux.18,19
In contrast, zinc signals (free Zn(II) changes) fulfill a dynamic role in communication between cells and in the conversion of the extracellular stimuli to intracellular responses, thereby gaining amplification of the signal.20,21 Moreover, intracellular free Zn(II) fluctuations have an impact on both functioning of genomic components, such as the transcription factor MTF-1, which changes the transcription rates of numerous genes,22,23 and much faster, protein-based pathways. It has been shown that an increase in cytosolic free Zn(II) upon extracellular stimulation occurs within minutes by an apparent release of Zn(II) called a “zinc wave” from the endoplasmic reticulum (ER) or Golgi24 and this Zn(II) release has downstream effects on cell signaling.25,26 An important component of Zn(II) action in cells is the ability to inhibit protein tyrosine phosphatase activity,27 resulting in activation of kinase-dependent signaling pathways.28,29 Free Zn(II) fluctuations can also be driven by the release of Zn(II) ions into the extracellular space, which modulates the process of synaptic transmission in the central nervous system,30,31 enables the egg-to-embryo transition with the so-called “zinc sparks”32 or suppresses insulin secretion from pancreatic β-cells.33
Apart from the exact local free Zn(II) concentration, there is an exact protein abundance within a cell needed to form biologically active zinc complexes. Nevertheless, the quantitative knowledge of the zinc proteome has not been fully considered yet. Even though absolute abundances of protein are deposited, the local protein concentrations may differ by ranges of magnitude. Cellular reaction rates have been additionally increased by reduction of dimensionality through membrane surfaces, where many cellular machineries carry out their function.34,35 Lipid rafts as regions of membranes with a distinct, characteristic structural composition appear to act as platforms to recruit and colocalize proteins involved in intracellular signaling pathways, such as Src family kinases, G proteins, growth factor receptors, mitogen-activated protein kinases (MAPK) and the protein kinase C family, facilitating their interaction36,37 or other cellular processes, such as membrane trafficking, cell adhesion or cytoskeletal rearrangement.38 Dynamic partitioning into rafts increases specific interprotein collision rates, and microdomains can readily operate as protein concentrators or isolators, but significant constraints on size and mobility appear.39
Regulation of Zn(II) ion fluctuations with the participation of many proteins is a key molecular function of Zn(II)-dependent machinery in biology, which impinges on their impact on cellular homeostasis and human health. Proteins can adopt various architectures of zinc binding, which can be either intra- or intermolecular.7,14 In consequence, Zn(II)-to-protein binding occurs not only equimolarly, but also two or more Zn(II) ions can coordinate to one protein molecule and, conversely, one Zn(II) ion can be bound to one or more protein molecules.5,6 The arising questions that we want to address here are: (i) how is the various stoichiometry of complex assembly connected with dynamic changes in available Zn(II) under physiological conditions, (ii) does protein saturation with Zn(II) depend on its concentration, and (iii) how are those factors connected with each other? At present, there is no available systematic research investigating the level of zinc protein assembly in light of the complexes’ stoichiometry and the access to free Zn(II) ions as well as protein cellular concentrations. Here, we provide new experimental insights into the interdependence between the parameters affecting zinc protein complex formation regarding different stoichiometries, protein subunit levels and free Zn(II) status.
Experimental
Materials
All chemicals were of the highest purity from Sigma-Aldrich, Bio-Rad, Carl Roth GmbH, New England Biolabs, Avantor Performance Materials, and Iris Biotech GmbH. All buffers used in the study were incubated with the metal-chelating resin Chelex 100 (Bio-Rad) prior to use.Peptide synthesis and concentration determination
The peptides used in the study – LIM (LIM domain of PDLIM1 protein), ZF (11th zinc finger of ZF133 protein), HK (minimal hook domain from Rad50), CD4 (cytoplasmic tail of cluster of differentiation 4), Lck (N-terminal domain of lymphocyte-specific protein tyrosine kinase) – were synthesized on a solid phase (SPPS) using a Liberty 1 microwave-assisted synthesizer (CEM) as described before (see Table S1 in the ESI† for amino acid sequences).40–42 Peptides were either acetylated (LIM) or N-terminally labeled with dansyl (ZF133, HK), FAM (CD4) or TAMRA (Lck) moieties. It should be noted that the Trp residue was placed at the C-terminus of HK and at the 14th position of the ZF as a Lys to Trp substitution in order to sense as a FRET donor.42,43 Modified peptides were cleaved using TFA-based mixtures with thiol protectants. Cleaved peptide crudes were purified on HPLC (Dionex Ultimate 3000) on a Phenomenex C18 column using a gradient of 0.1% TFA in water and ACN. The identity of peptides was finally confirmed by mass spectrometry using an API 2000 Applied Biosystems instrument. The concentration of peptides was estimated by measuring the thiolate or Zn(II) content at pH 7.4 using the DTNB, PAR or Zincon assays with a molar absorption coefficient of 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 150 M−1 cm−1 at 412 nm,44 70
150 M−1 cm−1 at 412 nm,44 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 M−1 cm−1 at 492 nm45 or 24
500 M−1 cm−1 at 492 nm45 or 24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 M−1 cm−1 at 618 nm,46 respectively. In addition, the concentration of modified peptides was determined by measuring FAM group absorbance at 492 nm in 0.1 M NaOH (ε = 79
200 M−1 cm−1 at 618 nm,46 respectively. In addition, the concentration of modified peptides was determined by measuring FAM group absorbance at 492 nm in 0.1 M NaOH (ε = 79![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 M−1 cm−1) or TAMRA group absorbance at 555 nm in 6 M GdnHCl at pH 8.0 (ε = 65
000 M−1 cm−1) or TAMRA group absorbance at 555 nm in 6 M GdnHCl at pH 8.0 (ε = 65![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 M−1 cm−1).47
000 M−1 cm−1).47
Expression, purification, and reconstitution of human MT2a
Human metallothionein, isoform 2a (MT) was prepared and reconstituted as described previously with some minor changes.48,49 Briefly, MT was expressed in BL21(DE3)pLysS E. coli cells as an intein fusion protein by the induction of 0.1 mM IPTG. Protein fusion was initially purified from bacteria cell extract using chitin resin and subsequently cleaved by 100 mM DTT. Eluted fractions of supernatant were acidified and purified using an SEC-70 column (Bio-Rad) equilibrated with 10 mM HCl to obtain apo-protein (T, thionein). Zinc MT2a reconstitution was performed by mixing T solution with 10 eq. of ZnSO4 in the presence of 2 mM TCEP under anaerobic conditions and the pH was adjusted to 8.6 followed by purification on an SEC70 column equilibrated with 20 mM Tris–HCl.49,50Metal buffers used for the Zn(II) competition
To determine the free Zn(II) range, denoted here as pZn (−log[Zn(II)]free), in which zinc domains fold, all peptides were incubated with competing chelators (complexones). In the case of the zinc finger, LIM and zinc hook domain, 1 mM EDTA and HEDTA were used to match their high stability. Tables S2–S4 (ESI†) show initial and corrected (see below) pZn values of each sample before and after incubation with different peptide concentrations (0.5, 1, 3 and 10 μM for ZF, 5, 10, 15 and 20 μM for the LIM domain, and 0.5, 1, 5 and 10 μM for ZH). For the zinc clasp (0.1, 0.25, 0.5 and 1 μM), the free Zn(II) buffering range was extended to EGTA, which competes for Zn(II) less strongly than EDTA and HEDTA (Table S5, ESI†).51,52 Briefly, 1 mM concentration of an appropriate chelator (EDTA, HEDTA, EGTA) was used with various concentrations of ZnSO4 (0.025–0.9 mM) in 50 mM HEPES, 100 mM NaCl, and 50 μM TCEP at pH 7.4 to maintain the free Zn(II) concentration in the range of 6.2 × 10−16 to 6.3 × 10−10 M. The sets of all peptides in metal buffers were equilibrated for 24 h prior to measurements to ensure that equilibration was reached.CD and fluorometric-monitored Zn(II) binding study
Circular dichroic (CD) spectra of equilibrated LIM samples at 200–260 nm were recorded on a Jasco 1100 spectropolarimeter at 25 °C in 1 cm quartz cuvette. Ellipticity values at 220 nm were taken for further data processing.41 In the case of ZF or HK peptides, Zn(II) binding was monitored fluorometrically in the range of 315–550 nm at 25 °C using a Jobin Yvon FluoroMax-3 spectrofluorometer (Horiba) when samples were excited at 280 nm.40,43 Intensity at 354 nm (donor maximum) was taken for data analysis to avoid inaccuracy of ratio analysis, as was recently pointed out.40 The formation of the clasp domain was monitored for an equimolar mixture of Lck and CD4 peptide. Samples were excited at 492 nm and fluorescence was monitored in the range of 498–650 nm. In this case, the intensity of the donor at 525 nm was also taken for Zn(II) binding analysis. Experiments were performed in triplicate.Zn(II) transfer from MT to CD4 and Lck peptide mixture
Purified reduced thionein (T) and Zn(II)-loaded metallothionein 2a (MT) were mixed in 50 mM HEPES, 100 mM NaCl, and 0.3 mM TCEP at pH 7.4 to prepare a set of buffers with various T/(T + MT) ratios. The T concentration varied within a range of 0–3 μM while MT remained constant at 2.5 μM. CD4 and Lck peptides were added at equimolar concentrations of 0.25 μM, 0.5 μM and 1 μM each and equilibrated for 12 h. FRET between FAM and TAMRA was measured as described above by exciting the sample at 492 nm.Results
To investigate the dependence of the Zn(II) binding mode and protein concentration on the level of saturation of zinc domains, we selected four structurally different domains to work with. The choice of zinc protein assemblies was dictated by parameters critical for monitoring their formation and concentration-dependent studies. To examine how Zn(II)-mediated protein folding is affected by changes in free Zn(II), small and stable zinc domains with a well-characterized structure and stoichiometry were chosen, where more than one Zn(II) ion is bound to the protein or when more than one protein molecule is coordinated with Zn(II) with the comparison of the most prevalent equimolar domains (Fig. 1). The first and most widespread zinc domain in biology that we aimed to analyze was a classical (ββα) zinc finger (ZF), which forms ZnP stoichiometry (Fig. 1a).42,53 Another selected motif was the LIM (Lin11, Isl-1 & Mec-3) domain from the PDLIM1 protein—an example of a double zinc finger domain that binds two Zn(II) ions (Zn2P) (Fig. 1b).41 The third and fourth zinc domains of our choice, characterized by completely different architecture, were the homodimeric zinc hook domain (ZH) from the Rad50 protein formed on the interface of the protein (ZnP2, Fig. 1c) and the heterodimeric zinc clasp domain (ZC) where Zn(II) is co-coordinated with CD4 and Lck proteins (ZnP1P2, Fig. 1d).54–56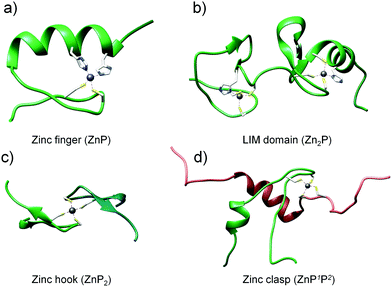 | ||
| Fig. 1 Structures of zinc protein domains examined in this work: classical zinc finger (PDB: 1VA3), LIM domain of PDLIM1 protein (PDB: 1X62), minimal hook domain of Rad50 protein (PDB: 1L8D) and zinc clasp domain being a complex of CD4 (P1) and Lck (P2) (PDB: 1Q68). | ||
Zn(II) binding and domain formation
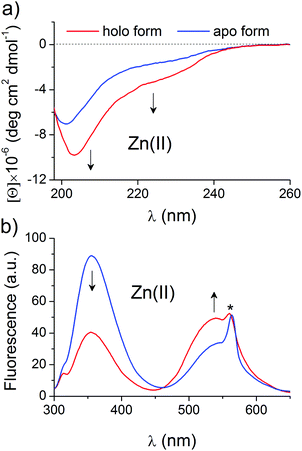
Selected and synthesized peptides were briefly characterized in terms of Zn(II) binding and spectral changes upon complexation to apply them to a systematic stability study. LIM peptide, without any fluorescent modification, was titrated spectropolarimetrically to examine the dynamic range of spectral changes associated with domain formation. Fig. 2a shows CD spectra of 5 μM metal-free and fully saturated LIM peptide at a Zn(II)-to-peptide ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, which indicates significant changes in peptide conformation. Metal-coupled folding of the zinc hook domain with dansyl and tryptophan residues results in FRET, which was monitored fluorometrically. Fig. 2b demonstrates spectra of metal-free and fully saturated HK peptide at a Zn(II)-to-peptide molar ratio of 1
1, which indicates significant changes in peptide conformation. Metal-coupled folding of the zinc hook domain with dansyl and tryptophan residues results in FRET, which was monitored fluorometrically. Fig. 2b demonstrates spectra of metal-free and fully saturated HK peptide at a Zn(II)-to-peptide molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2. The decrease in intensity of the Trp emission band at 354 nm is observed while the dansyl band at 540 nm increases, which indicates intramolecular FRET between Dns and Trp residues. The obtained results are consistent with previously published data.41,43 A similar approach for ZF peptide with the same set of fluorogenic residues, but differently placed, shows comparable changes in band intensities, which confirms stable domain formation as indicated by Lakowicz and co-workers (Fig. S1, ESI†).57 Saturation occurs at a 1
2. The decrease in intensity of the Trp emission band at 354 nm is observed while the dansyl band at 540 nm increases, which indicates intramolecular FRET between Dns and Trp residues. The obtained results are consistent with previously published data.41,43 A similar approach for ZF peptide with the same set of fluorogenic residues, but differently placed, shows comparable changes in band intensities, which confirms stable domain formation as indicated by Lakowicz and co-workers (Fig. S1, ESI†).57 Saturation occurs at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio of Zn(II) and ZF peptide. The formation of the clasp domain was monitored by intermolecular FRET between FAM and TAMRA. Fig. S2 (ESI†) shows fluorescence spectra of an equimolar (1 μM
1 molar ratio of Zn(II) and ZF peptide. The formation of the clasp domain was monitored by intermolecular FRET between FAM and TAMRA. Fig. S2 (ESI†) shows fluorescence spectra of an equimolar (1 μM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 μM) mixture of CD4 and Lck peptides without and with a Zn(II) ion at the saturation point that occurs at a Zn(II)-CD4-Lck molar ratio of 1
1 μM) mixture of CD4 and Lck peptides without and with a Zn(II) ion at the saturation point that occurs at a Zn(II)-CD4-Lck molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The spectra demonstrate a significant decrease in the fluorescence intensity of the FAM and TAMRA bands at 525 and 573 nm; however, the normalization of the intensities at 525 nm clearly shows gain of the fluorescence at 573 nm. The weak increase in TAMRA intensity is typical for low donor and acceptor concentrations and has been described previously.58,59 However, strong intensity changes at 525 nm allow quantitative monitoring of Zn(II) binding and domain formation.
1. The spectra demonstrate a significant decrease in the fluorescence intensity of the FAM and TAMRA bands at 525 and 573 nm; however, the normalization of the intensities at 525 nm clearly shows gain of the fluorescence at 573 nm. The weak increase in TAMRA intensity is typical for low donor and acceptor concentrations and has been described previously.58,59 However, strong intensity changes at 525 nm allow quantitative monitoring of Zn(II) binding and domain formation.
Domain formation in free Zn(II)-controlled media
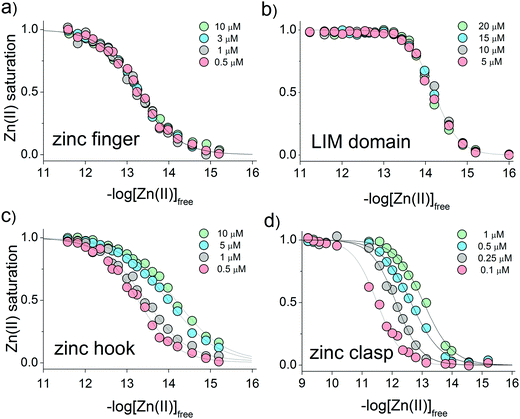
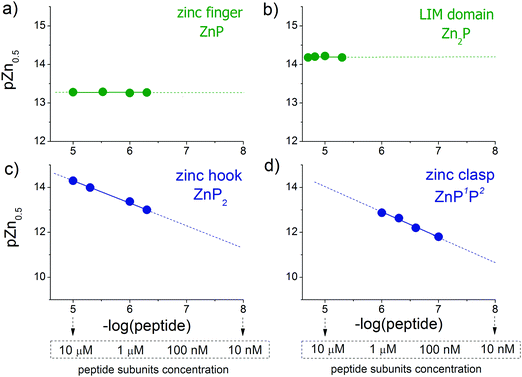
The results obtained for selected zinc domains show that they bind Zn(II) with high affinity at pH 7.4, although the stoichiometry of the formed complexes differs. In the next step of the study, peptides at various concentrations were incubated with metal buffer components to determine half saturation points (the point where half of the complex is present, also named the inflection point or pZn0.5) and quantitatively examine the relationship between the amount of formed complex and free zinc status. For each zinc domain, the experimental concentration was set individually taking into consideration the used methodology. The peptide subunits’ solubility and fluorophores’ non-specific interactions as well as the linearity of the fluorescence response were considered to obtain the most accurate readouts. The lowest concentration range was used in the case of the clasp domain due to non-specific interactions above 1 μM of reactants.CD ellipticities and fluorescence intensities obtained for each domain at a certain concentration in sets of metal buffers (raw data in Tables S2–S5, ESI†) were first normalized to the 0–1 range of molar fraction of the complex and then fitted to the logarithmic version of the Hill equation as described elsewhere.43 Then, initial pZn values of metal buffers were corrected for the amount of Zn(II) transferred from metal buffer components to the peptide after equilibration based on fractional saturation of a particular domain. Initial and corrected pZn values are presented in Tables S6–S9 (ESI†). Due to the low concentration of CD4 and Lck peptides, correction of initial pZn values was not necessary and was omitted in this case. Fig. 3 shows dependence of zinc domain saturation plotted against pZn values with appropriate Hill's fits for all studied zinc architectures regarding the concentration changes. The four isotherms obtained overlap regardless of peptide concentration with the averaged half saturation point at 13.28 ± 0.02 and 14.20 ± 0.02 for the ZF and LIM domains, respectively (Fig. 3a and b). Metal-coupled folding of interprotein domains (zinc hook and zinc clasp) occur in a significantly different manner compared with ZF and LIM. Their equilibration with various zinc buffers results in differences of a complex fraction at the same pZn when peptides differ in the concentration values between each other. Fig. 3c and d show that pZn at inflection points shifts when the concentration of domain components increases from 13.00 to 14.30 and from 11.80 to 12.87 for ZH and ZC, respectively. Fig. 4 demonstrates the relationship between pZn0.5 and concentration of four of the peptides displayed as −log([peptide]). For intraprotein metal binding modes ZnP (ZF) and Zn2P (LIM), the interdependence clearly shows a lack of domain saturation difference regardless of its total concentration (Fig. 4a and b). In contrast, interfacial zinc binding sites ZnP2 (ZH) and ZnP1P2 (ZC) show a linear function between pZn0.5 and −log([peptide]) values in the applied experimental range (Fig. 4c and d).
Clasp domain formation in metallothionein buffer
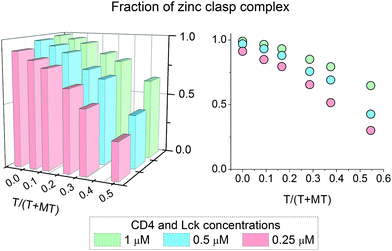
It has been shown above that the Zn(II)-dependent formation of the zinc clasp assembly is closely related to heterodimer component concentration. In the next step of the study, we examined how this domain is formed in the presence of metallothionein (MT), which functions as a key player in cellular zinc homeostasis,60 and how this formation depends on the presence or lack of thionein (apo-form). For that purpose 0.25, 0.5 and 1 μM zinc clasp components (CD4 and Lck peptides) were incubated with MT and amounts of T in the range of 0 to 0.5 of T/(T + MT) for 12 h. After this period, fluorescence at 525 nm was recorded and normalized to the fraction of the folded clasp domain. Fig. 5 shows normalized results for ZC domain formation in the changing T/(T + MT) ratio and domain concentration. With 100% of MT (no thionein present) the complex is mostly folded regardless of concentration; however, 100% was obtained only for the highest ZC concentration. The addition of thionein to a T/(T + MT) ratio of ∼0.3, which is the reference value for various tissues,61 causes a decrease in the zinc clasp fraction to ∼80% when both components are used at 1 μM concentration (CD4 + Lck). A four times lower concentration of CD4 and Lck peptides (0.25 μM) results in the formation of only 65% of the domain under the same ratio of T/(T/MT). Interestingly, a decreased concentration of clasp domain components results in incomplete complex formation, even in the absence of thionein.Discussion
General considerations
The first mathematical representation of chemical reactions was posited more than 150 years ago by Guldberg and Waage.62 Since that time, the canonical law of mass action has stated consistently that the velocity of the reaction is proportional to the product of the reactants. The proportionality of the reaction rate with the concentration of substrates comes from the higher encounter probability, leading to extensive complex formation. Consequently, the affinity concept, although subconsciously intuitive, is directly derived from the law of mass action as well. This conjecture furthermore describes the chemical reaction in the equilibrium state, where the concentrations of the chemicals involved bear a constant relationship to each other, which is described by an equilibrium constant (see below) that is valuable to compare the molecular complex affinities. While applied in a wide range of scientific areas, even to describe chaos and complicated oscillations, applications of the law of mass action in modeling of biological systems can be an extreme idealization, especially in terms of defining the parameters particularly sensitive or influential to the system, but not in terms of the definition of the law itself. The numerous tightly intermingled forces, factors and spatial aspects lead to the use of power-law formulations to model most of the biological systems to date,63 such as the Hill equation applied in this study. However, the foundation for a general modeling framework in biology, which will help uncover the structure of the regulatory functioning of cellular systems and their position in state space, is beyond the capacity of today's science.Proteins are nature's most widespread and versatile building blocks, maintaining dynamicity of complex cellular systems through a set of non-covalent interactions modulating the prevalence of essential assemblies. Cellular utilization of metal ion coordination in a protein interface design is derived from its wide availability in the early evolution stage, small interaction surface ensuring proteins’ proximity during optimization and additional gain of orientation specificity, which led to 30–40% of all proteins being associated transiently or permanently with metal ions.4,64 The construction of functional metal centers embraces the exquisite tuning of metal selectivity and reactivity through a combination of interactions65 as well as complex geometries with various binding architectures, which led to diversification of affinities providing different rates of their saturation under cellular conditions.
Although the position of Zn(II) in intraprotein assemblies as a structural and catalytic site is well established, the recognition of interprotein zinc sites is more sophisticated and thus escapes detection or focused attention. Zn(II)-stabilized permanent or transient interprotein interactions enable multisubunit complexes involved in cellular signaling to adopt distinct conformations in response to stimuli and perform challenging chemical reactions in consequence. Although obeying the law of mass action for zinc proteins is indisputable, we aimed to put it in the cellular context, where Zn(II) seems to play the role of a limitation factor demonstrated in its strict cellular buffering and where concentrations of proteins undergo alterations as well. To examine the role of parameters influential for Zn(II) complex formation (Zn(II) and protein concentration), we aimed to determine the assembly metalation rate in four examples of inter- and intraprotein Zn(II) complexes.
The relationship between intra- and interprotein complexes and free metal concentration
Taking into consideration the factor of Zn(II)-dependent changes in zinc domain formation, it is clearly seen that the intraprotein zinc complexes (classical zinc finger and LIM domain) demonstrate high similarity. However, the distinction can be seen in the case of the ZF complex, where the inflection points correspond exceptionally to a logarithmic value of its dissociation constants (−log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Kd) defined in eqn (1) and (2) due to its ZnP stoichiometry. Nevertheless, when the stoichiometry of the assembly under examination is different from 1
Kd) defined in eqn (1) and (2) due to its ZnP stoichiometry. Nevertheless, when the stoichiometry of the assembly under examination is different from 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, such as for the LIM domain with Zn2P stoichiometry, that dependence is no longer valid (eqn (3) and (4)). Regarding the affinity of the individual Zn(II) towards its site within the LIM domain, the dissociation constant is non-distinguishable for both zinc sites and is 14.2 (−log
1, such as for the LIM domain with Zn2P stoichiometry, that dependence is no longer valid (eqn (3) and (4)). Regarding the affinity of the individual Zn(II) towards its site within the LIM domain, the dissociation constant is non-distinguishable for both zinc sites and is 14.2 (−log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Kd) for each (Kd1 = Kd2),41 but if the definition is consistent with the law of mass action, the dissociation constant (−log
Kd) for each (Kd1 = Kd2),41 but if the definition is consistent with the law of mass action, the dissociation constant (−log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Kd) of the Zn2P complex will be 28.4 (Kd1 × Kd2). It is worth noting that this simplification is frequently used for zinc sites that bind more than one metal ion per protein and may lead to misunderstanding and cumulative errors.
Kd) of the Zn2P complex will be 28.4 (Kd1 × Kd2). It is worth noting that this simplification is frequently used for zinc sites that bind more than one metal ion per protein and may lead to misunderstanding and cumulative errors.| Zn(II)free + P ⇆ ZnP | (1) |
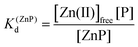 | (2) |
| 2Zn(II)free + P ⇆ Zn2P | (3) |
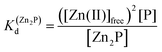 | (4) |
Metal-coupled folding of interprotein domains occurs in a significantly different manner when compared with ZF and LIM, showing strict concentration dependence of the protein subunits. It is worth noting that despite the variation in pZn0.5 when the concentration of peptides is changing, the dissociation constants (Kd) of the ZH and ZC complexes remain constant in agreement with the law of mass action (eqn (5)–(8)). The Zn(II)-dependent behavior of the zinc clasp is comparable to the zinc hook domain formation, but the functional pZn range is slightly shifted for the examined concentration range. Although the zinc clasp system indicates the same ternary binding mode as ZH, it should be underlined that ZC is a heterodimer (ZnP1P2) and the definition of its formation/dissociation equilibrium is quite different (eqn (7) and (8)). Within this approach, equal concentrations of CD4 (P1) and Lck (P2) were used, but it is worth being aware of their variation in cellular conditions and the limitation of the formed heterodimer fraction by the particular protein component of the system.
| Zn(II)free + 2P ⇆ ZnP2 | (5) |
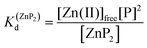 | (6) |
| Zn(II)free + P1 + P2 ⇆ ZnP1P2 | (7) |
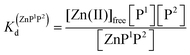 | (8) |
Demonstration of the strict dependence of Zn(II) status on the level of a functional complex in view of the protein architecture, according to the fundamental chemical principle, has a cellular impact on the proper functioning of the zinc metalloproteome. The relatively low concentration of available Zn(II), which is buffered under cellular conditions, makes it a limiting factor following the equilibrium state of the Zn(II) complex system. Even distinct free Zn(II) fluctuations may alter the outcome of the assembly, regardless of the binding stoichiometry. The prominent factor that can attenuate alteration of the metalation process is the affinity of the zinc binding site, discussed in more detail below. It is also important to be aware of both the metal binding mode and the definition of the dissociation constant used in its determination to valuably compare the Zn(II) complex affinities as long as the experimental conditions significantly diminish the Kd values as well.
Impact of the metal binding mode and protein concentration on zinc protein complex assembly
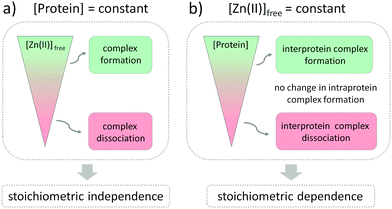
The influence of Zn(II) ions on zinc protein assembly formation indicates independence of the stoichiometry of investigated zinc binding sites (Fig. 6a). Nevertheless, the core point is that regulation of Zn(II) interprotein saturation is not only driven by changes in free Zn(II) but is also closely related to fluctuations in the protein subunit concentration (Fig. 6b), which makes it an additional factor differentiating the intra- and interprotein complexes. This seems reasonable as the formation of the intermolecular Zn(II) binding site requires association of two protomers, and larger amounts of the ternary complexes are formed when the subunit concentration is higher as well. However, when we observe it from the cellular perspective, a more interesting view appears. Modulation of the local protein abundance affects the free Zn(II) concentration needed to obtain the same level of a zinc complex. Thanks to the interdependence of free Zn(II) status and local protein concentration in complex formation effectiveness, the cell gains an additional way to modulate the necessary zinc assembly content. It seems to be especially advantageous when the spatiotemporal separation of processes is required, without disturbing the Zn(II) homeostasis within the cell. One way to increase the local abundance of interacting components is by embedding them in the membranes or even in the membrane platforms with higher specificity to enter, such as lipid rafts or nanoclusters. It has been shown for the zinc clasp domain that this interaction contributes to the enrichment of CD4 within lipid domains, prompting crucial cellular processes, but the role of Zn(II) has not been established yet.66,67The extremely high dynamic range of protein abundance in cells, from less than 50 to more than 106 molecules,68 is conducive to the potential regulatory role of intermolecular zinc sites. Moreover, local versus global concentrations of proteins display great diversity.69 The dynamic characteristic revealed in spatial distribution impinges on the higher encounter probability of interprotein interactions on the one hand and diminishes the Zn(II) ion concentration needed to form functional complexes on the other, thereby achieving a win–win situation. Although proteomic abundance profiling approaches have yielded valuable insights into changes in protein expression lately,70 they mostly lack the spatial dimension, which makes them useless in the prediction of interfacial zinc interactions. Global dynamic mapping of protein subcellular localization along with their concentration has just started to be performed,71 but the high variability of the investigated systems renders the identification of genuine organellar transitions difficult.72
Interprotein zinc domain formation in a metallothionein/thionein buffer system
The main proteins responsible for maintaining the free Zn(II) concentration in the required cellular ranges, its storage and redistribution are the family of metallothioneins (MT) capable of binding up to seven Zn(II) into two clusters.60,73 On the one hand, metallothionein functions as a storage protein of Zn(II) ions, while on the other it regulates the function of zinc proteins, serving as a donor and acceptor of Zn(II) ions. The flexibility of Zn(II) in the coordination sphere facilitates the diversification of Zn(II) ion binding affinities, thus enabling the presence of partially saturated Zn7−xMT species and wide cellular buffering properties in a pZn from the nanomolar to picomolar range.49 The level of the buffered zinc pool is frequently expressed in the amount of thionein within the T/(T + MT) ratio with the reference value ∼0.3 for various tissues.61,74 In our studies, we successfully utilized the high dynamics of this system and the quick equilibration of thionein to particular zinc species75 and showed that the regulation of Zn(II)-dependent ternary zinc clasp complex formation is indeed maintained under the metallothionein buffer environment mimicking the eukaryotic cellular Zn(II) availability. Re-equilibration of Lck and CD4 peptides with metallothionein displays the connection of functional zinc clasp domain formation with both T/(T + MT) ratios and peptide mixture concentration. Moreover, the local abundance of interacting interfaces and cellular transience of the pZn value mutually modulate the efficiency of functional complex formation. As a result, physiological changes in the T/MT equilibrium may influence the formation of the ternary zinc clasp complex of the N-terminal region of kinase Lck with T-cell CD4 coreceptors and consequently alter the T-cell receptor activation cascade.76Diversification of Zn(II) affinities is related to the regulatory role of zinc proteins
The zinc homeostatic machinery uses different strategies to maintain the Zn(II)-to-protein affinity including changes in binding mode architecture, the presence or lack of stabilizing interaction in the second coordination sphere, and the metal-coupled folding process. This maintenance is critical for protein saturation under physiological conditions, both constant and transient. High-affinity zinc sites in proteins are mostly structural and catalytic, such as zinc fingers in transcription factors and Zn(II) coordinating sites in enzymes at the head. Tightly bound Zn(II) ensures their constant saturation with it, so as to be available to perform protein cellular roles. Such proteins cannot afford to lose their Zn(II), because it would likely cause pathological states of the cell.77 However, within the zinc finger class of proteins, MTF-1 is an example where binding affinities are lowered by about 10–50 fold in contrast with the classical picomolar structural sites, even though the donor atoms are identical.78 Modulation of protein affinity to Zn(II) is one way to increase the regulatory role of zinc assembly, affecting its saturation and functionality in consequence. The indirect regulatory mechanisms include phosphorylation by several kinases or S-nitrosylation of thiols in zinc–sulfur clusters.79 The lower affinity prevents the Zn(II) from being constantly bound to the proteins and the Zn(II) ion fluctuations along with protein localization act as triggers to maintain the functional zinc domain formation. One may see the diversification of Zn(II)-to-protein affinities in the case of proteins detecting the zinc signals, thereby performing transient Zn(II)-dependent activities. The affinity of Zn(II)-to-protein tyrosine phosphatase (PTP) family members varies significantly, resulting in different patterns of reactivity toward free Zn(II) fluctuations.27,80 As a result, some of the cellular regulatory pathways, dependent on PTP activity, are not regulated by available Zn(II) in the physiological range.81 The modulation of zinc protein regulation may also involve its binding to the active site of the protein or its distinct parts with a different set of ligands, which has been reported for the caspases participating in the central cascades of apoptosis pathways.82,83 A multitude of factors determine Zn(II) regulation, including the physicochemical properties of protein, stability of interaction, on and off kinetics, stoichiometry of complexes, metal-to-ligand reactivity and protein structure. It is important to note that the mechanisms that alter the Zn(II)-to-protein binding properties mutually influence one another. Transient activation or deactivation of proteins is a central issue of Zn(II) regulatory function and zinc signaling. Increasingly, the amount of research on this topic shows that we are starting to understand the underlying principles of Zn(II)-dependent regulation of protein functions, revealing their great diversity.Significance of metallointeractome regulation
The formation of the functional zinc domains is regulated by two factors in the cellular-like environment: the free Zn(II) availability and the metal binding protein concentration (Fig. 6). According to the law of mass action, both of these shift equilibria in the direction of either domain formation or complex dissociation to its components depending on the collision rate probability. To what extent the parameters influence the regulation of zinc domain formation is mostly dictated by the zinc domain architecture. Fluctuations in free Zn(II) affect the whole zinc proteome regardless of the stoichiometry, so long as the range of pZn fluctuations covers the Zn(II)-to-protein affinity. Setting the protein affinities in particular ranges for the zinc fluxes to reach using different strategies of modification of the coordination and outer spheres is the prominent method for regulation of zinc protein functionality. However, when we examine the influence of the protein subunit concentration on the interfacial zinc domain assembly we can see that gaining the additional parameter regulating the net result of functional protein complex formation is valuable for the cell. Ternary-manner interactions through the metal interface possess their Zn(II) in a different way since the formation of a zinc binding site requires spatiotemporal association of at least three components, including Zn(II). The role of zinc interfaces, differing from most of the structural and catalytic zinc sites, seems to contribute significantly to zinc site regulation thanks to the behavior in a concentration-dependent manner. Although the number of identified interfacial zinc sites is not comparable to the intraprotein zinc domains, the difficulties with their identification, including the lack of a coordination sphere and geometry prediction, necessitate further exploration. In contrast, intraprotein zinc complexes, such as transcription factors (even though their role may be stated as regulatory as well) are resistant to the protein concentration-dependent factor, thus maintaining more constant saturation, and the exclusive Zn(II) dependence seems to be the right choice for their regulation.To gain even higher specificity of protein–protein interactions, cells willingly reduce the dimensionality by pursuing the interactions with the involvement of membranes. Lipid rafts facilitate distinct protein–protein interactions to build signaling complexes by increasing their local concentration and the probability of interaction with other molecules from the network. The role of membrane trafficking as well as the prominence of Zn(II) ions has been well established in the regulation of T lymphocytes and the entire immune system.84 The compactly folded zinc clasp domain (examined in this study), which renders the Lck recruitment enabling further phosphorylation events essential to initiate the signal transduction cascade in early T-cell signaling, contributes to the enrichment of CD4 in lipid rafts. This entails its ability to enhance receptor tyrosine phosphorylation67 as well as potentially altering the dose response of T cells to interleukin-7, which indicates the significance of metallointeractome in the major cellular processes.85 Another way to increase the specificity of Zn(II)-dependent interactions is the mediation of free Zn(II) changes by modification of cysteine residues in the Zn(II)-thiolate coordination environment by oxidative and nitrosative stress.79 Metallothionein clusters are prone to oxidation, which leads to Zn(II) accumulation and neuronal cell death.86 Several phosphatases are also redox sensitive and are inhibited by direct thiol oxidation.87 However, Zn(II) release in oxidative pathways involves normal physiological functions as well.88 Associations of redox and zinc signals are thus integrally coupled to cellular redox potential changes and serve as redox-sensitive switches for zinc signals.2,17
Conclusions
Overall, our results show that not only the availability of Zn(II) itself but also the local abundance of proteins from the zinc interactome network affect the formation of functional Zn(II) complexes involved in cellular homeostasis. Although the changes in free Zn(II) concentration influence the formation of complexes with no regard to their stoichiometry, the zinc binding mode seems to be crucial for the protein subunits’ concentration-dependent regulation. The important cellular context is that to gain the same level of intermolecular zinc complex saturation, a reduced free Zn(II) ion concentration is necessary along with an increasing amount of protein. The presence of interfacial Zn(II)-mediated interaction is then controlled in a dual, free Zn(II) and concentration-dependent manner under a cellular pZn range entailing major implications for the balance between health and disease. The high stability of certain zinc assemblies, especially zinc fingers, is required to keep them suitably folded at even a low free Zn(II) concentration, and their susceptibility to oxidation or modification is not dependent on protein concentration. If the affinity is high enough, those sites remain saturated, and this state is not dependent on zinc signals. Interprotein zinc binding sites are more exposed to dissociation under cellular free Zn(II) changes. This behavior is closely connected with the function and dynamics of Zn(II)-mediated complexes.Author contributions
A. Krężel designed the experiments. A. Kocyła and J. Adamczyk performed the experiments. A. Krężel, J. Adamczyk and A. Kocyła analyzed the data. A. Krężel and A. Kocyła wrote the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was financially supported by a grant from the National Science Centre (NCN) under Opus grant no. 2014/13/B/NZ1/00935. The authors thank Tomasz Kochańczyk, Michał Bojarski and Agnieszka Drozd for help with the spectrofluorometric experiments, protein preparation and helpful discussion. The research resulted from the networking efforts of the COST Action TD1304. The project is supported by the Wroclaw Centre of Biotechnology, Leading National Research Centre (KNOW) program for years 2014–2018.References
- J. A. Marsh and S. A. Teichmann, Structure, dynamics, assembly, and evolution of protein complexes, Annu. Rev. Biochem., 2015, 84, 551–575 CrossRef CAS PubMed.
- W. Maret, Zinc biochemistry: from a single zinc enzyme to a key element of life, Adv. Nutr., 2013, 4, 82–91 CrossRef CAS PubMed.
- J. E. Coleman, Zinc proteins: enzymes, storage proteins, transcription factors, and replication proteins, Annu. Rev. Biochem., 1992, 61, 897–946 CrossRef CAS PubMed.
- C. Andreini, L. Banci, I. Bertini and A. Rosato, Counting the zinc-proteins encoded in the human genome, J. Proteome Res., 2006, 5, 196–201 CrossRef CAS PubMed.
- W. Maret, Protein interface zinc sites: the role of zinc in the supramolecular assembly of proteins and in transient protein–protein interactions, in Handbook of Metalloproteins, ed. W. B. Albrecht Messerschmidt and M. Cygler, John Wiley& Sons, Ltd, 2004 Search PubMed.
- W. J. Song and F. A. Tezcan, A designed supramolecular protein assembly with in vivo enzymatic activity, Science, 2014, 346, 1525–1528 CrossRef CAS PubMed.
- D. S. Auld, Zinc coordination sphere in biochemical zinc sites, BioMetals, 2001, 14, 271–313 CrossRef CAS PubMed.
- Y. Li and W. Maret, Transient fluctuations of intracellular zinc ions in cell proliferation, Exp. Cell Res., 2009, 315, 2463–2470 CrossRef CAS PubMed.
- A. Krężel and W. Maret, Zinc-buffering capacity of a eukaryotic cell at physiological pZn, J. Biol. Inorg. Chem., 2006, 11, 1049–1062 CrossRef PubMed.
- R. A. Colvin, A. I. Bush, I. Volitakis, C. P. Fontaine, D. Thomas, K. Kikuchi and W. R. Holmes, Insights into Zn2+ homeostasis in neurons from experimental and modeling studies, Am. J. Physiol.: Cell Physiol., 2008, 45701, 726–742 CrossRef PubMed.
- J. L. Vinkenborg, T. J. Nicolson, E. A. Bellomo, M. S. Koay, G. A. Rutter and M. Merkx, Genetically encoded FRET sensors to monitor intracellular Zn2+ homeostasis, Nat. Methods, 2009, 6, 737–740 CrossRef CAS PubMed.
- A. M. Hessels, P. Chabosseau, M. H. Bakker, W. Engelen, G. A. Rutter, K. M. Taylor and M. Merkx, eZinCh-2: A Versatile, Genetically Encoded FRET Sensor for Cytosolic and Intraorganelle Zn2+ Imaging, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 7351–7356 CrossRef CAS PubMed; A. M. Hessels, P. Chabosseau, M. H. Bakker, W. Engelen, G. A. Rutter, K. M. Taylor and M. Merkx, ACS Chem. Biol., 2015, 10, 2126–2134 CrossRef PubMed.
- W. Maret and Y. Li, Coordination dynamics of zinc in proteins, Chem. Rev., 2009, 109, 4682–4707 CrossRef CAS PubMed.
- T. Kochańczyk, A. Drozd and A. Krężel, Relationship between the architecture of zinc coordination and zinc binding affinity in proteins – insights into zinc regulation, Metallomics, 2015, 7, 244–257 RSC.
- R. Bozym, F. Chimienti, L. J. Giblin, G. W. Gross, I. Korichneva, Y. Li, S. Libert, W. Maret, M. Parviz, C. J. Frederickson and R. B. Thompson, Free zinc ions outside a narrow concentration range are toxic to a variety of cells in vitro, Exp. Biol. Med., 2010, 235, 741–750 CrossRef CAS PubMed.
- A. Krężel and W. Maret, Different redox states of metallothionein/thionein in biological tissue, Biochem. J., 2007, 402, 551–558 CrossRef PubMed.
- A. Krężel, Q. Hao and W. Maret, The zinc/thiolate redox biochemistry of metallothionein and the control of zinc ion fluctuations in cell signaling, Arch. Biochem. Biophys., 2007, 463, 188–200 CrossRef PubMed.
- R. A. Colvin, W. R. Holmes, C. P. Fontaine and W. Maret, Cytosolic zinc buffering and muffling: their role in intracellular zinc homeostasis, Metallomics, 2010, 2, 306–317 RSC.
- W. Maret, Metals on the move: zinc ions in cellular regulation and in the coordination dynamics of zinc proteins, BioMetals, 2011, 24, 411–418 CrossRef CAS PubMed.
- T. Hirano, M. Murakami, T. Fukada, K. Nishida, S. Yamasaki and T. Suzuki, Roles of zinc and zinc signaling in immunity: zinc as an intracellular signaling molecule, Adv. Immunol., 2008, 97, 149–176 CAS.
- J. Kaltenberg, L. M. Plum, J. L. Ober-Blobaum, A. Honscheid, L. Rink and H. Haase, Zinc signals promote IL-2-dependent proliferation of T cells, Eur. J. Immunol., 2010, 40, 1496–1503 CrossRef CAS PubMed.
- F. Radtke, R. Heuchel, O. Georgiev, M. Hergersberg, M. Gariglio, Z. Dembic and W. Schaffner, Cloned transcription factor MTF-1 activates the mouse metallothionein I promoter, EMBO J., 1993, 12, 1355–1362 CAS.
- C. Hogstrand, D. Zheng, G. Feeney, P. Cunningham and P. Kille, Zinc-controlled gene expression by metal-regulatory transcription factor 1 (MTF1) in a model vertebrate, the zebrafish, Biochem. Soc. Trans., 2008, 36, 1252–1257 CrossRef CAS PubMed.
- S. Yamasaki, K. Sakata-Sogawa, A. Hasegawa, T. Suzuki, K. Kabu, E. Sato, T. Kurosaki, S. Yamashita, M. Tokunaga, K. Nishida and T. Hirano, Zinc is a novel intracellular second messenger, J. Cell Biol., 2007, 177, 637–645 CrossRef CAS PubMed.
- H. Haase, J. L. Ober-Blobaum, G. Engelhardt, S. Hebel, A. Heit, H. Heine and L. Rink, Zinc signals are essential for lipopolysaccharide-induced signal transduction in monocytes, J. Immunol., 2008, 181, 6491–6502 CrossRef CAS.
- S. Yamasaki, A. Hasegawa, S. Hojyo, W. Ohashi, T. Fukada, K. Nishida and T. Hirano, A novel role of the L-type calcium channel α1D subunit as a gatekeeper for intracellular zinc signaling: zinc wave, PLoS One, 2012, 7, e39654 CAS.
- E. Bellomo, K. B. Singh, A. Massarotti, C. Hogstrand and W. Maret, The metal face of protein tyrosine phosphatase 1B, Coord. Chem. Rev., 2016, 327–328, 70–83 CrossRef CAS PubMed.
- Y.-M. Kim, W. Reed, W. Wu, P. A. Bromberg, L. M. Graves and J. M. Samet, Zn2+-induced IL-8 expression involves AP-1, JNK, and ERK activities in human airway epithelial cells, Am. J. Physiol.: Lung Cell. Mol. Physiol., 2006, 290, L1028–L1035 CrossRef CAS PubMed.
- K. M. Taylor, S. Hiscox, R. I. Nicholson, C. Hogstrand and P. Kille, Protein kinase CK2 triggers cytosolic zinc signaling pathways by phosphorylation of zinc channel ZIP7, Sci. Signaling, 2012, 5(210), ra11 Search PubMed.
- C. J. Frederickson, J.-Y. Koh and A. I. Bush, The neurobiology of zinc in health and disease, Nat. Rev. Neurosci., 2005, 6, 449–462 CrossRef CAS PubMed.
- S. L. Sensi, P. Paoletti, A. I. Bush and I. Sekler, Zinc in the physiology and pathology of the CNS, Nat. Rev. Neurosci., 2009, 10, 780–791 CrossRef CAS PubMed.
- E. L. Que, R. Bleher, F. E. Duncan, B. Y. Kong, S. C. Gleber, S. Vogt, S. Chen, S. A. Garwin, A. R. Bayer, V. P. Dravid, T. K. Woodruff and T. V. O'Halloran, Quantitative mapping of zinc fluxes in the mammalian egg reveals the origin of fertilization-induced zinc sparks, Nat. Chem., 2014, 7, 130–139 CrossRef PubMed.
- M. Tamaki, Y. Fujitani, A. Hara, T. Uchida, Y. Tamura, K. Takeno, M. Kawaguchi, T. Watanabe, T. Ogihara, A. Fukunaka, T. Shimizu, T. Mita, A. Kanazawa, M. O. Imaizumi, T. Abe, H. Kiyonari, S. Hojyo, T. Fukada, T. Kawauchi, S. Nagamatsu, T. Hirano, R. Kawamori and H. Watada, The diabetes-susceptible gene SLC30A8/ZnT8 regulates hepatic insulin clearance, J. Clin. Invest., 2013, 123, 4513–4524 CAS.
- D. Wang, S. Y. Gou and D. Axelrod, Reaction rate enhancement by surface diffusion of adsorbates, Biophys. Chem., 1992, 43, 117–137 CrossRef CAS PubMed.
- J. Gureasko, W. J. Galush, S. Boykevisch, H. Sondermann, D. Bar-Sagi, J. T. Groves and J. Kuriyan, Membrane-dependent signal integration by the Ras activator Son of sevenless, Nat. Struct. Mol. Biol., 2008, 15, 452–461 CAS.
- K. Simons and D. Toomre, Lipid rafts and signal transduction, Nat. Rev. Mol. Cell Biol., 2000, 1, 31–39 CrossRef CAS PubMed.
- P. C. Calder and P. Yaqoob, Lipid rafts – composition, characterization, and controversies, J. Nutr., 2007, 137, 545–547 Search PubMed.
- D. A. Brown and E. London, Structure and function of sphingolipid- and cholesterol-rich membrane rafts, J. Biol. Chem., 2000, 275, 17221–17224 CrossRef CAS PubMed.
- D. V. Nicolau, K. Burrage, R. G. Parton and J. F. Hancock, Identifying optimal lipid raft characteristics required to promote nanoscale protein-protein interactions on the plasma membrane, Mol. Cell. Biol., 2006, 26, 313–323 CrossRef CAS PubMed.
- A. Pomorski, T. Kochańczyk, A. Miłoch and A. Krężel, Method for accurate determination of dissociation constants of optical ratiometric systems: chemical probes, genetically encoded sensors, and interacting molecules, Anal. Chem., 2013, 85, 11479–11486 CrossRef CAS PubMed.
- M. Sikorska, A. Krężel and J. Otlewski, Femtomolar Zn2+ affinity of LIM domain of PDLIM1 protein uncovers crucial contribution of protein–protein interactions to protein stability, J. Inorg. Biochem., 2012, 115, 28–35 CrossRef CAS PubMed.
- A. Miłoch and A. Krężel, Metal binding properties of the zinc finger metallome – insights into variations in stability, Metallomics, 2014, 6, 2015–2024 RSC.
- T. Kochańczyk, P. Jakimowicz and A. Krężel, Femtomolar Zn(II) affinity of minimal zinc hook peptides-a promising small tag for protein engineering, Chem. Commun., 2013, 49, 1312–1314 RSC.
- P. Eyer, F. Worek, D. Kiderlen, G. Sinko, A. Stuglin, V. Simeon-Rudolf and E. Reiner, Molar absorption coefficients for the reduced Ellman reagent: reassessment, Anal. Biochem., 2003, 312, 224–227 CrossRef CAS PubMed.
- A. Kocyła, A. Pomorski and A. Krężel, Molar absorption coefficients and stability constants of metal complexes of 4-(2-pyridylazo)resorcinol (PAR): revisiting common chelating probe for the study of metalloproteins, J. Inorg. Biochem., 2015, 152, 82–92 CrossRef PubMed.
- A. Kocyła, A. Pomorski and A. Krężel, Molar absorption coefficients and stability constants of Zincon metal complexes for determination of metal ions and bioinorganic applications, J. Inorg. Biochem., 2017, 176, 53–65 CrossRef PubMed.
- R. W. Sabnis, Handbook of fluorescent dyes and probes, Wiley, 2015 Search PubMed.
- N. E. Wezynfeld, E. Stefaniak, K. Stachucy, A. Drozd, D. Płonka, S. Drew, A. Krężel and W. Bal, Resistance of Cu(Aβ4–16) to copper capture by Metallothionein-3 supports a function for the Aβ4–42 peptide as a synaptic Cu(II) scavenger, Angew. Chem., Int. Ed., 2016, 55, 8235–8238 CrossRef CAS PubMed.
- A. Krężel and W. Maret, Dual nanomolar and picomolar Zn(II) binding properties of metallothionein, J. Am. Chem. Soc., 2007, 129, 10911–10921 CrossRef PubMed.
- A. Krężel, R. Latajka, G. D. Bujacz and W. Bal, Coordination properties of tris(2-carboxyethyl)phosphine, a newly introduced thiol reductant, and its oxide, Inorg. Chem., 2003, 42, 1994–2003 CrossRef PubMed.
- A. Krężel and W. Maret, The biological inorganic chemistry of zinc ions, Arch. Biochem. Biophys., 2016, 611, 3–19 CrossRef PubMed.
- A. E. Martell and R. M. Smith, Critical Stability Constants, Plenum Press, 1974 Search PubMed.
- A. Klug and D. Rhodes, Zinc fingers: a novel protein fold for nucleic acid recognition, Cold Spring Harbor Symp. Quant. Biol., 1987, 52, 473–482 CrossRef CAS PubMed.
- T. Kochańczyk, M. Nowakowski, D. Wojewska, A. Ejchart, W. Koźmiński and A. Krężel, Metal-coupled folding as the driving force for the extreme stability of Rad50 zinc hook dimer assembly, Sci. Rep., 2016, 6, 36346 CrossRef PubMed.
- K. P. Hopfner, L. Craig, G. Moncalian, R. A. Zinkel, T. Usui, B. A. Owen, A. Karcher, B. Henderson, J. L. Bodmer, C. T. McMurray, J. P. Carney, J. H. Petrini and J. A. Tainer, The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair, Nature, 2002, 418, 562–566 CrossRef CAS PubMed.
- P. W. Kim, Z.-Y. J. Sun, S. C. Blacklow, G. Wagner and M. J. Eck, A zinc clasp structure tethers Lck to T cell coreceptors CD4 and CD8, Science, 2003, 301, 1725–1728 CrossRef CAS PubMed.
- P. S. Eis, J. Kuśba, M. L. Johnson and J. R. Lakowicz, Distance distributions and dynamics of a zinc finger peptide from fluorescence resonance energy transfer measurements, J. Fluoresc., 1993, 3, 23–31 CrossRef CAS PubMed.
- Y. B. Park, M. Hohl, M. Padjasek, E. Jeong, K. S. Jin, A. Krężel, J. H. Petrini and Y. Cho, Eukaryotic Rad50 functions as a rod-shaped dimer, Nat. Struct. Mol. Biol., 2017, 24, 248–257 CAS.
- C. Berney and G. Danuser, FRET or No FRET: A Quantitative Comparison, Biophys. J., 2003, 84, 3992–4010 CrossRef CAS PubMed.
- A. Krężel and W. Maret, The functions of metamorphic metallothioneins in zinc and copper metabolism, Int. J. Mol. Sci., 2017, 18, E1237 CrossRef PubMed.
- Y. Yang, W. Maret and B. L. Vallee, Differential fluorescence labeling of cysteinyl clusters uncovers high tissue levels of thionein, Proc. Natl. Acad. Sci. U. S. A., 2001, 98, 5556–5559 CrossRef CAS PubMed.
- C. M. Guldberg and P. Waage, Studier i affiniteten (translation: Studies on affinities), Videnskabs-Selskabet i Christiana, 1864, 35.
- E. O. Voit, H. A. Martens and S. W. Omholt, 150 years of the Mass Action Law, PLoS Comput. Biol., 2015, 11, e1004012 Search PubMed.
- W. J. Song, P. A. Sontz, X. I. Ambroggio and F. A. Tezcan, Metals in protein–protein interfaces, Annu. Rev. Biophys., 2014, 43, 409–431 CrossRef CAS PubMed.
- E. N. Salgado, J. D. Brodin, M. M. To and F. A. Tezcan, Templated construction of a Zn-selective protein dimerization motif, Inorg. Chem., 2011, 50, 6323–6329 CrossRef CAS PubMed.
- M. Foti, M. Phelouzat, A. Holm, B. J. Rasmusson and J. Carpentier, p56 Lck anchors CD4 to distinct microdomains on microvilli, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 2008–2013 CrossRef CAS PubMed.
- R. Fragoso, D. Ren, X. Zhang, M. W. Su, S. J. Burakoff and Y. J. Jin, Lipid raft distribution of CD4 depends on its palmitoylation and association with Lck, and evidence for CD4-induced lipid raft aggregation as an additional mechanism to enhance CD3 signaling, J. Immunol., 2003, 170, 913–921 CrossRef CAS.
- S. Ghaemmaghami, W. K. Huh, K. Bower, R. W. Howson, A. Belle, N. Dephoure, E. K. O'Shea and J. S. Weissman, Global analysis of protein expression in yeast, Nature, 2003, 425, 737–741 CrossRef CAS PubMed.
- J. Wu and T. D. Pollard, Counting cytokinesis proteins globally and locally in fission yeast, Science, 2012, 310, 310–314 CrossRef PubMed.
- S. J. Deeb, S. Tyanova, M. Hummel, M. Schmidt-Supprian, J. Cox and M. Mann, Machine learning-based classification of diffuse large B-cell lymphoma patients by their protein expression profiles, Mol. Cell. Proteomics, 2015, 14, 2947–2960 CAS.
- D. N. Itzhak, S. Tyanova, J. Cox and G. H. H. Borner, Global, quantitative and dynamic mapping of protein subcellular localization, eLife, 2016, 5, e16950 Search PubMed.
- L. Gatto, L. M. Breckels, T. Burger, D. J. Nightingale, A. J. Groen, C. Campbell, N. Nikolovski, C. M. Mulvey, A. Christoforou, M. Ferro and K. S. Lilley, A foundation for reliable spatial proteomics data analysis, Mol. Cell. Proteomics, 2014, 13, 1937–1952 CAS.
- C. Blindauer and O. I. Leszczyszyn, Metallothioneins: unparalleled diversity in structures and functions for metal ion homeostasis and more, Nat. Prod. Rep., 2010, 27, 720–741 RSC.
- A. Pattanaik, C. F. Shaw, D. H. Petering, J. Garvey and A. J. Kraker, Basal metallothionein in tumors: widespread presence of apoprotein, J. Inorg. Biochem., 1994, 54, 91–105 CrossRef CAS PubMed.
- A. Krężel and W. Maret, Thionein/metallothionein control Zn(II) availability and the activity of enzymes, J. Biol. Inorg. Chem., 2008, 13, 401–409 CrossRef PubMed.
- J. M. Rice, A. Zweifach and M. A. Lynes, Metallothionein regulates intracellular zinc signaling during CD4+ T cell activation, BMC Immunol., 2016, 17, 13 CrossRef PubMed.
- H. Haase, D. J. Mazzatti, A. White, K. H. Ibs, G. Engelhardt, S. Hebel, J. R. Powell and L. Rink, Differential gene expression after zinc supplementation and deprivation in human leukocyte subsets, Mol. Med., 2007, 13, 362–370 CAS.
- J. H. Laity and G. K. Andrews, Understanding the mechanisms of zinc-sensing by metal-response element binding transcription factor-1 (MTF-1), Arch. Biochem. Biophys., 2007, 463, 201–210 CrossRef CAS PubMed.
- K. D. Kroncke, Zinc Finger proteins as molecular targets for nitric oxide-mediated gene regulation, Antioxid. Redox Signaling, 2001, 3, 565–575 CrossRef CAS PubMed.
- M. Wilson, C. Hogstrand and W. Maret, Picomolar concentrations of free zinc(II) ions regulate receptor protein–tyrosine phosphatase β activity, J. Biol. Chem., 2012, 287, 9322–9326 CrossRef CAS PubMed.
- L. M. Plum, A. Brieger, G. Engelhardt, S. Hebel, A. Nessel, M. Arlt, J. Kaltenberg, U. Schwaneberg, M. Huber, L. Rink and H. Haase, PTEN-inhibition by zinc ions augments interleukin-2-mediated Akt phosphorylation, Metallomics, 2014, 6, 1277–1287 RSC.
- K. L. Huber and J. A. Hardy, Mechanism of zinc-mediated inhibition of caspase-9, Protein Sci., 2012, 21, 1056–1065 CrossRef CAS PubMed.
- E. M. Velázquez-Delgado and J. A. Hardy, Zinc-mediated allosteric inhibition of caspase-6, J. Biol. Chem., 2012, 287, 36000–36011 CrossRef PubMed.
- M. A. Alonso and J. Millán, The role of lipid rafts in signalling and membrane trafficking in T lymphocytes, J. Cell Sci., 2001, 114, 3957–3965 CAS.
- C. Kittipatarin, N. Tschammer and A. R. Khaled, The interaction of LCK and the CD4 co-receptor alters the dose response of T-cells to interleukin-7, Immunol. Lett., 2010, 131, 170–181 CrossRef CAS PubMed.
- J. Y. Lee, J. H. Kim, R. D. Palmiter and J. Y. Koh, Zinc released from metallothionein-III may contribute to hippocampal CA1 and thalamic neuronal death following acute brain injury, Exp. Neurol., 2003, 184, 337–347 CrossRef CAS PubMed.
- J. J. Tanner, Z. D. Parsons, A. H. Cummings, H. Zhou and K. S. Gates, Redox Regulation of Protein Tyrosine Phosphatases: Structural and Chemical Aspects, Antioxid. Redox Signaling, 2011, 15, 77–97 CrossRef CAS PubMed.
- P. J. Bernal, et al., Nitric oxide mediated zinc release contributes to hypoxic regulation of pulmonary vascular tone, Circ. Res., 2008, 102, 1575–1583 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7mt00301c |
| This journal is © The Royal Society of Chemistry 2018 |