Extracting pulmonary surfactants to form inverse micelles on suspended graphene nanosheets†
Zhen
Luo
a,
Shixin
Li
a,
Yan
Xu
a,
Hao
Ren
 a,
Xianren
Zhang
a,
Xianren
Zhang
 b,
Guoqing
Hu
cd,
Fang
Huang
a and
Tongtao
Yue
b,
Guoqing
Hu
cd,
Fang
Huang
a and
Tongtao
Yue
 *a
*a
aState Key Laboratory of Heavy Oil Processing, Center for Bioengineering and Biotechnology, China University of Petroleum (East China), Qingdao 266580, China. E-mail: yuett@upc.edu.cn; Tel: +86 53286981567
bState Key Laboratory of Organic–Inorganic Composites, Beijing University of Chemical Technology, Beijing 100029, China
cState Key Laboratory of Nonlinear Mechanics (LNM), Institute of Mechanics, Chinese Academy of Sciences, Beijing 100190, China
dSchool of Engineering Science, University of Chinese Academy of Sciences, Beijing 100049, China
First published on 9th November 2017
Abstract
The increasing risk of human exposure to emerging nanoparticles (NPs) has created a wide concern about their inhalation toxicity. Once NPs are inhaled, they get through the branching airway to deposit in the alveoli, where a thin pulmonary surfactant (PS) layer acts as the first barrier against them entering the deep lung. However, in terms of the inhalation toxicity of NPs, the mechanisms by which inhaled NPs interact with the PS layer and how the morphological change of NPs due to the PS layer interactions influences the subsequent fate of NPs are still elusive. By using molecular dynamics simulations, we investigate the interactions between graphene nanosheets (GNs) and the PS layer. It is found that when GNs suspend above or slowly penetrate the PS layer, PS molecules can spontaneously be extracted from the layer and made to form inverse micelles via cooperative molecular motion and rearrangement on the GN surfaces. We demonstrate that the PS layer tension and GN properties like size, oxidation ratio and curvature significantly affect the extraction dynamics. Notably, for curved GNs, only the concave surface can vigorously extract PS molecules, showing that the dispersive adhesion between GN and PS dominates the extracting and rearranging process. Our results propose new mechanisims for the experimental findings on the inhalation toxicity of graphene-family nanomaterials. Moreover, bearing in mind that the surface properties of GNs have been masked by adsorbed PS molecules, the newly formed structure may act like a corona that influences the biological identity of GNs. Therefore, anyone either evaluating inhalation toxicity or promoting biomedical applications of GNs should consider the first contact with PS at the air–water interface in alveoli.
Environmental significanceGraphene-family nanomaterials display novel properties that endow them with promising applications in science and technology. However, the increasing use of graphene also causes a general biosafety concern. As a portal of entry, the human lung can easily be accessed by graphene nanosheets (GNs). Once inhaled, a large portion of GNs get deposited in the alveoli where a thin pulmonary surfactant (PS) layer acts as the first barrier against GNs entering the deep lung. As an effort to understand the inhalation toxicity of GNs, we studied the interactions between GNs and the PS layer using molecular dynamics (MD) simulation. Our central finding is that GNs suspended in alveoli show size-, oxidation- and curvature-dependent performance on extracting PS molecules to form inverse micelles on GN surfaces. Two-fold environmental significances can be reflected. First, the inhalation toxicity of GNs could be derived from PS extraction, which induces severe PS depletion and biophysical inhibition. Second, the PS extraction represents the initial nano-bio interaction for GNs upon entering the alveoli, and the formed structures act like a corona that certainly influences the subsequent fate of GNs in the lungs. |
Introduction
Nanotechnology has been widely applied in different areas of science and technology. Most applications rely on the use of nanoparticles (NPs) that have unique performance due to their ultrasmall size.1 For example, NPs with different physicochemical properties have been intelligently designed and synthesized for use in bioimaging and drug delivery.2–5 Meanwhile, the growing use of diverse emerging NPs in the daily lives of humans has caused a general concern about their biosafety.6–8Advancing biomedical applications and evaluating the toxicity of NPs require a complete understanding of their interactions with living organisms.9 Once inside the body, NPs, on account of their small size and high surface free energy, can immediately adsorb biomolecules to form the so-called corona,10 which is believed to mask the physicochemical properties of NPs and change their biological identity to a large extent.11 Therefore, studies of nano-bio interactions should consider the influence of the corona.12,13 In general, the detailed organization and compositions of the corona are determined by the properties of NPs,11,14 but also dependent on which biological fluid NPs first encounter upon entering the biological system. Specifically, NPs entering the body via pulmonary inhalation get through the respiratory tract to reach the alveolar surface and first interact with a thin pulmonary surfactant (PS) layer. The natural PS is composed of approximately 90% lipids and 10% proteins in weight and plays a dual role in reducing the surface tension and acting as the first barrier against inhaled NPs or pathogens entering the deep lung.15 The detailed interactions of inhaled NPs with the PS layer are expected to influence their subsequent fate and toxicity.16–18 Recently, Hu et al. simulated formation of the PS corona on two spherical NPs in alveolar fluid.19 Other NPs were found to reside in, suspend above, or aggregate in the PS layer,20–23 thus taking much more time to penetrate through it. In this context, we hypothesize that both corona formation and NP invasion should start upon first contact with the PS located at the alveolar air–water interface.
Graphene is an emerging two-dimensional NP with a single layer of carbon atoms.24 It has fascinating mechanical, electrical, optical and thermal properties, which make it a desirable material for a wide range of applications, including solar cells,25 light-emitting diodes,26 bioimaging and drug delivery.27 In spite of its excellent performance and wide potential applications, the toxicity of graphene has been extensively debated.28–30 Here we used coarse-grained molecular dynamics (CGMD) simulation to study the interactions between graphene nanosheets (GNs) and the PS layer. A central finding is that under certain conditions of GN suspension and slow PS layer translocation,20,21 PS molecules can be spontaneously extracted from the PS layer and made to form inverse micelles stably adsorbed onto the GN surfaces. Two-fold implications of this finding can be considered. First, both in vivo and in vitro experiments have reported the inhalation toxicity of GN-based nanomaterials, while the underlying mechanisms still remain elusive.31 Our simulations suggest that the inhalation toxicity of GNs could be derived from PS extraction, which induces severe PS depletion and biophysical inhibition. Second, extraction of PS molecules by suspended GNs represents the initial nano-bio interaction upon entering the alveoli, and the newly formed structures can be regarded as a corona that certainly influences the subsequent fate of GNs in the lungs.
Experimental
Models and system setup
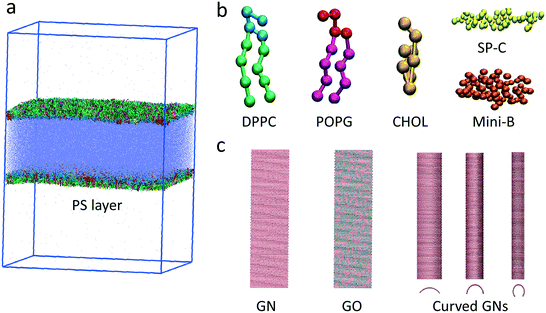
To simulate the GN–PS interactions in adequate temporal and spatial scales, we adopted the widely used MARTINI CG force field.32 The system setup included a water slab in vacuum with two symmetric PS layers at the two air–water interfaces (Fig. 1a). Each PS layer contained 2266 dipalmitoylphosphatidylcholine (DPPC), 704 palmitoyloleoylphosphatidylglycerol (POPG) and 704 cholesterol molecules (Fig. 1b). Two types of hydrophobic PS-associated proteins, including 13 SP-B and 13 SP-C, were included in the PS system (Fig. 1b). The middle fluid contains 238![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 858 water beads and 1148 Na+ ions to neutralize the system. CG models for both proteins were derived from their all-atom models in the protein data bank. Mini-B (PDB ID: 2DWF) is a 34-residue peptide composed of the C- and N-terminal helical regions of the full-length 79-residue SP-B. It has been shown that Mini-B retains major properties of the full-length SP-B.33 The SP-C model (PDB ID: 1SPF) has two palmitoyl tails that are critical for its chemical properties.34
858 water beads and 1148 Na+ ions to neutralize the system. CG models for both proteins were derived from their all-atom models in the protein data bank. Mini-B (PDB ID: 2DWF) is a 34-residue peptide composed of the C- and N-terminal helical regions of the full-length 79-residue SP-B. It has been shown that Mini-B retains major properties of the full-length SP-B.33 The SP-C model (PDB ID: 1SPF) has two palmitoyl tails that are critical for its chemical properties.34
In the CG model of GN, the honeycomb atomic structure was reduced to a triangular lattice of SC4-type beads, where every three carbons in the all-atom GN were modeled as one bead (Fig. 1c).35 The angular force constant and the equilibrium angle are κangle = 700 kcal mol−1 rad−2 and θ = 60°, respectively. The bond force constant and equilibrium bond length are κbond = 700 kcal mol−1 and l = 0.3684 nm, respectively. The dihedral angle force constant, multiplicity and equilibrium angle are κχ = 3.1 kcal mol−1, n = 2 and δ = 180°, respectively. To study the influence of oxidation ratio of GNs on the PS extraction, a defined number of beads were randomly selected and replaced with the hydrophilic Nda type beads. We also prepared GNs with defined curvature to explore the effect on PS extraction.
Process of the CGMD simulations
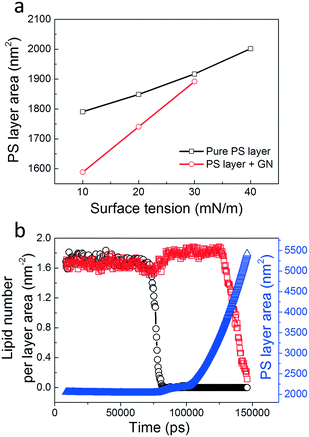
Our simulations were based on the fact that high inhalation concentration of GNs could lead to a portion of them being suspended in the alveolar cavity, thus having a high probability to obtain contact with and extract PS molecules. Before simulations of GN–PS interactions, the PS layer system was equilibrated under four different surface tensions of 10 mN m−1, 20 mN m−1, 30 mN m−1 and 40 mN m−1. Fig. S1† shows evolutions of the PS layer area under different tensions. After that, two identical GNs were placed above and below the two symmetric PS layers in the simulation box. The initial suspension distance from the layer was found to not critically affect the simulation results. After multiple simulations, we found that a slight initial contact between the proximal edge of GNs and the PS layer always led to the spontaneous extraction of PS molecules. To maintain suspension of GNs above and below the PS layers, the distal edges were frozen during the whole simulation process.36–38 If GN edges were freed, the rectangular GNs were found to lie down and finally adhere onto the PS layer, similar to our previous simulation results (data not shown).21All simulations were performed with constant particle number, PS layer tension and temperature. The surface tension, which is given by the formula γm = (Pz − Pxy)Lz/2, was kept constant using the semi-isotropic Berendsen barostat with a coupling constant of τP = 4 ps. The system compressibility was set to be 5 × 10−5 bar−1 in the x–y plane and 0 in the z-direction. The temperature was maintained at 310 K by Berendsen coupling with a coupling constant of τT = 1 ps. Periodic boundary conditions were applied in all three directions. The time step of simulations was 20 fs, and the neighbor list was updated every 10 steps. All simulations were performed using GROMACS 4.6.7.39 Snapshots were rendered by VMD.40
Results and discussion
Kinetics for extracting PS molecules to form inverse micelles on GN surfaces
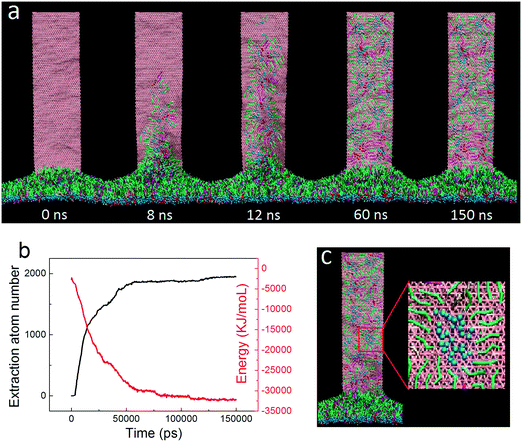
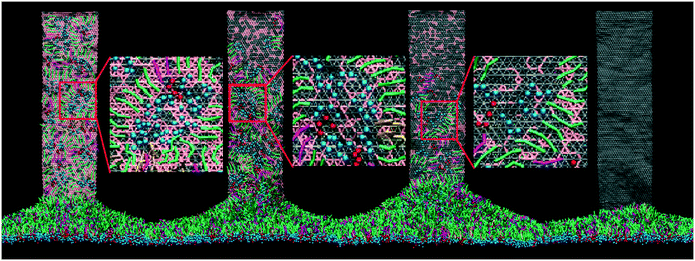
For the initial simulation, a rectangular GN with a lateral size of 36.8 nm × 9.2 nm was positioned vertically with its proximal short edge in close contact with the PS layer. Surface tension of the PS layer was fixed to 30 mN m−1 to model the inhalation condition, under which the layer was expanded.15 One typical simulated extraction event is depicted in Fig. 2a (the dynamic process of extraction can be found in Video S1 of the ESI†). It is clear that adjacent PS molecules started to be extracted upon contacting the suspended GNs. In the early stage, the extracted PS molecules stacked loosely on the GN surface. As more PS molecules were extracted, previously extracted molecules were repelled upwards to form a more compact PS arrangement. Fig. 2b shows evolutions of the extraction number and the GN–PS interaction energy. Clearly, the extraction number first increased rapidly from zero to about 1500, then increased at a slower rate to the final value of 2000, and finally stayed nearly unchanged in the rest of the simulation, suggesting that the whole process can roughly be divided into the rapid extracting and the slow rearranging stages. We observed a corresponding decrease of the GN–PS interaction energy, showing that the rapid extraction is driven by the favorable GN–PS adhesive interactions. Moreover, different types of PS molecules were found to interact with GNs in different strengths (Fig. S2†). Specifically, similar binding strengths were obtained for DPPC and POPG molecules, whereas the lower value was for the cholesterol molecule, indicating that GNs could preferentially extract DPPC and POPG molecules rather than cholesterol molecules. A local enlarged ultrastructure is displayed in Fig. 2c, showing that the PS molecules rearranged themselves to form inverse micellar structures where the polar headgroups were centralized with tails scattering to maximize interactions with the GN.We focused on how the extracted PS molecules rearrange themselves to form the final inverse micellar structures on the GN surfaces. By tracing formation of each inverse micelle during the simulation, we identified three typical pathways, depending on the onset time of PS extraction. For the PS molecules extracted in the early stage (Fig. 3a), they first distributed dispersively on the GN surface (8–12 ns). As more molecules were extracted, they were repelled upwards to occupy a more crowded space (16 ns). To create space for subsequent extraction, they gradually approached each other to form the final inverse micellar structure (47 ns). Fig. 3b shows the second pathway for the PS molecules being extracted in the middle stage. At t = 12 ns, these molecules started to climb the GN surface and rapidly formed several small inverse micelles due to existing PS molecules that made the GN surface crowded. Subsequently, these small inverse micelles fused with each other as they were further repelled by newly extracted PS molecules. For the PS molecules being extracted in the late stage, there was no space for them to further climb the GN surface. They thus remained at the bottom of the GN surface, locally rearranging themselves to form a large inverse micelle (Fig. 3c).
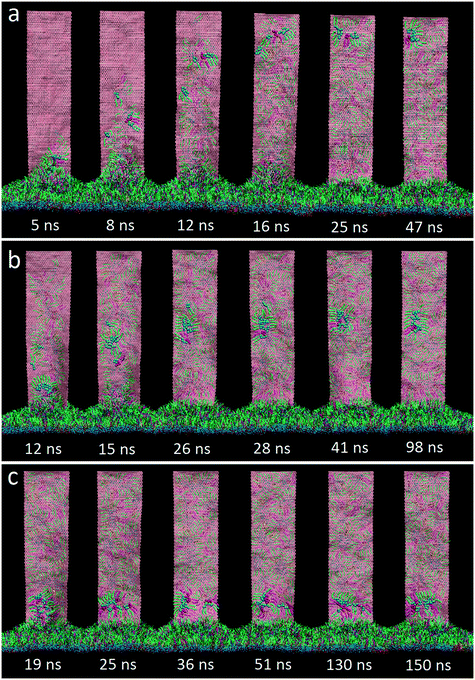 | ||
| Fig. 3 Three typical pathways of the inverse micelle assembly for extracted PS molecules, depending on the onset time of extraction: early stage (a), middle stage (b) and late stage (c). | ||
Effect of PS layer tension on extraction
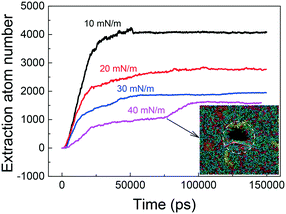
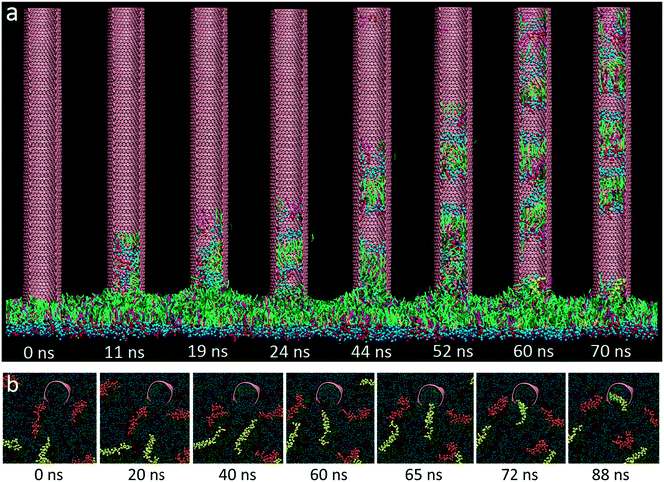
In cycles of respiration, the PS layer tension varies from tens mN m−1 to near zero, during which transitions between liquid-expanded and liquid-condensed phases occur repeatedly.41 Thermodynamically, the extraction of PS molecules is driven by favorable adhesive interactions with GNs, but hindered by PS–PS interactions inside the layer. It was expected that the PS extraction could be affected by changing the PS layer tension due to the change in intralayer PS–PS interactions. Before introducing GNs, we first equilibrated the PS layer under different tensions of 10 mN m−1, 20 mN m−1, 30 mN m−1 and 40 mN m−1, respectively (Fig. S1†). Then we simulated the PS extraction by GNs under different tensions. As expected, more PS molecules were extracted by GN under lower surface tension (Fig. 4). The evolutions of GN–PS interaction energy under different tensions were also provided (Fig. S3†), further verifying that the tension effect on PS extraction is energetically relevant. Even SP-C proteins could be extracted and stably attached on the GN surface under lower tension of 10 mN m−1 (Fig. S4†). In contrast, the PS extraction was suppressed by increasing the PS layer tension, reflected by an obvious decrease of the extraction number (Fig. 4). Moreover, the PS layer underwent rupture once a number of PS molecules were extracted under a higher tension of 40 mN m−1 (inset of Fig. 4; for the dynamic process, see Video S2†). Our simulations indicate that PS extraction can induce severe depletion from the layer and cause mechanical inhibition and ultrastructure perturbation, as has been experimentally observed when placing GNs on cellular membranes.36Effects of GN size and oxidation ratio on extraction
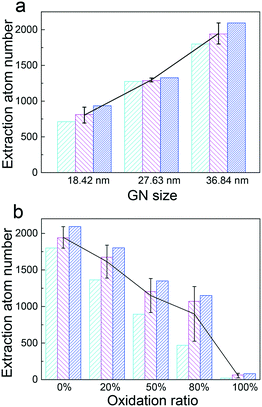
Among physicochemical properties of GNs, both size and oxidation ratio should have profound effects on the PS extraction. For the size effect, three GNs with the same width (D = 9.2 nm) but different lengths (L = 18.42 nm, 27.63 nm and 36.84 nm) were compared for their interactions with the PS layer. To increase the statistical quality, three independent simulations were performed for each GN size. The PS layer tension was fixed to 30 mN m−1, and the results are displayed in Fig. 5a. Simply, the final average extraction number increased roughly linearly with the GN size. No obvious change of the PS arrangement (inverse micelle) was observed for GNs with different sizes (Fig. S5†), except that shorter GNs reached the extraction saturation earlier than longer ones (Fig. S6a†). Our results suggest that larger GNs might be more toxic due to the more acute PS depletion, which is in line with previous in vitro experimental results. For example, using a constrained drop surfactometer method, Valle et al. have measured a higher PS inhibition by larger suspended graphenes.42Graphene oxide (GO) is a derivative of graphene containing different levels of reactive oxygen functional groups. It has attracted substantial interest due to its advantages for many biological applications such as drug delivery, diagnostics and bioimaging.43 Accordingly, the risk of human exposure to GO has been increasing and generating a general biosafety concern. Here, we studied PS extraction by GNs with different oxidation ratios. The average extraction numbers as a function of the GN oxidation ratio are given in Fig. 5b. It is clear that PS extraction can be retarded by increasing the oxidation ratio, further infering that the driving force for extraction is derived from adhesive interactions between PS tails and GN surface. Moreover, the inverse micellar structures of PS molecules were perturbed by increasing the oxidation ratio. For the randomly oxidized GNs, the extracted PS molecules rearranged their polar headgroups in contact with the oxidized region, leaving their tails covering the non-oxidized region (Fig. 6). Specifically for the GN with 100% oxidation, almost no PS molecules were extracted during the simulation (Fig. 5b and S6b†). Note that there have been both cell and mice experiments reporting acute lung injury and pulmonary fibrosis induced by inhaled GOs.44,45 Along this line, our simulations suggest that the inhalation toxicity of GOs might not derive from the PS extraction, but may be ascribed to other mechanisms. For example, Chang et al. and Mittal et al. both reported that GOs can be internalized by human lung cells (A549) and induce oxidative stress mediated cytotoxicity.46,47 When incubated with 10% fetal bovine serum, the concentration-dependent toxicity of GOs could be largely attenuated.12 More recently, Zhu et al. found that GOs can induce plasma injuries and cytoskeletal damages by interacting with integrin to perturb signal transduction, thus helping kill cancer cells.48 The same research group also reported the size dependence of GO in stimulating inflammatory responses in cells.49
Role of GN curvature
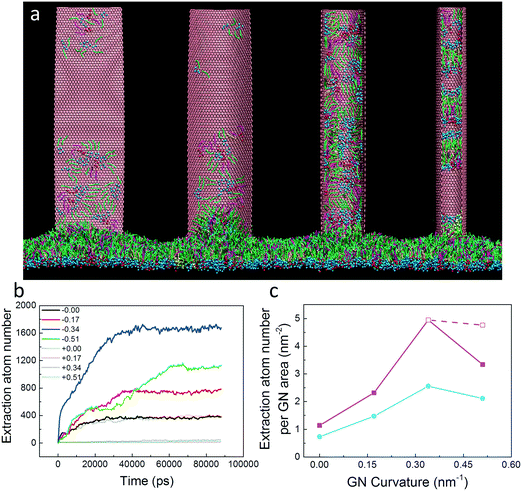
Though considered one of the strongest materials ever tested, monolayer GN can easily undergo deformation under external shearing or pressing force.50 In our simulations, we observed a wrinkling in GNs when interacting with the PS layer. Intuitively, such GN deformation was explained as a result of asymmetric forces exerted by PS molecules interacting with two different GN surfaces. More importantly, the generated GN curvature further led to the asynchronous extraction of PS molecules by its two surfaces (Fig. S7a†). That is, more PS molecules were first rapidly extracted and climbed the GN surface with a negative curvature, whereas the extraction by the convex surface obviously lagged behind (Fig. S7b†). This finding indicates the importance of GN curvature in PS extraction.To further understand the influence of GN curvature on PS extraction, we prepared four GNs with the same lateral size (36.8 nm × 9.2 nm) but different curvatures. To maintain constant GN curvature during the simulations, each bead of GNs was fixed. The final configurations are displayed in Fig. 7a, in which the influence of GN curvature is evidenced. Our simulations showed that the PS molecules were extracted significantly by the GN surface having a negative curvature, while almost no PS molecules were extracted by the convex GN surface. Reminiscent of previous simulations on lipid extraction by curved GNs from bacterial cell membranes,51 preferred PS extraction by the concave GN surface can similarly be understood as a wetting process and is mainly driven by the enthalpic interactions between PS molecules and the GN surface. We should note that the PS extraction was somewhat restrained by freezing the GN, suggesting the importance of GN undulation in interacting with the PS molecules.
By further increasing the GN curvature (0.51 nm−1), however, we observed a decrease in the extraction number (Fig. 7b and c). Unlike the inverse micellar structures of PS molecules adsorbing onto the flat and slight concave GN surfaces, PS molecules were found to form separate hollow bicellar structures on the deeper concave GN surface. Combining evolutions of the extraction number and time sequence of typical snapshots (Fig. 7b and 8a), the extraction process seems to be stepwise. Especially in the late stage, the PS extraction was blocked by a SP-C protein (Fig. 8b; for the dynamic process, see Video S3†). By artificially removing this protein from the layer, the final extraction number was increased, but it was still less than that for the GN having a smaller curvature (Fig. 7c). Another explanation of the decreased extraction is the hindered diffusion of PS molecules from the bulk to the hemi-closed GN inner surface. To test it, we fixed the curvature and decreased the GN width from 9.2 nm to 5.5 nm. In each case, the average extraction number per GN surface area was decreased (Fig. 7c). Also, both inverse micellar and bicellar structures were perturbed by decreasing the GN width (Fig. S8†), albeit with no change of the optimal GN curvature (0.34 cm−1) for maximum extraction. We speculate that increasing the GN curvature can enhance adhesive interactions with PS molecules, but also affect the PS assembly pattern, thus collectively modulating the extraction performance.
Thermodynamic analysis
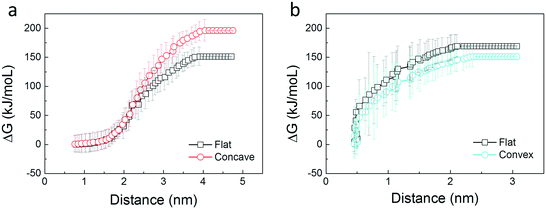
We present a simple energetic analysis to thermodynamically understand the spontaneous extraction of PS molecules by GNs. First, a DPPC molecule was stably adsorbed onto the flat, convex, and concave GN surfaces, respectively. Then a spring force with a constant of 200 kJ mol−1 nm−2 was exerted on COM of the lipid along the GN surface to pull it apart from the GN at a slow rate of 0.01 nm ns−1. Evolutions of the pulling force are given in Fig. S9a,† which illustrates the preferred adsorption of PS molecules on the concave GN surface. Note that when a moving lipid was approaching the convex GN rim, a sudden lipid flipping toward the opposite concave surface was observed (Fig. S9b†), accompanied by a sudden increase of the pulling force (Fig. S9a†). After that, the evolution of the pulling force resembles that for the concave GN surface. To compare PS extraction performance for the three GN surfaces more accurately, we performed two more pulling simulations for both flat and convex GN surfaces, in which the spring force was exerted along the GN normal direction. After that, we estimated the free energy change based on the Jarzynski equality52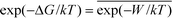 , where ΔG denotes the free energy change and W represents the work exerted on the moving lipid. According to Fig. 9a, transferring a lipid from adsorbtion on the concave GN surface to vacuum is more energetically unfavorable than from the flat GN surface. In the other group, we observed a higher free energy increase for the flat GN surface than the convex one (Fig. 9b). Combining the above simulation results, it is convincing that PS molecules can spontaneously be extracted by GNs, and this effect is further enhanced by GN surfaces having negative curvature. In contrast, the convex GN surface provides less adhesive interactions, thus having a lower PS extraction performance.
, where ΔG denotes the free energy change and W represents the work exerted on the moving lipid. According to Fig. 9a, transferring a lipid from adsorbtion on the concave GN surface to vacuum is more energetically unfavorable than from the flat GN surface. In the other group, we observed a higher free energy increase for the flat GN surface than the convex one (Fig. 9b). Combining the above simulation results, it is convincing that PS molecules can spontaneously be extracted by GNs, and this effect is further enhanced by GN surfaces having negative curvature. In contrast, the convex GN surface provides less adhesive interactions, thus having a lower PS extraction performance.
PS depletion and mechanical inhibition
In vitro experiments showed that NPs with increasing hydrophobicity cause a higher degree of PS inhibition, likely due to increased NP retention and PS depletion.53,54 The strong correlation between NP hydrophobicity and pulmonary toxicity was confirmed by subsequent in vivo experiments in mice.55 Cell experiments also confirmed that phospholipids can be extracted by hydrophobic GNs from bacterial cell membranes, inducing serious membrane stress and significantly reducing cell viability.36,56 Our simulations are in agreement with those experiments, suggesting that PS extraction is probably destructive and related to the inhalation toxicity of graphene-based nanomaterials. To quantify how the suspended GNs induce PS layer stress, we calculated the average PS layer area as a function of the PS layer tension (Fig. 10a). For the pure PS layer system, the area increased roughly linearly with the tension, with the area compressibility, defined as κ = (1/A)(∂A/∂γ), estimated as less than 0.01 m mN−1, similar to that measured by in vitro exepriments using the constrained drop surfactometer method.42 After introducing the suspended GNs, PS molecules were extracted, causing a reduction of PS layer area under the same tensions. The area reduction was more evident under lower tension due to the higher extent of PS extraction. Apparently, the PS layer compressibility was increased. Note that the natural PS layer should have very low compressibility contributing to lung recoil.15 The increase in compressibility by suspended GNs is thus an important criterion for evaluating biophysical inhibition of the PS. Our simulations suggest a new mechanism for PS inhibition, that is, PS molecules can be extracted from the layer by suspended GNs that cause severe PS depletion and biophysical inhibition.Under a higher tension of 40 mN m−1, we observed a PS layer rupture from interaction with GNs (Fig. 4 and Video S2†). Fig. 10b shows evolutions of the local lipid densities in both adjacent and distant regions with respect to the GN invasion site. At t = 75 ns, the striking decrease of the local lipid density shows the sudden layer rupture close to the GN. A simultaneous slight increase of lipid density in the distant region suggests a transient release of the layer stress. As the simulation proceeded, the rupture rapidly expanded to the whole region, reflected by a subsequent decrease of lipid density in the distant region. Correspondingly, the box lateral size continued to increase under the high tension. We next increased the GN size (36.8 nm × 18.4 nm) and found a PS layer rupture under a lower tension of 30 mN m−1 (Fig. S10a†). Even under a low tension of 10 mN m−1, local rupture occurred around the GN, albeit with no rupture expansion being observed (Fig. S10b†). Similar results were also observed for larger curved GNs, where a hydrophilic pore was formed inside the GN cavity (Fig. S11†). These findings suggest that the PS layer integrity can be perturbed by GNs through destructive PS extraction.
Generation of so-called PS corona
To date, numerous studies have highlighted the importance of the corona that determines the biological identity of inhaled NPs in interacting with the pulmonary system.16,57,58 By contrast, the mechanism of how the structure of PS corona is formed still remains elusive, mainly due to the limited spatiotemporal resolution for current experimental methods.59 Our simulations can provide molecular insights into the formation of PS corona on inhaled GNs. Since the PS layer forms the first alveolar capillary barrier against inhaled particles, the PS corona formation should start from the air–water interface through interaction with the PS layer. In this context, the pre-adsorption of PS molecules may play a role as the “precursor” that further influences subsequent biomolecule adsorption to form the final “hard” or “soft” corona layers.60,61 Very recently, Hu et al. simulated formation of PS corona on spherical NPs in the alveolar fluid,19 different from our findings simulated at the air–water interface. To find whether and how the formed structure evolves in the alveolar fluid, we transferred the GN–PS complex into a water box. It was found that the preformed inverse micellar structures shrank and transformed to micellar structures with the hydrophobic PS tails shielded inside a shell composed of PS headgroups (Fig. S12†). This structure minimizes the system free energy by increasing interactions with GN and preventing water molecules from contacting the PS tail moities. Note that in real alveolar fluid, a variety of biomolecules, including PS molecules and PS-associated proteins, exist to possibly interact with inhaled NPs. It is suggested by our simulations that the final composition of the PS corona should consider the pre-adsorption of PS molecules onto the NP surface upon contact with the PS layer. Along this line, further investigations should be conducted in the future to elucidate how the pre-formed structures influence subsequent adsorption of biomolecules to form the final corona structures.Conclusions
In summary, we have presented the simulation investigation on how inhaled GNs interact with the complex PS layer at the air–water interface, mainly focusing on the spontaneous extraction of PS molecules by suspended GNs. We demonstrated that the vigorous extraction is driven by strong dispersive interactions between GNs and PS molecules. To maximize interactions with GNs, the extracted PS molecules cooperatively move and rearrange themselves to form inverse micellar structures on the GN surfaces. By varying the GN size, oxidation ratio and curvature, both the extraction performance and assembly pattern can be significantly manipulated. For curved GNs, only the concave surface can vigorously extract PS molecules, while weaker or no extraction was observed for the convex surface. The PS extraction mediated by GNs is convincing according to our energetic analysis.Our simulations have implications for understanding both inhalation toxicity of GN-family nanomaterials and formation of PS corona structures. First, the inhalation toxicity of GNs could derive from the destructive PS extraction that causes severe depletion and biophysical inhibition of the PS. Even PS layer rupture can be prompted by interaction with larger GNs, suggesting the size-dependent inhalation toxicity of GNs. Second, bearing in mind that the PS layer, located at the air–water interface of lung alveoli, behaves as the first barrier against inhaled particles, our findings imply that the formation of so-called corona should start from the alveolar air–water interface by extracting PS molecules and further influence the toxicity and subsequent fate of GNs in the lungs.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work is financially supported by National Natural Science Foundation of China (21303269, 91543125), Science and Technology Major Project of Shandong Province (2016GSF117033), Qingdao Science and Technology Project (no. 16-5-1-73-jch), the CAS Key Research Program of Frontier Sciences (QYZDB-SSW-JSC036), and the CAS Strategic Priority Research Program (XDB22040403). The support from the Open Fund of LNM is also gratefully acknowledged.References
- V. M. Rotello, Nanoparticles: building blocks for nanotechnology, Springer Science & Business Media, 2004 Search PubMed.
- P. Sharma, S. Brown, G. Walter, S. Santra and B. Moudgil, Adv. Colloid Interface Sci., 2006, 123, 471–485 CrossRef PubMed.
- Z. Liu, S. Tabakman, K. Welsher and H. Dai, Nano Res., 2009, 2, 85–120 CrossRef CAS PubMed.
- S. Huo, S. Jin, X. Ma, X. Xue, K. Yang, A. Kumar, P. C. Wang, J. Zhang, Z. Hu and X.-J. Liang, ACS Nano, 2014, 8, 5852–5862 CrossRef CAS PubMed.
- L. Wang, Q. Sun, X. Wang, T. Wen, J.-J. Yin, P. Wang, R. Bai, X.-Q. Zhang, L.-H. Zhang and A.-H. Lu, J. Am. Chem. Soc., 2015, 137, 1947–1955 CrossRef CAS PubMed.
- Y. Zhao, G. Xing and Z. Chai, Nat. Nanotechnol., 2008, 3, 191–192 CrossRef CAS PubMed.
- A. D. Maynard, R. J. Aitken, T. Butz, V. Colvin, K. Donaldson, G. Oberdörster, M. A. Philbert, J. Ryan, A. Seaton and V. Stone, Nature, 2006, 444, 267–269 CrossRef CAS PubMed.
- J. W. Bennett, C. Allen, S. Pramanik, M. J. Gallagher, N. V. Hudson-Smith, D. Jones, M. O. Krause and S. E. Mason, Environ. Sci.: Nano, 2017, 4, 1428–1432 RSC.
- A. E. Nel, L. Mädler, D. Velegol, T. Xia, E. M. Hoek, P. Somasundaran, F. Klaessig, V. Castranova and M. Thompson, Nat. Mater., 2009, 8, 543–557 CrossRef CAS PubMed.
- S. Tenzer, D. Docter, J. Kuharev, A. Musyanovych, V. Fetz, R. Hecht, F. Schlenk, D. Fischer, K. Kiouptsi and C. Reinhardt, Nat. Nanotechnol., 2013, 8, 772–781 CrossRef CAS PubMed.
- M. Lundqvist, J. Stigler, G. Elia, I. Lynch, T. Cedervall and K. A. Dawson, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 14265–14270 CrossRef CAS PubMed.
- W. Hu, C. Peng, M. Lv, X. Li, Y. Zhang, N. Chen, C. Fan and Q. Huang, ACS Nano, 2011, 5, 3693–3700 CrossRef CAS PubMed.
- S. Lin, M. Mortimer, R. Chen, A. Kakinen, J. Riviere, T. P. Davis, F. Ding and P. C. Ke, Environ. Sci.: Nano, 2017, 4, 1433–1454 RSC.
- S. Tenzer, D. Docter, S. Rosfa, A. Wlodarski, J. R. Kuharev, A. Rekik, S. K. Knauer, C. Bantz, T. Nawroth and C. Bier, ACS Nano, 2011, 5, 7155–7167 CrossRef CAS PubMed.
- Y. Y. Zuo, R. A. Veldhuizen, A. W. Neumann, N. O. Petersen and F. Possmayer, Biochim. Biophys. Acta, Biomembr., 2008, 1778, 1947–1977 CrossRef CAS PubMed.
- A. J. Thorley, P. Ruenraroengsak, T. E. Potter and T. D. Tetley, ACS Nano, 2014, 8, 11778–11789 CrossRef CAS PubMed.
- A. A. Kapralov, W. H. Feng, A. A. Amoscato, N. Yanamala, K. Balasubramanian, D. E. Winnica, E. R. Kisin, G. P. Kotchey, P. Gou and L. J. Sparvero, ACS Nano, 2012, 6, 4147–4156 CrossRef CAS PubMed.
- G. Hu, B. Jiao, X. Shi, R. P. Valle, Q. Fan and Y. Y. Zuo, ACS Nano, 2013, 7, 10525–10533 CrossRef CAS PubMed.
- Q. Hu, X. Bai, G. Hu and Y. Y. Zuo, ACS Nano, 2017, 11, 6832–6842 CrossRef CAS PubMed.
- Q. Hu, B. Jiao, X. Shi, R. P. Valle, Y. Y. Zuo and G. Hu, Nanoscale, 2015, 7, 18025–18029 RSC.
- T. Yue, X. Wang, X. Zhang and F. Huang, RSC Adv., 2015, 5, 30092–30106 RSC.
- T. Yue, Y. Xu, S. Li, X. Zhang and F. Huang, Phys. Chem. Chem. Phys., 2016, 18, 18923–18933 RSC.
- C. C. Chiu, W. Shinoda, R. H. DeVane and S. O. Nielsen, Soft Matter, 2012, 8, 9610–9616 RSC.
- A. K. Geim and K. S. Novoselov, Nat. Mater., 2007, 6, 183–191 CrossRef CAS PubMed.
- M. Zhong, D. Xu, X. Yu, K. Huang, X. Liu, Y. Qu, Y. Xu and D. Yang, Nano Energy, 2016, 28, 12–18 CrossRef CAS.
- J. Lee, T.-H. Han, M.-H. Park, D. Y. Jung, J. Seo, H.-K. Seo, H. Cho, E. Kim, J. Chung and S.-Y. Choi, Nat. Commun., 2016, 7, 11791 CrossRef CAS PubMed.
- K. Yang, L. Feng and Z. Liu, Adv. Drug Delivery Rev., 2016, 105, 228–241 CrossRef CAS PubMed.
- A. Bianco, Angew. Chem., Int. Ed., 2013, 52, 4986–4997 CrossRef CAS PubMed.
- P. Chen and L.-T. Yan, J. Mater. Chem. B, 2017, 5, 4290–4306 RSC.
- J. K. Kim, J. H. Shin, J. S. Lee, J. H. Hwang, J. H. Lee, J. E. Baek, T. G. Kim, B. W. Kim, J. S. Kim and G. H. Lee, Nanotoxicology, 2016, 10, 891–901 CrossRef CAS PubMed.
- L. Ou, B. Song, H. Liang, J. Liu, X. Feng, B. Deng, T. Sun and L. Shao, Part. Fibre Toxicol., 2016, 13, 57 CrossRef PubMed.
- S. J. Marrink, H. J. Risselada, S. Yefimov, D. P. Tieleman and A. H. De Vries, J. Phys. Chem. B, 2007, 111, 7812–7824 CrossRef CAS PubMed.
- M. Sarker, A. J. Waring, F. J. Walther, K. M. Keough and V. Booth, Biochemistry, 2007, 46, 11047–11056 CrossRef CAS PubMed.
- F. Baumgart, O. L. Ospina, I. Mingarro, I. Rodríguez-Crespo and J. Pérez-Gil, Biophys. J., 2010, 99, 3234–3243 CrossRef CAS PubMed.
- A. V. Titov, P. Král and R. Pearson, ACS Nano, 2009, 4, 229–234 CrossRef PubMed.
- Y. Tu, M. Lv, P. Xiu, T. Huynh, M. Zhang, M. Castelli, Z. Liu, Q. Huang, C. Fan and H. Fang, Nat. Nanotechnol., 2013, 8, 594–601 CrossRef CAS PubMed.
- L. Zhang, B. Xu and X. Wang, J. Phys. Chem. B, 2016, 120, 957–964 CrossRef CAS PubMed.
- Z. Gu, Z. Yang, B. Luan, X. Zhou, L. Hong, H. Zhou, J. Luo and R. Zhou, J. Phys. Chem. C, 2017, 121, 2444–2450 CAS.
- B. Hess, C. Kutzner, D. Van Der Spoel and E. Lindahl, J. Chem. Theory Comput., 2008, 4, 435–447 CrossRef CAS PubMed.
- W. Humphrey, A. Dalke and K. Schulten, J. Mol. Graphics Modell., 1996, 14, 33–38 CrossRef CAS.
- Y. Y. Zuo, R. Chen, X. Wang, J. Yang, Z. Policova and A. W. Neumann, Langmuir, 2016, 32, 8501–8506 CrossRef CAS PubMed.
- R. P. Valle, T. Wu and Y. Y. Zuo, ACS Nano, 2015, 9, 5413–5421 CrossRef CAS PubMed.
- X. Sun, Z. Liu, K. Welsher, J. T. Robinson, A. Goodwin, S. Zaric and H. Dai, Nano Res., 2008, 1, 203–212 CrossRef CAS PubMed.
- M. C. Duch, G. S. Budinger, Y. T. Liang, S. Soberanes, D. Urich, S. E. Chiarella, L. A. Campochiaro, A. Gonzalez, N. S. Chandel and M. C. Hersam, Nano Lett., 2011, 11, 5201–5207 CrossRef PubMed.
- B. Li, J. Yang, Q. Huang, Y. Zhang, C. Peng, Y. Zhang, Y. He, J. Shi, W. Li and J. Hu, NPG Asia Mater., 2013, 5, e44 CrossRef CAS.
- S. Mittal, V. Kumar, N. Dhiman, L. K. S. Chauhan, R. Pasricha and A. K. Pandey, Sci. Rep., 2016, 6, 39548 CrossRef CAS PubMed.
- Y. Chang, S.-T. Yang, J.-H. Liu, E. Dong, Y. Wang, A. Cao, Y. Liu and H. Wang, Toxicol. Lett., 2011, 200, 201–210 CrossRef CAS PubMed.
- J. Zhu, M. Xu, M. Gao, Z. Zhang, Y. Xu, T. Xia and S. Liu, ACS Nano, 2017, 11, 2637–2651 CrossRef CAS PubMed.
- J. Ma, R. Liu, X. Wang, Q. Liu, Y. Chen, R. P. Valle, Y. Y. Zuo, T. Xia and S. Liu, ACS Nano, 2015, 9, 10498–10515 CrossRef CAS PubMed.
- K. Min and N. Aluru, Appl. Phys. Lett., 2011, 98, 013113 CrossRef.
- B. Luan, T. Huynh and R. Zhou, Nanoscale, 2016, 8, 5750–5754 RSC.
- C. Jarzynski, Phys. Rev. Lett., 1997, 78, 2690 CrossRef CAS.
- Q. Fan, Y. E. Wang, X. Zhao, J. S. Loo and Y. Y. Zuo, ACS Nano, 2011, 5, 6410–6416 CrossRef CAS PubMed.
- R. P. Valle, C. L. Huang, J. S. Loo and Y. Y. Zuo, ACS Sustainable Chem. Eng., 2014, 2, 1574–1580 CrossRef CAS.
- L. A. Dailey, R. Hernández-Prieto, A. M. Casas-Ferreira, M.-C. Jones, Y. Riffo-Vasquez, E. Rodríguez-Gonzalo, D. Spina, S. A. Jones, N. W. Smith and B. Forbes, Nanotoxicology, 2015, 9, 106–115 CrossRef CAS PubMed.
- S. Liu, T. H. Zeng, M. Hofmann, E. Burcombe, J. Wei, R. Jiang, J. Kong and Y. Chen, ACS Nano, 2011, 5, 6971–6980 CrossRef CAS PubMed.
- C. A. Ruge, U. F. Schaefer, J. Herrmann, J. Kirch, O. Canadas, M. Echaide, J. Pérez-Gil, C. Casals, R. Müller and C.-M. Lehr, PLoS One, 2012, 7, e40775 CAS.
- S. S. Raesch, S. Tenzer, W. Storck, A. Rurainski, D. Selzer, C. A. Ruge, J. Perez-Gil, U. F. Schaefer and C.-M. Lehr, ACS Nano, 2015, 9, 11872–11885 CrossRef CAS PubMed.
- M. Kokkinopoulou, J. Simon, K. Landfester, V. Mailänder and I. Lieberwirth, Nanoscale, 2017, 9, 8858–8870 RSC.
- M. I. Setyawati, C. Y. Tay, D. Docter, R. H. Stauber and D. T. Leong, Chem. Soc. Rev., 2015, 44, 8174–8199 RSC.
- S. Milani, F. Baldelli Bombelli, A. S. Pitek, K. A. Dawson and J. Rädler, ACS Nano, 2012, 6, 2532–2541 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: For additional simulation results, including twelve figures and three videos. See DOI: 10.1039/c7en00843k |
| This journal is © The Royal Society of Chemistry 2018 |