Translesion DNA synthesis across double-base lesions derived from cross-links of an antitumor trinuclear platinum compound: primer extension, conformational and thermodynamic studies†
O.
Novakova
a,
N. P.
Farrell
 b and
V.
Brabec
b and
V.
Brabec
 *a
*a
aInstitute of Biophysics, Czech Academy of Sciences, Kralovopolska 135, CZ-61265 Brno, Czech Republic. E-mail: brabec@ibp.cz
bDepartment of Chemistry, Virginia Commonwealth University, 1001 W. Main Street, Richmond, VA 23284-2006, USA
First published on 28th November 2017
Abstract
Polynuclear platinum complexes represent a unique structural class of DNA-binding agents of biological significance. They contain at least two platinum coordinating units bridged by a linker, which means that the formation of double-base lesions (cross-links) in DNA is possible. Here, we show that the lead compound, bifunctional [{trans-PtCl(NH3)2}2μ-trans-Pt(NH3)2{H2N(CH2)6NH2}2]4+ (Triplatin or BBR3464), forms in DNA specific double-base lesions which affect the biophysical and biochemical properties of DNA in a way fundamentally different compared to the analogous double-base lesions formed by two adducts of monofunctional chlorodiethylenetriamineplatinum(II) chloride (dienPt). We find concomitantly that translesion DNA synthesis by the model A-family polymerase, the exonuclease deficient Klenow fragment, across the double-base lesions derived from the intrastrand CLs of Triplatin was markedly less extensive than that across the two analogous monofunctional adducts of dienPt. Collectively, these data provide convincing support for the hypothesis that the central noncovalent tetraamine platinum linker of Triplatin, capable of hydrogen-bonding and electrostatic interactions with DNA and bridging the two platinum adducts, represents an important factor responsible for the markedly lowered tolerance of DNA double-base adducts of Triplatin by DNA polymerases.
Significance to metallomicsThe most studied antitumor polynuclear platinum complex, Triplatin, forms in DNA specific double-base lesions which affect the biophysical and biochemical properties of DNA differently from the analogous double-base lesions formed by two adducts of monofunctional chlorodiethylenetriamineplatinum(II) chloride. The data demonstrate that the central noncovalent tetraamine platinum linker of Triplatin represents an important factor responsible for the markedly lowered tolerance of DNA double-base adducts of Triplatin by DNA polymerases. The results may be helpful in the future design of polynuclear metal-based compounds whose biological action is connected with a new DNA binding mode mediated by the linkers connecting DNA binding units. |
Introduction
Polynuclear platinum compounds comprise a unique class of anticancer platinum agents with distinct chemical and biological properties different from the mononuclear platinum drugs.1 The concept of development of these drugs was based on the assumption that alteration of DNA binding mode in comparison to that of mononuclear platinum cytostatics would induce differential downstream effects with respect to the processes leading to cell death. The lead compound, [{trans-PtCl(NH3)2}2μ-trans-Pt(NH3)2{H2N(CH2)6NH2}2]4+ (Triplatin or BBR3464) (Scheme 1), is a trinuclear, bifunctional DNA binding agent with an overall charge of 4+, in which the bridging between the two platinating units is made by the [{trans-Pt(NH3)2}μ-{H2N(CH2)6NH2}2] linker. This bridging central unit does not contribute to coordinative DNA binding but incorporates into the linker backbone a charge and hydrogen-bonding capacity which dramatically increase the DNA affinity, affect the charge/lipophilicity balance, and further increase the distance between the two Pt–DNA-binding coordination spheres.2–4 The predominant DNA adducts of Triplatin are long-range {Pt,Pt} cross-links (CLs) distinctly different from short-range CLs of cisplatin5 and its mononuclear derivatives. Preclinical studies indicated that Triplatin is markedly more cytotoxic than cisplatin and its antitumor derivatives used in the clinic1,6,7 and retains activity against cell lines and tumors resistant to cisplatin in vitro and in vivo.8,9 Several reviews on various and unique aspects of the cytotoxicity of Triplatin and other polynuclear platinum compounds are available.10–12 Triplatin underwent Phase I and II human clinical trials but has not yet advanced to full use.13–15 Nonetheless, the contributions that studies of the mechanism of action of Triplatin have made to the delineation of novel concepts of antitumor platinum drug design are unquestionable.There is a large body of experimental evidence that the success of platinum complexes in killing tumor cells results from their ability to form on DNA various types of covalent adducts.5,12 The interactions of antitumor polynuclear platinum compounds with target DNA are distinct from the mononuclear-based cisplatin family and, indeed, unlike those of any DNA-damaging agent in clinical use. The ability of Triplatin to induce DNA adducts such as long-range intra- and interstrand CLs,2 which are not produced by conventional mononuclear platinum compounds, suggests that Triplatin may escape, at least in part, the classical mechanism of cisplatin resistance related to DNA damage recognition and repair [the platinated sites in long-range CLs are separated by two or more base pairs (bps)]. Importantly, the CLs of Triplatin bind damaged-DNA binding-proteins and are processed by enzymes differently than the DNA adducts of cisplatin.16–18
DNA adducts of antitumor platinum drugs, such as cisplatin, are believed to exert their cytotoxic effects by inhibiting both replication and/or transcription and inducing programmed cell death (apoptosis) or necrosis.19 On the other hand, in vitro bypass by replicative DNA polymerases in a process termed translesion DNA synthesis (TLS) by helix distorting DNA lesions including DNA adducts of some antitumor platinum compounds has been observed as well.18,20–24 Tolerance of DNA adducts, a property linked in part to the development of cellular resistance to cisplatin and creation of more aggressive cancer,25 may be considered to be altered through the formation of different DNA adduct structures of Triplatin by circumventing the cellular processes associated with cisplatin–DNA adduct recognition and repair.12,26,27 In order to learn more about the molecular basis of the replication of DNA modified by Triplatin and tolerance of its DNA adducts, we investigated in the present work DNA polymerization using the DNA templates site-specifically modified with two types of CLs formed in DNA by Triplatin. Namely, by intrastrand CLs, such as a 1,2-GG CL in which two neighboring guanine residues were cross-linked or by a long-range 1,5-GTTTG intrastrand CL in which two crosslinked guanine residues in one strand were separated by three intervening nucleotides.
An important feature of bifunctional Triplatin is that two reactive groups in this complex are linked with a relatively long tetraamine platinum unit linker (Scheme 1). To investigate whether this linker affects the processing of the intrastrand CLs of Triplatin by DNA polymerases, we also examined DNA polymerization using the template containing two isolated monofunctional adducts of chlorodiethylenetriamineplatinum(II) chloride (dienPt) formed at the guanine residues in one strand which were platinated by Triplatin in the 1,2-GG or 1,5-GTTTG intrastrand CLs. Thus, the two platinum adducts of monofunctional dienPt formed in one DNA strand, adjacent or separated by three nucleotides, mimic the effects of 1,2- or 1,5 intrastrand CLs of Triplatin eliminating the effect of the linker.
The DNA polymerization was examined using two DNA polymerases, Klenow fragment of DNA polymerase I and reverse transcriptase of human immunodeficiency virus type 1 (RT HIV-1). We chose these DNA polymerases to provide a basis for making similar comparisons with other prokaryotic or eukaryotic polymerases. The extensive genetic, biochemical, and structural studies that have been carried out on Klenow fragment make it an ideal model system for investigating the molecular mechanism of template-directed DNA synthesis and the way in which the polymerases interact with DNA.28 Thus, the DNA polymerase I class of enzymes have served as the prototype for the studies on the structural and biochemical mechanisms of DNA replication.29,30 The exonuclease deficient Klenow fragment (KF−) was selected here because the translesion synthesis proficient DNA polymerases of the X or Y families share some common properties including the lack of associated 3′ to 5′ exonuclease proofreading activity and not least the proofreading mechanism itself may introduce effects more dependent on the adduct type.31 In these studies, we also used RT HIV-1 which also possesses the DNA template-dependent DNA polymerase activity, but relatively low processivity and fidelity.32
The goal of this study, performed in cell-free media, was to show whether there are correlations between the biophysical properties (conformational, thermal and thermodynamic characteristics) of the DNA intrastrand CLs of Triplatin on one hand and the efficiency of these adducts to block DNA polymerization on the other hand. In fact, the CLs of bifunctional polynuclear Triplatin are formally equivalent with two monofunctional platinum adducts connected with the charged “central noncovalent” tetraamine platinum linker, trans-H2N(CH2)6NH2Pt(NH3)2(CH2)6NH2, capable of hydrogen-bonding and electrostatic interactions with DNA.1 Therefore, to investigate the role of the linker in the Triplatin molecule, we performed the experiments, identical to those with the CLs of Triplatin, also with two monofunctional platinum adducts of dienPt formed to mimic the CLs of Triplatin, but to eliminate the effects of its tetraamine platinum linker.
Experimental
Chemicals
Triplatin was prepared by published methods. DienPt was a generous gift of professor G. Natile from the University of Bari. The stock solutions of platinum compounds were prepared at a concentration of 5 × 10−4 M in deionized water and stored at room temperature in the dark. Synthetic oligodeoxyribonucleotides were purchased from VBC-Genomics (Vienna, Austria) and were purified as described previously.33 KF− and T4 polynucleotide kinase were purchased from New England Biolabs (Beverly, MA). Acrylamide, bis(acrylamide), urea, and NaCN were from Merck KgaA (Darmstadt, Germany). Dimethyl sulfate (DMS), KMnO4, diethyl pyrocarbonate (DEPC), KBr and KHSO5 were from Sigma (Prague, Czech Republic). Radioactive products were from MP Biomedicals, LLC (Irvine, CA).Platination of oligonucleotides
Oligonucleotides containing single, 1,2-GG or 1,5 intrastrand CL of Triplatin or the two monofunctional adducts of dienPt were prepared as described.17,33 Briefly, the single-stranded oligonucleotides (23mer or 25mer shown in Fig. 1A and B, respectively) were reacted in stoichiometric amounts with Triplatin or with two equivalents of dienPt in H2O at 310 K for 24 h. The platinated oligonucleotides were repurified by ion-exchange HPLC. It was verified by platinum graphite furnace atomic absorption spectrophotometry (GF-AAS) and by the measurements of the optical density that the modified oligonucleotides contained two platinum atoms. It was also verified using DMS footprinting of platinum on DNA34 that, in the platinated strands, the N7 position of the two guanine residues was not accessible for the reaction with DMS.Inhibition of DNA polymerization
The 25mer or 23mer templates (see Fig. 1A and B) containing a single intrastrand CL of Triplatin or two monofunctional adducts of dienPt were prepared as described above. An 8mer DNA primer (its sequence is shown in Fig. 1A and B) was complementary to the 3′ termini of the 23mer or 25mer templates. DNA substrates (5 × 10−8 M) were formed by annealing the templates and 5′-end-labeled 8mer primers at a molar ratio of 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. All the experiments using KF− were performed at 298 K in 50 μl of a buffer containing 50 mM Tris–HCl (pH 7.4), 10 mM MgCl2, 0.1 mM DTT, 50 μg mL−1 BSA with KF− (0.5 U) in the presence of all four deoxyribonucleotide 5′-triphosphates (dNTPs, 25 μM). The reactions were terminated by the addition of EDTA so that its resulting concentration was 20 mM and heating at 373 K for 30 s. The products were resolved by denaturing 20% polyacrylamide (PAA)/8 M urea gel and visualized by autoradiography. The amount of TLS was defined as the amount of radioactivity corresponding to the products 21–25 nucleotides long on the 5′ side of the template strand and beyond divided by the total radioactivity in the lane. Other details have been published previously.35,36
1. All the experiments using KF− were performed at 298 K in 50 μl of a buffer containing 50 mM Tris–HCl (pH 7.4), 10 mM MgCl2, 0.1 mM DTT, 50 μg mL−1 BSA with KF− (0.5 U) in the presence of all four deoxyribonucleotide 5′-triphosphates (dNTPs, 25 μM). The reactions were terminated by the addition of EDTA so that its resulting concentration was 20 mM and heating at 373 K for 30 s. The products were resolved by denaturing 20% polyacrylamide (PAA)/8 M urea gel and visualized by autoradiography. The amount of TLS was defined as the amount of radioactivity corresponding to the products 21–25 nucleotides long on the 5′ side of the template strand and beyond divided by the total radioactivity in the lane. Other details have been published previously.35,36
Chemical probes of DNA conformation
The 25mer or 23mer (the sequences are shown in Fig. 1A and B, respectively) nonmodified or containing the two monofunctional adducts of dienPt was allowed to anneal with a nonplatinated complementary strand in 0.02 M NaClO4 and used immediately in further experiments. The top (platinated) or bottom (complementary) strands of the duplex were 5′-end-labeled with [γ-32P]ATP. This annealing procedure included a rapid heating of the mixture of the complementary oligonucleotides to 328 K followed by incubation at 298 K for 2 h. It was verified that under these conditions, the monofunctional adducts of dienPt were stable for at least 24 h. The modifications of double-stranded oligonucleotides by KMnO4, DEPC, and KBr/KHSO5 were performed as described previously.37–40 In the case of the platinated oligonucleotides, the platinum complex was removed after the reaction of the DNA with the probe by incubation with 0.2 M NaCN (pH 11) at 318 K for 10 h in the dark.Differential-scanning calorimetry
The 15-bp duplexes nonmodified or containing the double-base adducts of Triplatin or dienPt used in the DSC experiments (ESI,† Fig. S3A) were prepared in the same way as the substrates used in the experiments in which the inhibition of DNA polymerization was examined. The only exception was the 15-bp duplex with the central sequence TGTTTGT in the top strand [duplex TGTTTGT(15) shown in the ESI,† Fig. S3A] containing a unique adduct of dienPt formed at the 5′-G. In the pyrimidine-rich strand containing the central sequence TGTTTGT, 3′-G was replaced by 7-deazaguanine (7-deazaG). The 7-deazaG is an isosteric analog of native guanine in which the aromatic N7 atom is replaced by C–H, which accurately mimics the properties of the natural base. As the guanine N7 is the site at which platinum complexes preferentially react in DNA, 7-deazaG is incapable of forming platinum adducts.41 Thus, 7-deazaG makes it possible to prepare a single monofunctional adduct of dienPt uniquely at the 5′-G in a TGTTTGT sequence context of pyrimidine-rich oligodeoxyribonucleotides, that is, 3′-G is 7-deaza-G.Thermograms depicting excess heat capacity (ΔCp) versus temperature profiles for the thermally induced transitions of non-modified duplexes (for their sequences, see ESI,† Fig. S3) or duplexes containing the adducts of dienPt or Triplatin were obtained in the same way as described in our previous articles.42–44 Briefly, the thermograms were obtained using a Nano DSC Calorimeter (Nano DSC, TA Instruments); the concentration of the duplexes was 6.6 μM, the heating rate was 1 K min−1, and the maximum temperature was 360 K. After reaching the maximum temperature, the samples were cooled at the same rate to the starting temperature of 283 K. The enthalpy (ΔH) and entropy (ΔS) values of duplex melting were calculated using the ORIGIN v.5.0 software (Microcal). The free energy of duplex dissociation at 298 K (ΔG0298) was calculated using the standard thermodynamic relationship given in [eqn (1)] and the corresponding ΔH and ΔS values:
| ΔG0298 = ΔH − 298.15ΔS | (1) |
Polymerase dissociation experiments
The reaction conditions and DNA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KF− ratios were the same as those described above for the experiments demonstrating the inhibition of DNA polymerization by KF−. Other conditions were also the same as described in a previously published article.50 Briefly, we used short duplex substrates (17mer primer/30mer template) and long duplex substrates (40mer primer/53mer template) shown in Fig. 3A and B. The ability of KF− to equally recognize and bind both the short and long duplex substrates was verified by mixing defined ratios of the short and long duplexes in the same way as described in the previously published article.50 The results (not shown) demonstrated that our short and long duplexes were equal substrates for KF−. In the experiments to determine the preference of KF− for either short or long substrates, the total primer/template concentration remained constant, 55 nM, with 10 nM being 32P 5′ end labeled. The ratio of short
KF− ratios were the same as those described above for the experiments demonstrating the inhibition of DNA polymerization by KF−. Other conditions were also the same as described in a previously published article.50 Briefly, we used short duplex substrates (17mer primer/30mer template) and long duplex substrates (40mer primer/53mer template) shown in Fig. 3A and B. The ability of KF− to equally recognize and bind both the short and long duplex substrates was verified by mixing defined ratios of the short and long duplexes in the same way as described in the previously published article.50 The results (not shown) demonstrated that our short and long duplexes were equal substrates for KF−. In the experiments to determine the preference of KF− for either short or long substrates, the total primer/template concentration remained constant, 55 nM, with 10 nM being 32P 5′ end labeled. The ratio of short![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) long duplexes tested was 1
long duplexes tested was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10. When KF− was tested for dissociation from the short duplex substrates, KF− (0.007 U) was prebound to the short substrates (5.8 nM). The reactions were initiated by the addition of dNTPs together with the long duplex substrate in 10-fold molar excess (50 nM with 10% end labeled). The reactions were incubated at 310 K for the indicated times and quenched by the addition of an equal volume of stop dye.
10. When KF− was tested for dissociation from the short duplex substrates, KF− (0.007 U) was prebound to the short substrates (5.8 nM). The reactions were initiated by the addition of dNTPs together with the long duplex substrate in 10-fold molar excess (50 nM with 10% end labeled). The reactions were incubated at 310 K for the indicated times and quenched by the addition of an equal volume of stop dye.
Other physical methods
Absorption spectra were measured using a Beckmann DU-7400 spectrophotometer. GF-AAS measurements were carried out using a Varian AA240Z Zeeman atomic absorption spectrometer equipped with a GTA 120 graphite tube atomizer. For GF-AAS analysis, the modified oligonucleotides were dialyzed against deionized water. Purification of oligonucleotides with the aid of HPLC was carried out on a Waters HPLC system consisting of Waters 262 Pump, Waters 2487 UV detector and Waters 600S Controller with MonoQ HR 5/5 column. Gels were visualized using a BAS 2500 FUJIFILM bioimaging analyzer, and the radioactivities associated with the bands were quantitated with the AIDA image analyzer software (Raytest, Germany).Results and discussion
DNA synthesis by KF− and RT HIV-1 across the double-base adducts
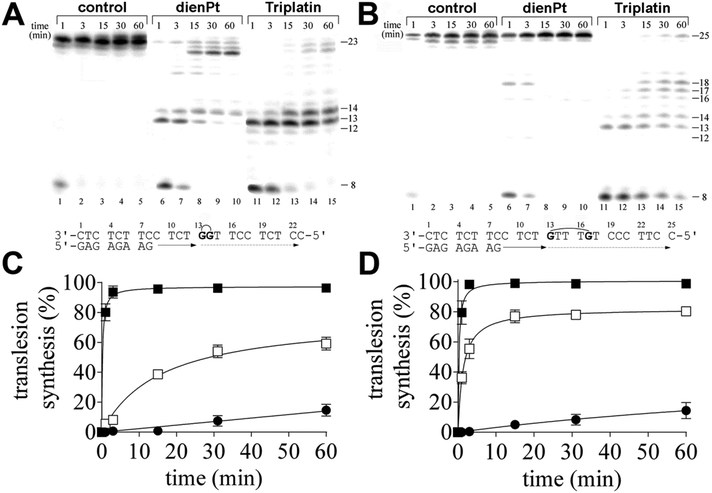
In the present work, we investigated DNA polymerization by KF− using the templates site-specifically modified by the double-base adducts derived from the intrastrand CLs of trinuclear bifunctional Triplatin. The results of these experiments were compared with DNA polymerization using the same templates also modified by the double-base adducts, but site-specifically modified by the two monofunctional adducts formed by mononuclear and monofunctional dienPt at the same sites (guanine residues) as those platinated by Triplatin in the intrastrand CLs. Thus, the important difference between the double-base lesions of Triplatin and dienPt lies in the absence of the tetraamine platinum linker covalently bridging the two monofunctional adducts at guanine residues in the case of the double-base lesions formed by dienPt. The comparative studies of the double-base DNA lesions of Triplatin and dienPt may help better understand the role of the linker in DNA adducts of Triplatin in creating unique distortions and subsequent downstream cellular processes leading to the death of cancer cells. KF− was used as a model enzyme frequently in the studies aimed at understanding the processes in which nucleic acid polymerases take part.In order to assess the translesion replication capacity of KF−, we investigated its capacity to elongate a 5′ 32P labeled 8mer primer annealed to untreated 23 or 25mer templates (Fig. 1A and B, lanes 1–5) or 23mer or 25mer templates containing the 1,2-GG or 1,5-GTTTG intrastrand CL of Triplatin, respectively depicted in Fig. 1A and B, lanes 11–15 (the platinated nucleotides in these templates are marked by bold letters throughout the text). The 3′ guanine involved in the 1,2-GG or 1,5-GTTTG CL on the template strand was located at its 13th position from the 3′ terminus (positioning the 3′-end of the primer five bases before the first platinated base in the template strand) (Fig. 1A and B). The newly synthesized DNA products were resolved by denaturing PAA gel electrophoresis and visualized by radiography.
DNA polymerization through the single 1,2-GG or 1,5-GTTTG intrastrand CL of Triplatin on the template by KF− in the presence of all four dNTPs was stopped at various time intervals, and the products were resolved by a sequencing gel (Fig. 1A and B, lanes 11–15). Polymerization proceeded rapidly up to the nucleotide corresponding to the sites opposite to the 1,2- or 1,5-intrastrand CLs, such that the 13mer and 14mer or 13–17mer products, respectively, accumulated to a significant extent (Fig. 1A and B, lanes 11–15), albeit in the case of the polymerization across the 1,2 intrastrand CL, some low level of incorporation was observed opposite to the base preceding the lesion (Fig. 1A, lanes 11–15). Interesting results were also obtained when polymerization across the 1,5-GTTTG intrastrand CL was examined. There was also an accumulation of a larger DNA intermediate, 18mer product (Fig. 1B, lanes, 11–15) and the amount of the shorter intermediates (within those 13–18 nucleotides long) decreased with the incubation time with the concomitant increase in the amount of longer intermediates. Also interestingly, DNA polymerization was arrested pronouncedly more at the sites opposite to the platinated guanines in the 1,5-GTTTG intrastrand CL than at those opposite to the base (thymine) residues between these platinated residues. In Fig. 1A and B, lanes 1–5 also show that KF− efficiently replicated the untreated template, and no intermediate products were seen with the 23mer or 25mer control template as the full-length product was being formed. There was no accumulation of other shorter or larger DNA intermediates. Interestingly, the full-length products were also noticed with the 23mer or 25mer template containing the CLs of Triplatin at longer times of incubation of the templates with KF− (15–60 min), although in a markedly smaller amount than with the nonmodified template; the amount of the full-length products accumulated approximately in the same amount with the templates containing both 1,2-GG and 1,5-GTTTG CLs (Fig. 1A and B, lanes 11–15 and Fig. 1C and D).
In contrast, under the same experimental conditions, DNA polymerization by KF− using the template containing two isolated monofunctional adducts of dienPt formed at the same sites (guanine residues) as those platinated by Triplatin in the 1,2-GG or 1,5-GTTTG intrastrand CLs proceeded in a distinctly different way. The amount of the full-length or almost full-length products (23–25 nucleotide long) accumulated markedly more with these templates than with those containing the intrastrand CLs of Triplatin. In addition, there was a difference between DNA polymerization through the two monofunctional adducts of dienPt at the guanine residues depending on whether they were located in one strand separated by three nucleotides or between the neighboring guanine residues (Fig. 1A and B, lanes 6–10 and Fig. 1C and D). Polymerization using the template containing two neighboring monofuctional adducts of dienPt proceeded rapidly up to the nucleotides opposite to the platinated guanine residues, such that mainly the 13mer and 14mer accumulated (shown in Fig. 1A, lanes 6–10), i.e. the result was qualitatively similar to that when the template containing 1,2-GG intrastrand CL of Triplatin was used. However, in contrast, polymerization using the template containing the two monofunctional adducts of dienPt separated by three nucleotides was prematurely terminated only very slightly and only at shorter times of incubation of the templates with KF− (1–3 min). The only minor, intermediate product observed at these short incubations was a 18mer product, i.e., at the site immediately following that opposite to the monofunctionally adducted 5′ guanine. Collectively, TLS by KF− across the double-base lesions derived from the intrastrand CLs of Triplatin was markedly less extensive than that across the two analogous monofunctional adducts of dienPt. In other words, this report suggests that the Triplatin noncovalent tetraamine platinum linker, capable of hydrogen-bonding and electrostatic interactions with DNA, connecting the two platinum adducts appears to be a major factor responsible for the markedly lowered tolerance of the DNA double-base adducts of Triplatin by KF−. A gently tuned catalytically active structure of the ternary DNA polymerase-primer/template DNA-incoming dNTP complex is a prerequisite for the correct course of the DNA polymerization reaction. In regard to the different effects of the DNA double-base lesions of Triplatin and dienPt on the KF− polymerization reaction (Fig. 1), the formation of this ternary complex can be disturbed by (i) different conformational distortions imposed on the template DNA by the double-base lesions in which the single lesions were connected and were not connected by the tetraamine platinum linker complexes and/or (ii) additional steric clashes between the linker connecting the double-base lesions derived from the interstrand CLs of Triplatin and other components of the ternary complex, which do not occur in the case of double-base lesions of dienPt.
Interestingly, the two monofunctional adducts formed by dienPt at the neighboring G residues represented a stronger block for KF− than the two monofunctional adducts formed by dienPt at the G residues separated by three nucleotides. It is possible that the monofunctional adducts of the platinum drugs formed at the adjacent nucleotides affect DNA conformation and its processing by DNA enzymes synergistically in contrast to the same distant adducts separated by several base pairs.
We have also examined the effects of the DNA adducts of Triplatin and dienPt on polymerization by RT HIV-1 which operates through a mechanism different from KF−. In these studies, the elongation of the 17mer/30mer primer–template duplexes was tested in the same way as in the experiments performed with KF−. The results (ESI,† Fig. S1) analogous to those obtained with KF− were obtained (ESI,† Fig. S1), i.e., the double-base lesions derived from the CLs of Triplatin represented a stronger block to DNA polymerase than the analogous double-base lesions formed by dienPt. The only substantial difference between the polymerization by KF− and RT HIV-1 is the significantly lower efficiency of RT HIV-1 to discriminate between the adducts of dienPt and Triplatin (cf.Fig. 1C, D and Fig. S1C, D, ESI†). The different processing of the double-base lesions of Triplatin or dienPt by RT HIV-1 and KF− may be due to the fact that RT HIV-1 has a sterically more flexible active site.51 Hence, we propose that, when compared to other DNA polymerases, RT HIV-1 can more easily overcome interferences imposed by the tetraamine linker of Triplatin upon the processes connected with the creation of the catalytically active structure of the ternary DNA polymerase primer/template DNA incoming dNTP complex, which is a prerequisite for the correct course of the DNA polymerization reaction.
Chemical probes of conformational alterations induced by double-base adducts
As mentioned above, the different effects of the DNA double-base lesions of Triplatin and dienPt on the KF− polymerization reaction can also be connected with different conformational distortions imposed on the template DNA by the double-base lesions in which the single lesions were connected or were not connected by the noncovalent tetraamine linker. In order to shed light on the characteristics of the damage induced by the double-base lesions derived from the intrastrand CLs of Triplatin and from the two monofunctional adducts of dienPt formed at the sites identical to those involved in the CLs of Triplatin, further studies of the present work were focused on analysis of the distortions of the double-base lesions with the aid of chemical probes of DNA conformation. The platinated duplexes were treated with several chemical agents that are used as tools for monitoring the existence of conformations other than canonical B-DNA. These agents include KMnO4, DEPC, and bromine. They react preferentially with single-stranded DNA and distorted double-stranded DNA.37–40,52 Thus, this method allows the determination of the extent and localization of conformational distortions. In the present work, the duplexes containing two monofunctional adducts of dienPt were only analyzed and compared with the results of the identical experiments performed with the same duplexes, but containing 1,2- and 1,5-intrastrand CLs of Triplatin published in our previous article.17 Autoradiograms of the gels used to analyze the piperidine-induced specific strand cleavage at KMnO4-, DEPC-, and KBr/KHSO5-modified bases in the duplexes unplatinated or containing the two monofunctional adducts of dienPt at the guanine residues are shown in the ESI,† Fig. S2. The pattern and degree of reactivity toward the chemical probes found for the double-base lesions of Triplatin and dienPt are summarized in Fig. 2. It has been shown33 that the distortion induced by the single monofunctional adduct of dienPt is mainly localized at the base pair containing a base flanking the platinated guanine on its 5′ side. The qualitative evaluation of the results (Fig. 2) suggests that distortions of the duplexes due to the formation of the two monofunctional adducts of dienPt represent an approximate sum of two distortions of single monofunctional adducts. In addition, the double-base lesion formed by dienPt at two neighboring G residues induced a stronger local conformation distortion mainly at the 5′ side of the lesion than that represented by two, more distant monofunctional adducts of dienPt (Fig. 2). Interestingly, the distortions induced by the intrastrand CLs of Triplatin were somewhat more pronounced than those due to the two monofunctional adducts of dienPt formed at the sites identical to those involved in the intrastrand CLs of the polynuclear platinum drug.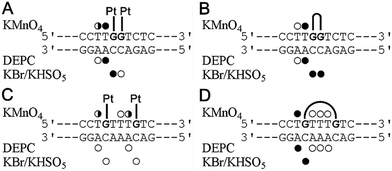 | ||
| Fig. 2 Summary of the reactivity of chemical probes with the 21-bp duplexes containing the adducts of dienPt or Triplatin. (A) The duplex containing two monofunctional adducts of dienPt at the neighboring guanine residues at the center of the duplex. (B) The duplex containing the 1,2-GG intrastrand CL of Triplatin. (C) The duplex containing the two monofunctional adducts of dienPt at the guanine residues separated by three intervening nucleotides. (D) The duplex containing the 1,5-GTTTG intrastrand CL of Triplatin. The duplexes containing the central sequences are shown in each panel. Closed, half-closed and open circles designate the strong, medium or weak reactivity, respectively. Data for the adducts of Triplatin (Fig. 3B and D) were taken from our previously published work.17 | ||
Alterations in energetics of DNA duplexes induced by double-base adducts
In our quest to further understand why the damage induced by the double-base lesions derived from the intrastrand CLs of Triplatin and the two monofunctional adducts of dienPt affects TLS by KF− or RT HIV-1 differently, we also used DSC to investigate the effects of these double-base lesions formed in the 15-bp DNA duplexes on their energetics. These oligodeoxyribonucleotide duplexes (their nucleotide sequences are shown in the ESI,† Fig. S3A) were nonmodified or contained the adducts in the top strands formed by the platinum complexes at the guanine residues in two different sequence contexts, TGGT or TGTTTGT (at these sequences the two monofunctional adducts of dienPt or the single 1,2 or 1,5-intrastrand CLs of Triplatin were formed at the guanine residues). Thermograms of these duplexes are shown in the ESI,† Fig. S3B and C. Importantly, no changes in the heat capacities between the initial and final states were observed, and the denaturation and renaturation curves were identical (not shown), which is consistent with the reversibility of the melting equilibrium. Thus, our data obtained from the DSC measurements can be evaluated assuming that the thermodynamic parameters for the melting of the duplexes can be attributed to the differences in the initial duplex states. This implies that the final single-stranded states should be thermodynamically equivalent at the elevated temperatures at which they are formed. This assumption was verified (not shown) similarly to earlier reports46–48,53,54 by recording identical CD spectra for samples of duplexes heated at 360 K. Collectively, our DSC measurements provide meaningful thermodynamic data.Analysis of thermograms yielded the thermodynamic parameters, referring to the duplex dissociation process, summarized in Table 1. We found that the formation of the intrastrand CLs of Triplatin or the two isolated monofunctional adducts of dienPt at the same sites (guanine residues) as those platinated by Triplatin in its intrastrand CLs reduced the thermal stability of the duplexes. The intrastrand CL of Triplatin formed between two neighboring G residues (1,2-intrastrand CLs) reduced the duplex thermal stability (Tm) considerably more (by 3.1 K) than the two neighboring monofunctional adducts of dienPt formed at the same sites as those platinated by Triplatin in its 1,2-intrastrand CL. Similarly, the 1,5-intrastrand CL of Triplatin reduced Tm even more efficiently (by 12.3 K) than the two monofunctional adducts formed by dienPt at the same G residues as those platinated by Triplatin in its 1,5-intrastrand CL.
| T m (K) | ΔHb (kJ mol−1) | ΔSb (kJ K−1 mol−1) | ΔG0298![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) b (kJ mol−1) b (kJ mol−1) |
K D | |
|---|---|---|---|---|---|
a The platinated nucleotides in the top strand of these duplexes are marked by bold letters. Their nucleotide sequences are shown in the ESI Fig. S3A.
b The ΔH and ΔS values are the averages derived from three independent experiments. The experimental uncertainties of the parameters are as follows: Tm (±0.5 K), ΔH (±2%), ΔS (±3%), ΔG0298 (±3%). “ΔΔ” parameters are in parentheses (these parameters are computed by subtracting the appropriate value measured for the control, non-modified duplex from the value measured for the duplex containing the single, site-specific platinum adduct).
c
K
D denotes the dissociation constant for strand dissociation [ΔG0298 = −RT![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ln KD; T is the temperature in Kelvin, and R is the universal gas constant (8.314472 J K−1 mol−1)].
d The duplex TGTTTGT(15) containing a single monofunctional adduct of dienPt at 5′-G in the top strand in which 3′-G was replaced by 7-deazaG (G*) (for details about this duplex, see the Materials and methods section). ln KD; T is the temperature in Kelvin, and R is the universal gas constant (8.314472 J K−1 mol−1)].
d The duplex TGTTTGT(15) containing a single monofunctional adduct of dienPt at 5′-G in the top strand in which 3′-G was replaced by 7-deazaG (G*) (for details about this duplex, see the Materials and methods section).
|
|||||
| Non-modified duplexesa | |||||
| TGGT(15) | 332.6 | 357 | 1.072 | 37.5 | 0.27 μM |
| TGTTTGT(15) | 334.2 | 349 | 1.043 | 38.2 | 0.20 μM |
| Duplexes containing 1,2 or 1,5 cross-link of Triplatin | |||||
| TGGT(15) | 315.6 (−17.0) | 196 (−163) | 0.620 (−0.452) | 11.0 (−26.5) | 11.80 mM |
| TGTTTGT(15) | 311.9 (−22.3) | 225 (−124) | 0.724 (−0.319) | 9.2 (−29.0) | 24.40 mM |
| Duplexes containing two adducts of dienPt | |||||
| TGGT(15) | 321.7 (−13.9) | 298 (−99) | 0.948 (−0.124) | 15.4 (−22.1) | 2.00 mM |
| TGTTTGT(15) | 322.2 (−10.0) | 296 (−53) | 0.924 (−0.119) | 20.6 (−17.6) | 0.25 mM |
| Duplex containing one adduct of dienPt | |||||
| TGTTTG*T(15)d | 328.3 (−5.9) | 318 (−31) | 0.969 (−0.074) | 29.2 (−9.0) | 7.66 μM |
Changes in the melting temperature of DNA have often been taken as parameters related to the effectiveness of DNA lesions to distort conformation of this nucleic acid.55,56 This approach is not, however, quite accurate since a simple correspondence between the changes in Tm and the free energy of duplex dissociation (ΔG0) due to the DNA damage has not always been observed.57,58 This is because the Tm values reflect the properties of DNA at high temperature whereas the free-energy changes can be calculated for a low-temperature standard state, typically 298 or 310 K; these low-temperature standard states correspond to the range of temperatures of the processes relevant to the in vivo situation. Therefore, in the present work, we compared the effects of the adducts of Triplatin or dienPt on the DNA stability with an emphasis on the thermodynamic origins of that stability. The formation of the double-base lesions by Triplatin and dienPt led to the changes of the free energy of duplex dissociation at 298 K (ΔG0298) (Table 1) indicating thermodynamic destabilization of the duplexes due to the formation of the double-base lesions which was enthalpic in origin. In this regard, the double-base lesions derived from the intrastrand CLs of Triplatin were markedly more effective than the corresponding double-base lesions represented by the two monofunctional adducts of dienPt. The double-base lesions derived from the 1,5-intrastrand CLs of Triplatin were more effective in the thermodynamic destabilization of the duplex than the lesions derived from the 1,2-intrastrand CLs. In contrast, the double-base lesions represented by the two distant monofunctional adducts of dienPt were less effective in the thermodynamic destabilization of the duplex than the same lesions formed between the neighboring base residues. Interestingly, the thermodynamic destabilization of the duplex by the two distant adducts of monofunctional dienPt was approximately double of that observed for the destabilization by one molecule of dienPt. On the other hand, the destabilization of the duplex by two molecules of monofunctional dienPt bound to the neighboring base residues was greater although less than that induced by the 1,2-intrastrand CL of Triplatin.
The results obtained with the aid of DSC analyses (Table 1) yielded more accurate and quantitative information based on thermodynamic data on the extent of conformational distortions induced in DNA by the double-base lesions of Triplatin and dienPt. These results (Table 1) were basically consistent with the results obtained with the aid of chemical probes of DNA conformation (Fig. 3). It has been shown that the distortion induced in DNA by the monofunctional adduct of dienPt or [Pt(NH3)3Cl]Cl disturbs the stacking interactions in double-helical DNA, makes the bases around the adduct more accessible and slightly unwinds the DNA double helix.33,59 The results obtained with the aid of chemical probes (Fig. 2) and DSC (Table 1) indicate that the thermodynamic destabilization of the duplexes due to the formation of the two distant monofunctional adducts of dienPt represents an approximate sum of two distortions of single monofunctional adducts. On the other hand, the double-base lesion formed by dienPt at two neighboring G residues induced a stronger, more destabilizing conformation distortion. Notably, the two monofunctional adducts formed by dienPt at the neighboring G residues also represented a stronger block for KF− than the two distant adducts of dienPt (Fig. 1A and B, lanes 6–10). It is, therefore, possible to suggest that the monofunctional adducts of the mononuclear platinum(II) drugs formed at the adjacent nucleotides affect the DNA conformation and consequently its processing by DNA enzymes synergistically in contrast to the same distant adducts separated by several base pairs.
The distortions induced by the intrastrand CLs of Triplatin destabilized the DNA more than those induced by the two monofunctional adducts formed at the identical sites by dienPt. Thus, the results are consistent with the thesis that the stronger conformational distortions imposed on the template DNA by the double-base lesions of Triplatin correlate with their more pronounced inhibition effects on the KF− polymerization reaction. This correlation suggests an important role of the tetraamine linker of the Triplatin molecule connecting the platinum adducts in the double-base lesions of this trinuclear platinum complex. It has been shown17 that intrastrand CLs of Triplatin create a local conformational distortion. The principal feature of the conformational distortions induced in DNA by the intrastrand CLs of Triplatin is that they increase the flexibility of DNA duplexes. The conceivable reasons for greater DNA destabilizing effects of the two monofunctional platinum units of Triplatin may consist in their limited freedom to bind DNA as independent monofunctional and mononuclear platinum complexes. Hence, the creation of two monofunctional adducts by the two monofunctional platinum units in Triplatin may require a larger conformational distortion in DNA making accommodation in the DNA double helix of two monofunctional adducts connected by the linker possible. It has been suggested17 that the length of the Triplatin linker, its charge, and hydrogen-bonding capacity also produce a bulkier double-base adduct in contrast to the same double-base lesions formed by dienPt. It is reasonable to assume that these bulkier ligands of Triplatin also represent a stronger steric block for KF− than two monofunctional adducts formed by dienPt.
Dissociation/reassociation equilibrium of KF− during DNA synthesis across the double-base adducts
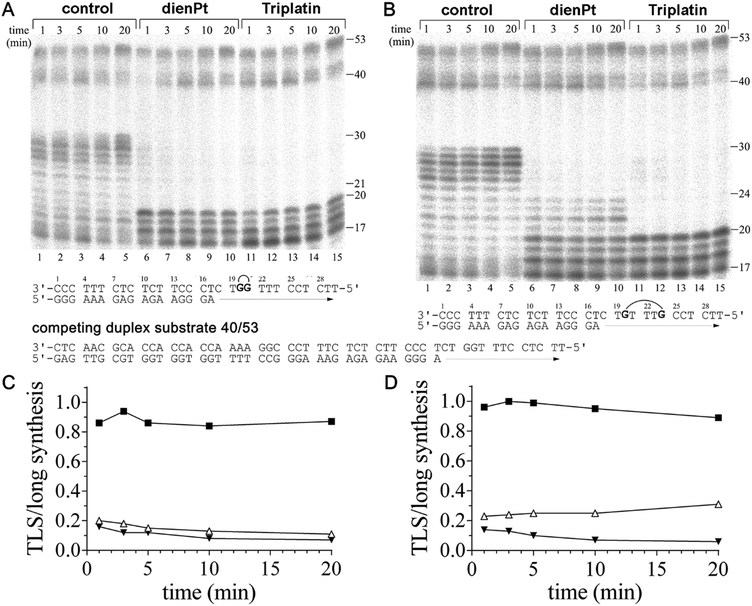
It has been shown that the duplex substrate interactions of KF− with the DNA minor groove are critical for maintaining the DNA polymerase complex during TLS.50 This implies that a factor also contributing to the decrease in the efficiency of DNA polymerases including KF− to bypass various DNA lesions is the dissociation of DNA polymerase from the substrate. Therefore, a dissociation assay was employed such that the dissociation/reassociation equilibrium of KF− during DNA synthesis could be determined. The KF− was prebound to the short duplex substrates (17mer primer/30mer template). The synthesis reactions were initiated by the simultaneous addition of dNTPs and a 10-fold molar excess of the long duplex substrate (40mer primer/53mer template), to act as a competitor for free KF− (Fig. 3A and B). If KF− dissociated from the short substrate, it is likely that it reassociated with the long duplex (in excess) and began synthesis using this long substrate.Differences were observed in the amount of long substrate synthesis completed after prebinding of KF− to the short substrates containing the adducts of Triplatin and dienPt (Fig. 3). This observation indicates the differences in the dissociation equilibrium of KF− during the DNA synthesis on the short substrates containing different platinum double-base lesions. The pattern of long synthesis for each short duplex substrate (containing double-base lesions of dienPt or Triplatin at the GG or GTTTG sequence in the template strand) is similar. On the short duplex substrate, TLS beyond the double-base adducts was observed in particular for the reactions lasting only for short times of up to 3–5 min, as compared to the experiments in which the primer extension activity of KF− was investigated in the absence of a competing duplex substrate (Fig. 1). In the latter experiments, the efficiency of KF− to bypass these lesions increased with increasing time of the reaction.
Quantitative analyses of the gels in Fig. 3A and B were performed to determine the ratio of TLS on the short complex to the total synthesis on the long complex, which is a measure of polymerase dissociation.50 On the control, unplatinated templates, this ratio increased with the reaction time for the reactions lasting up to 3 min (Fig. 3C and D), which implies that the synthesis on the short duplex substrates is greater than the synthesis on the long complex, consistent with the low dissociation from the undamaged short duplex substrate. The smaller ratios observed for the duplex substrates containing the adducts of Triplatin, relative to those containing the analogous adducts of dienPt, reflect the decreased affinity of KF− for the substrates modified by the trinuclear platinum complex. On the platinated short duplex substrates, the TLS/long synthesis ratio decreased with time for the short duplex substrates containing the 1,2- or 1,5 CLs of Triplatin and double-base lesion of dienPt at the GG sequence. This is consistent with the KF− dissociation from the short complex prior to TLS, followed by the reassociation and synthesis on the long duplex substrate. The reason why the TLS/long synthesis ratio increased with time for the short duplex substrate containing the two distant monofunctional adducts of dienPt (at the GTTTG sequence) (Fig. 3D) is very likely the fact that the amount of TLS was measured as defined in the Experimental part, i.e. as the amount of radioactivity corresponding to the incorporation across the nucleotide immediately flanking the second platinated G on the 5′ side of the template strand and beyond. However, most TLS was observed already after the nucleotide immediately flanking the first platinated G on the 3′ side of the double-base lesion formed by the two adducts of dienPt (Fig. 3B, lanes 6–10). Thus, this amount of TLS was not included into the yield of the total amount of TLS across the double-base lesion represented by the two adducts of dienPt. In contrast, negligible TLS was observed after the nucleotide immediately flanking the second platinated G on the 3′ side of the double-base lesion derived from the 1,5-intrastrand CL of Triplatin (Fig. 3B, lanes 11–15). In this case, most TLS was observed only after the nucleotide immediately flanking the second platinated G on the 5′ side of the template strand and beyond.
The results of the dissociation experiments (Fig. 3) indicate that the KF− dissociation contributes to the inhibition of DNA synthesis observed at the double-base platinum lesions. The extent of the dissociation of KF− from the template substrates containing a variety of double-base platinum lesions correlated with the extent of conformational distortions induced by these lesions determined with the aid of chemical probes and DSC (Fig. 2 and Table 1). Thus, the conformational distortions induced in the template substrates by the double-base lesions of the tested platinum complexes apparently decreased the affinity of the DNA polymerase for the DNA template substrate. In this regard, the adducts of Triplatin were more effective, which points out the importance of the tetraamine platinum linker of Triplatin for processing its double-base adducts by DNA polymerases.
Conclusions
In this work, DNA polymerization using DNA templates site-specifically modified with double-base platinum lesions formed by Triplatin and dienPt was examined. We used the model A-family polymerase, KF−, which has been extensively used as a model DNA polymerase for examining lesion bypass since it shares common properties of other TLS proficient DNA polymerases of the X and Y families. The goal of this study was to contribute to understanding the role of the “central noncovalent” tetraamine linker, which connects the platinum adducts in the double-base lesions (CLs) of Triplatin, in the mediation of inhibition of DNA polymerization. It has been shown that the CLs of Triplatin coordinatively bind to the N7 atoms of two guanine residues and simultaneously noncovalently to the central tetraamine platinum linker anchored in the minor groove.60 The interactions of the KF− with the minor groove of the DNA duplex substrate are critical for catalyzing the template-directed addition of a dNTP to a growing DNA strand.61 It is, therefore, conceivable to suggest that the presence of the linker in the minor groove in the double-base adducts of Triplatin can obstruct the formation of productive contacts of KF− with the minor groove of the DNA duplex substrates thereby contributing to DNA synthesis inhibition. The results demonstrate that the role of the central tetraamine platinum linker in the DNA double-base adducts of Triplatin in the mechanism of the blockage of KF− is multifactorial. The linker in Triplatin increases the size or bulkiness of DNA adducts, which become stronger blocks for DNA synthesis by KF−. The linker is also responsible for the formation of unique, destabilizing conformational distortions in DNA also decreasing the affinity of the KF− for the DNA template substrate, which contributes to the inhibition effects on the KF− polymerization reaction as well; if the DNA double-base adducts (CLs) of Triplatin are formed, the linker binds to the minor groove of the DNA substrate, which can also impede the productive binding of double-base adducts of Triplatin at the polymerase site of KF−. Collectively, the data help explain the role of the linker in Triplatin in the process of diminution of tolerance of its DNA adducts, i.e., in the process related to the development of cellular resistance to conventional platinum anticancer drugs.Although Triplatin has not yet advanced to full clinical use as of now, it exhibits high cytotoxic potency allied to its capability to form in DNA unusual double-base adducts connected by a tetraamine linker. Our findings are consistent with a model in which the linkers in bifunctional polynuclear platinum complexes are responsible for the specific properties of the DNA adducts of this class of antitumor platinum complexes. The results also help understand some factors responsible for these specific properties which potentiate the capability of bifunctional polynuclear platinum complexes to inhibit DNA synthesis. We suggest that the information contained in this communication may be helpful in the future design of polynuclear metal-based compounds whose biological significance is connected with a new DNA binding mode mediated by linkers connecting DNA binding metal units.
Conflicts of interest
There are no conflicts of interest.Acknowledgements
This work was supported by the Czech Science Foundation [17-09436S].References
- V. Brabec, J. Kasparkova, V. Menon and N. P. Farrell, in Metal Ions in Life Sciences, ed. A. Sigel, H. Sigel, E. Freisinger and R. K. O. Sigel, Walter de Gruyter, GmbH, Berlin, Germany, 2018, vol. 18, Metallo-Drugs: Development and Action of Anticancer Agents, pp. 43–68 Search PubMed.
- V. Brabec, J. Kasparkova, O. Vrana, O. Novakova, J. W. Cox, Y. Qu and N. Farrell, DNA modifications by a novel bifunctional trinuclear platinum Phase I anticancer agent, Biochemistry, 1999, 38, 6781–6790 CrossRef CAS PubMed.
- T. D. McGregor, A. Hegmans, J. Kasparkova, K. Neplechova, O. Novakova, H. Penazova, O. Vrana, V. Brabec and N. Farrell, A comparison of DNA binding profiles of dinuclear platinum compounds with polyamine linkers and the trinuclear platinum phase II clinical agent BBR3464, J. Biol. Inorg. Chem., 2002, 7, 397–404 CrossRef CAS PubMed.
- J. J. Moniodis, D. S. Thomas, M. S. Davies, S. J. Berners-Price and N. P. Farrell, Competitive formation of DNA linkage isomers by a trinuclear platinum complex and the influence of pre-association, Dalton Trans., 2015, 44, 3583–3593 RSC.
- T. C. Johnstone, K. Suntharalingam and S. J. Lippard, The next generation of platinum drugs: Targeted Pt(II) agents, nanoparticle delivery, and Pt(IV) prodrugs, Chem. Rev., 2016, 116, 3436–3486 CrossRef CAS PubMed.
- C. Billecke, S. Finniss, L. Tahash, C. Miller, T. Mikkelsen, N. P. Farrell and O. Boegler, Polynuclear platinum anticancer drugs are more potent than cisplatin and induce cell cycle arrest in glioma, Neuro-Oncology, 2006, 8, 215–226 CrossRef CAS PubMed.
- J. B. Mangrum and N. P. Farrell, Excursions in polynuclear platinum DNA binding, Chem. Commun., 2010, 46, 6640–6650 RSC.
- G. Pratesi, P. Perego, D. Polizzi, S. C. Righetti, R. Supino, C. Caserini, C. Manzotti, F. C. Giuliani, G. Pezzoni, S. Tognella, S. Spinelli, N. Farrell and F. Zunino, A novel charged trinuclear platinum complex effective against cisplatin-resistant tumours: hypersensitivity of p53-mutant human tumour xenografts, Br. J. Cancer, 1999, 80, 1912–1919 CrossRef CAS PubMed.
- A. Riccardi, D. Meco, C. Ferlini, T. Servidei, G. Carelli, G. Segni, C. Manzotti and R. Riccardi, In vitro and in vivo antitumor activity of the novel trinuclear platinum complex BBR 3464 in neuroblastoma, Cancer Chemother. Pharmacol., 2001, 47, 498–504 CrossRef CAS PubMed.
- N. J. Wheate and J. G. Collins, Multi-nuclear platinum complexes as anti-cancer drugs, Coord. Chem. Rev., 2003, 241, 133–145 CrossRef CAS.
- N. P. Farrell, Progress in platinum-derived drug development, Drugs Future, 2012, 37, 795–806 CrossRef CAS.
- V. Brabec, O. Hrabina and J. Kasparkova, Cytotoxic platinum coordination compounds. DNA binding agents, Coord. Chem. Rev., 2017, 351, 2–31 CrossRef CAS.
- C. Gourley, J. Cassidy, C. Edwards, L. Samuel, D. Bisset, G. Camboni, A. Young, D. Boyle and D. Jodrell, A phase I study of the trinuclear platinum compound, BBR 3464, in combination with protracted venous infusional 5-fluorouracil in patients with advanced cancer, Cancer Chemother. Pharmacol., 2004, 53, 95–101 CrossRef CAS PubMed.
- D. I. Jodrell, T. R. J. Evans, W. Steward, D. Cameron, J. Prendiville, C. Aschele, C. Noberasco, M. Lind, J. Carmichael, N. Dobbs, G. Camboni, B. Gatti and F. De Braud, Phase II studies of BBR3464, a novel tri-nuclear platinum complex, in patients with gastric or gastro-oesopliageal adenocarcinoma, Eur. J. Cancer, 2004, 40, 1872–1877 CrossRef CAS PubMed.
- T. A. Hensing, N. H. Hanna, H. H. Gillenwater, M. G. Camboni, C. Allievi and M. A. Socinski, Phase II study of BBR 3464 as treatment in patients with sensitive or refractory small cell lung cancer, Anti-Cancer Drugs, 2006, 17, 697–704 CrossRef CAS PubMed.
- J. Kasparkova, J. Zehnulova, N. Farrell and V. Brabec, DNA interstrand cross-links of the novel antitumor trinuclear platinum complex BBR3464. Conformation, recognition by high mobility group domain proteins, and nucleotide excision repair, J. Biol. Chem., 2002, 277, 48076–48086 CrossRef CAS PubMed.
- J. Zehnulova, J. Kasparkova, N. Farrell and V. Brabec, Conformation, recognition by high mobility group domain proteins, and nucleotide excision repair of DNA intrastrand cross-links of novel antitumor trinuclear platinum complex BBR3464, J. Biol. Chem., 2001, 276, 22191–22199 CrossRef CAS PubMed.
- V. Brabec, J. Kasparkova, H. Kostrhunova and N. P. Farrell, Inhibition of nuclear factor kappaB proteins-platinated DNA interactions correlates with cytotoxic effectiveness of the platinum complexes, Sci. Rep., 2016, 6, 28474 CrossRef CAS PubMed.
- M. A. Fuertes, J. Castilla, C. Alonso and J. M. Perez, Cisplatin biochemical mechanism of action: From cytotoxicity to induction of cell death through interconnections between apoptotic and necrotic pathways, Curr. Med. Chem., 2003, 10, 257–266 CrossRef CAS PubMed.
- Z. Suo, S. Lippard and K. Johnson, Single d(GpG)/cis-diammineplatinum(II) adduct-induced inhibition of DNA polymerization, Biochemistry, 1999, 38, 715–726 CrossRef CAS PubMed.
- O. Novakova, J. Kasparkova, J. Malina, G. Natile and V. Brabec, DNA-protein cross-linking by trans-[PtCl2(E-iminoether)2]. A concept for activation of the trans geometry in platinum antitumor complexes, Nucleic Acids Res., 2003, 31, 6450–6460 CrossRef CAS PubMed.
- V. Marini, P. Christofis, O. Novakova, J. Kasparkova, N. Farrell and V. Brabec, Conformation, protein recognition and repair of DNA interstrand and intrastrand cross-links of antitumor trans-[PtCl2(NH3)(thiazole)], Nucleic Acids Res., 2005, 33, 5819–5828 CrossRef CAS PubMed.
- V. Brabec, J. Malina, N. Margiotta, G. Natile and J. Kasparkova, Thermodynamic and mechanistic insights into translesion DNA synthesis catalyzed by Y-family DNA polymerase across a bulky double-base lesion of an antitumor platinum drug, Chem. – Eur. J., 2012, 18, 15439–15448 CrossRef CAS PubMed.
- S. Malvezzi, T. Angelov and S. J. Sturla, Minor Groove 3-Deaza-Adenosine Analogues: Synthesis and Bypass in Translesion DNA Synthesis, Chem. – Eur. J., 2017, 23, 1101–1109 CrossRef CAS PubMed.
- J. S. Choi, S. Kim, E. Motea and A. Berdis, Inhibiting translesion DNA synthesis as an approach to combat drug resistance to DNA damaging agents, Oncotarget, 2017, 8, 40804–40816 Search PubMed.
- V. Brabec and J. Kasparkova, Modifications of DNA by platinum complexes: Relation to resistance of tumors to platinum antitumor drugs, Drug Resist. Updates, 2005, 8, 131–146 CrossRef CAS PubMed.
- V. Brabec, DNA modifications by antitumor platinum and ruthenium compounds: their recognition and repair, Prog. Nucleic Acid Res. Mol. Biol., 2002, 71, 1–68 CAS.
- R. M. Turner, N. D. F. Grindley and C. M. Joyce, Interaction of DNA polymerase I (Klenow fragment) with the single-stranded template beyond the site of synthesis, Biochemistry, 2003, 42, 2373–2385 CrossRef CAS PubMed.
- W. C. Lam, E. J. C. Van der Schans, L. C. Sowers and D. P. Millar, Interaction of DNA polymerase I (Klenow fragment) with DNA substrates containing extrahelical bases: Implications for proofreading of frameshift errors during DNA synthesis, Biochemistry, 1999, 38, 2661–2668 CrossRef CAS PubMed.
- P. H. Patel, M. Suzuki, E. Adman, A. Shinkai and L. A. Loeb, Prokaryotic DNA polymerase I: Evolution, structure, and “base flipping” mechanism for nucleotide selection, J. Mol. Biol., 2001, 308, 823–837 CrossRef CAS PubMed.
- G. Villani, N. T. Le Gac, L. Wasungu, D. Burnouf, R. P. Fuchs and P. E. Boehmer, Effect of manganese on in vitro replication of damaged DNA catalyzed by the herpes simplex virus type-1 DNA polymerase, Nucleic Acids Res., 2002, 30, 3323–3332 CrossRef CAS PubMed.
- K. A. Johnson, Conformational coupling in DNA-polymerase fidelity, Annu. Rev. Biochem., 1993, 62, 685–713 CrossRef CAS PubMed.
- V. Brabec, J. Reedijk and M. Leng, Sequence-dependent distortions induced in DNA by monofunctional platinum(II) binding, Biochemistry, 1992, 31, 12397–12402 CrossRef CAS PubMed.
- V. Brabec and M. Leng, DNA interstrand cross-links of trans-diamminedichloroplatinum(II) are preferentially formed between guanine and complementary cytosine residues, Proc. Natl. Acad. Sci. U. S. A., 1993, 90, 5345–5349 CrossRef CAS.
- J. Kasparkova, O. Novakova, V. Marini, Y. Najajreh, D. Gibson, J.-M. Perez and V. Brabec, Activation of trans geometry in bifunctional mononuclear platinum complexes by a piperidine ligand: Mechanistic studies on antitumor action, J. Biol. Chem., 2003, 278, 47516–47525 CrossRef CAS PubMed.
- O. Novakova, H. Chen, O. Vrana, A. Rodger, P. J. Sadler and V. Brabec, DNA interactions of monofunctional organometallic ruthenium(II) antitumor complexes in cell-free media, Biochemistry, 2003, 42, 11544–11554 CrossRef CAS PubMed.
- V. Brabec, M. Sip and M. Leng, DNA conformational distortion produced by site-specific interstrand cross-link of trans-diamminedichloroplatinum(II), Biochemistry, 1993, 32, 11676–11681 CrossRef CAS PubMed.
- C. Bailly, D. Gentle, F. Hamy, M. Purcell and M. J. Waring, Localized chemical reactivity in DNA associated with the sequence-specific bisintercalation of echinomycin, Biochem. J., 1994, 300, 165–173 CrossRef CAS PubMed.
- S. A. Ross and C. J. Burrows, Cytosine-specific chemical probing of DNA using bromide and monoperoxysulfate, Nucleic Acids Res., 1996, 24, 5062–5063 CrossRef CAS PubMed.
- C. Bailly and M. J. Waring, in Drug-DNA Interaction Protocols, ed. K. R. Fox, Humana Press Inc., Totowa/NJ, 1997, pp. 51–79 Search PubMed.
- S. M. Cohen, Y. Mikata, Q. He and S. J. Lippard, HMG-Domain protein recognition of cisplatin 1,2-intrastrand d(GpG) cross-links in purine-rich sequence contexts, Biochemistry, 2000, 39, 11771–11776 CrossRef CAS PubMed.
- J. Malina and V. Brabec, Thermodynamic impact of abasic sites on simulated translesion DNA synthesis, Chem. – Eur. J., 2014, 20, 7566–7570 CrossRef CAS PubMed.
- J. Malina, O. Novakova, G. Natile and V. Brabec, The thermodynamics of translesion DNA synthesis past major adducts of enantiomeric analogues of antitumor cisplatin, Chem. – Asian J., 2012, 7, 1026–1031 CrossRef CAS PubMed.
- J. Florian, J. Kasparkova, N. Farrell and V. Brabec, Thermodynamic stability and energetics of DNA duplexes containing major intrastrand cross-links of second-generation antitumor dinuclear PtII complexes, J. Biol. Inorg. Chem., 2012, 17, 187–196 CrossRef CAS PubMed.
- V. Brabec, in Platinum-based Drugs in Cancer Therapy, ed. L. R. Kelland and N. P. Farrell, Humana Press Inc, Totowa/NJ, 2000, pp. 37–61 Search PubMed.
- C. Hofr and V. Brabec, Thermal and thermodynamic properties of duplex DNA containing site-specific interstrand cross-link of antitumor cisplatin or its clinically ineffective trans isomer, J. Biol. Chem., 2001, 276, 9655–9661 CrossRef CAS PubMed.
- C. Hofr, N. Farrell and V. Brabec, Thermodynamic properties of duplex DNA containing a site-specific d(GpG) intrastrand crosslink formed by an antitumor dinuclear platinum complex, Nucleic Acids Res., 2001, 29, 2034–2040 CrossRef CAS PubMed.
- V. Bursova, J. Kasparkova, C. Hofr and V. Brabec, Effects of monofunctional adducts of platinum(II) complexes on thermodynamic stability and energetics of DNA duplexes, Biophys. J., 2005, 88, 1207–1214 CrossRef CAS PubMed.
- J. H. Murphy and T. L. Trapane, Concentration and extinction coefficient determination for oligonucleotides and analogs using a general phosphate analysis, Anal. Biochem., 1996, 240, 273–282 CrossRef CAS PubMed.
- E. E. Gestl and K. A. Eckert, Loss of DNA minor groove interactions by exonuclease-deficient Klenow polymerase inhibits O6-methylguanine and abasic site translesion synthesis, Biochemistry, 2005, 44, 7059–7068 CrossRef CAS PubMed.
- A. P. Silverman and E. T. Kool, RNA probes of steric effects in active sites: High flexibility of HIV-1 reverse transcriptase, J. Am. Chem. Soc., 2007, 129, 10626–10627 CrossRef CAS PubMed.
- P. E. Nielsen, Chemical and photochemical probing of DNA complexes, J. Mol. Recognit., 1990, 3, 1–24 CrossRef CAS PubMed.
- N. Poklar, D. S. Pilch, S. J. Lippard, E. A. Redding, S. U. Dunham and K. J. Breslauer, Influence of cisplatin intrastrand crosslinking on the conformation, thermal stability, and energetics of a 20-mer DNA duplex, Proc. Natl. Acad. Sci. U. S. A., 1996, 93, 7606–7611 CrossRef CAS.
- D. S. Pilch, S. U. Dunham, E. R. Jamieson, S. J. Lippard and K. J. Breslauer, DNA sequence context modulates the impact of a cisplatin 1,2-d(GpG) intrastrand cross-link an the conformational and thermodynamic properties of duplex DNA, J. Mol. Biol., 2000, 296, 803–812 CrossRef CAS PubMed.
- S. Ghosh and M. M. Greenberg, Correlation of thermal stability and structural distortion of DNA interstrand cross-links Produced from oxidized abasic sites with their selective formation and repair, Biochemistry, 2015, 54, 6274–6283 CrossRef CAS PubMed.
- T. Inde, Y. Masaki, A. Maruyama, Y. Ito, N. Makio, Y. Miyatake, T. Tomori, M. Sekine and K. Seio, Synthesis of oligonucleotides containing 2-N-heteroarylguanine residues and their effect on duplex/triplex stability, Org. Biomol. Chem., 2017, 15, 8371–8383 CAS.
- D. S. Pilch, G. E. Plum and K. J. Breslauer, The thermodynamics of DNA structures that contain lesions or guanine tetrads, Curr. Opin. Struct. Biol., 1995, 5, 334–342 CrossRef CAS PubMed.
- G. E. Plum, C. A. Gelfand and K. J. Breslauer, in Exocyclic DNA Adducts in Mutagenesis and Carcinogenesis, ed. B. Singer and H. Bartsch, International Agency for Research on Cancer, Lyon, 1999, vol. 150, pp. 169–177 Search PubMed.
- V. Brabec, V. Boudny and Z. Balcarova, Monofunctional adducts of platinum(II) produce in DNA a sequence-dependent local denaturation, Biochemistry, 1994, 33, 1316–1322 CrossRef CAS PubMed.
- A. Hegmans, S. J. Berners-Price, M. S. Davies, D. Thomas, A. Humphreys and N. Farrell, Long range 1,4 and 1,6-interstrand cross-links formed by a trinuclear platinum complex. Minor groove pre-association affects kinetics and mechanism of cross-link formation as well as adduct structure, J. Am. Chem. Soc., 2004, 126, 2166–2180 CrossRef CAS PubMed.
- R. G. Federley and L. J. Romano, DNA polymerase: Structural homology, conformational dynamics, and the effects of carcinogenic DNA adducts, J. Nucleic Acids, 2010, 15 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7mt00266a |
| This journal is © The Royal Society of Chemistry 2018 |