Integrating proteomics, metabolomics and typical analysis to investigate the uptake and oxidative stress of graphene oxide and polycyclic aromatic hydrocarbons†
Xiaokang
Li
a,
Li
Mu
b and
Xiangang
Hu
 *a
*a
aKey Laboratory of Pollution Processes and Environmental Criteria (Ministry of Education), Tianjin Key Laboratory of Environmental Remediation and Pollution Control, College of Environmental Science and Engineering, Nankai University, Tianjin 300071, China. E-mail: huxiangang@nankai.edu.cn; Fax: +86 022 85358121; Tel: +86 022 85358121
bTianjin Key Laboratory of Agro-Environment and Safe-Product, Key Laboratory for Environmental Factors Control of Agro-Product Quality Safety (Ministry of Agriculture), Institute of Agro-Environmental Protection, Ministry of Agriculture, Tianjin 300191, China
First published on 9th November 2017
Abstract
Graphene oxide (GO) is an emergent engineered nanomaterial that shows great potential for use in agricultural applications, such as promoting crop production and controlling insect pests. Polycyclic aromatic hydrocarbons (PAHs) are widely distributed and can enter crops via contaminated water, soil and air. Therefore, it is crucial to understand the potential risks of the co-exposure of crops, such as rice, to GO and PAHs. However, information regarding the effects of GO on PAH toxicity and the specific molecular responses to GO is lacking. The present work revealed that GO significantly enhanced the accumulation of PAHs by 26.4–92.5% in rice and that PAH accumulation was also influenced by temperature and glycerol. GO further promoted increased aryl hydrocarbon receptor (AhR) and cytochrome P450 levels, which are induced by PAHs. The altered proteins, including ascorbate peroxidase (APX), aquaporins and those involved in ATP synthesis, were mainly associated with oxidative stress and transmembrane transport. Amino acid metabolism was the primary metabolic pathway influenced by GO and PAHs. Arabinose and pentanoic acid were positively associated with the uptake of PAHs and oxidative stress, respectively, during co-exposure to GO. The abovementioned results highlight the potential risks and specific molecular mechanisms of emergent engineered nanomaterials during co-exposure with traditional organic pollutants.
Environmental significanceGraphene oxide (GO) is an emergent engineered nanomaterial that is widely applied in various fields, and polycyclic aromatic hydrocarbons (PAHs) are traditional pollutants that are widely distributed in the environment. GO combining with PAHs is inevitable when it is released into the environment; however, information regarding the effects of GO on the toxicity of the PAHs and its specific molecular responses is lacking. The present work first reveals that GO exposure enhances the uptake of PAHs and oxidative stress in rice through omics and regular analysis. This study provides new implications for the potential environmental risks of emergent engineered nanomaterials during co-exposure with traditional organic pollutants. |
Introduction
Graphene oxide (GO), which is an emergent engineered nanomaterial, has attracted considerable attention in various fields (e.g., agriculture, chemistry, medicine, biology and environmental protection) due to its extraordinary properties.1–4 The applications of GO, such as promoting production and controlling insect pests, have attracted much attention in agriculture.5,6 Due to the widespread applications of GO, it is inevitably released into the environment during manufacture, use and disposal of GO-based products.7–9 Therefore, understanding the potential environmental risks of GO is essential. Natural environments contain multiple pollutants, and co-exposure to nanomaterials and other pollutants is more likely to occur than exposure to the nanomaterial alone.10,11 Recent studies that have investigated GO nanotoxicity mainly focused on exposure to GO alone and have reported several important findings, including the observations that GO inhibits or promotes cell growth, induces oxidative stress and alters the expression of genes or proteins.12–14 However, the environmental risks and molecular mechanisms of GO co-exposure with widely distributed organic pollutants remain largely unknown.15,16Plants are the primary producers in ecological systems and provide the main food source for humans.17 Previous studies assessing the risk of nanomaterials to plants have concentrated on alterations in reactive oxygen species (ROS), antioxidant enzymes, plant growth, and a few genes or proteins.18–20 The accumulation of contaminants and oxidative stress are two basic parameters used to assess the co-exposure to nanomaterials and other contaminations.21,22 Fullerene significantly reduced the uptake of dichlorodiphenyldichloroethylene (p,p′-DDE) in zucchinis, soybeans and tomatoes.23 Carbon nanotubes inhibited the uptake of pesticides in crop species24 but enhanced the toxicity of diuron in Chlorella vulgaris.25 Our recent work showed that GO amplified the phytotoxicity of arsenic in wheat and mediated the biocompatibility of antibiotics and resistance genes.11,26 However, the molecular mechanisms of phytotoxicity during co-exposure to carbon-based nanomaterials and other pollutants are unclear due to the lack of detailed studies and effective methods. The development of omics techniques provides new opportunities to discover molecular mechanisms from global perspectives.27 The field of proteomics explores the qualitative and quantitative changes that occur to the proteins in cells or an organism under certain conditions.28,29 Metabolites serve as direct signatures of biochemical activities and are easy to correlate with cellular biochemistries and biological phenomena.30 To achieve a solid conclusion, multiple tools and approaches should be applied to explore a particular question.31 The integration of proteomic, metabolomic and regular analyses (e.g., plant growth, oxidative stress and nanomaterial uptake) may reflect specific biological mechanisms more accurately than a single omics analysis or regular analyses alone.
Polycyclic aromatic hydrocarbons (PAHs) have attracted considerable public attention due to their wide distribution and carcinogenic, mutagenic, and teratogenic properties.32,33 In addition, PAH exposure poses high risks in crops.34–36 Importantly, GO can strongly adsorb PAHs by π–π and hydrogen bonds via their similar six-carbon rings and other groups.37,38 Because the six-carbon rings are comparable between the two materials, the effects of GO on the phytotoxicity of PAHs and its specific molecular mechanisms are applicable to both GO and PAH. Moreover, because plants are essential components in the ecosystem and rice is the most important global agricultural crop, an understanding of the uptake and toxicity of PAHs with GO in rice is crucial for assessing food safety.39,40 First, the effects of GO on the specific uptake pathways of PAHs and the typical responses of rice were studied in the present work. Then, to explore the underlying molecular mechanisms in the abovementioned phenomena, the significantly changed proteins and metabolites were identified using proteomic and metabolomic approaches, respectively. Moreover, the connections among the proteomic, metabolomic and regular analyses were analyzed using protein–protein interactions and orthogonal projections to latent structures discriminant analyses (OPLS-DA). These studies elucidated the specific responses and mechanisms in plants exposed to GO and PAHs by integrating proteomic, metabolomic and regular analyses to provide an understanding of the potential risks of emergent nanomaterials.
Results and discussion
Nanomaterial characterization
TEM and AFM were conducted to determine the morphology of GO. As shown in Fig. 1a, TEM imaging clearly showed that GO had a transparent sheet morphology with few-layer stacking, as denoted by the black arrow.16,41 AFM imaging also demonstrated that GO exhibited a nanosheet-like morphology with sheet thicknesses of 2.0 ± 0.5 nm and lateral lengths of approximately 0.5–5 μm, as shown in Fig. 1b. The thickness in the AFM image was consistent with the few-layer morphology (2–3 layers, approximately 0.8 nm per layer) of GO in the TEM image.42 The peak in the XRD spectra was centered at 2θ = 10.2° (Fig. 1c), which corresponded to the typical peak of GO.43 The XPS spectra showed that GO was composed of 68.73% C 1s and 31.27% O 1s, as shown in Fig. 1d. The C 1s showed four peaks at 284.7, 286.7, 287.1 and 288.7 eV, which represented the C–C (49%), C–O (35%), C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O (12%) and O
O (12%) and O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–C (4%) bonds, respectively. As shown in Fig. 1e, the zeta potentials of GO and PG ranged from approximately −30 mV to −45 mV at pH 5–9 and revealed a relatively stable negatively charged surface at the tested pH. The Hd was used to investigate the aggregation of GO,44 as shown in Fig. 1f. During the test (48 h), no significant alterations in the Hd were observed, but the Hd increased significantly from 226 to 405 nm with increasing concentration. After the adsorption of PAHs, the Hd of PG showed obvious time- and concentration-dependent alterations, suggesting that PAHs reduced the aggregation of GO.
C–C (4%) bonds, respectively. As shown in Fig. 1e, the zeta potentials of GO and PG ranged from approximately −30 mV to −45 mV at pH 5–9 and revealed a relatively stable negatively charged surface at the tested pH. The Hd was used to investigate the aggregation of GO,44 as shown in Fig. 1f. During the test (48 h), no significant alterations in the Hd were observed, but the Hd increased significantly from 226 to 405 nm with increasing concentration. After the adsorption of PAHs, the Hd of PG showed obvious time- and concentration-dependent alterations, suggesting that PAHs reduced the aggregation of GO.
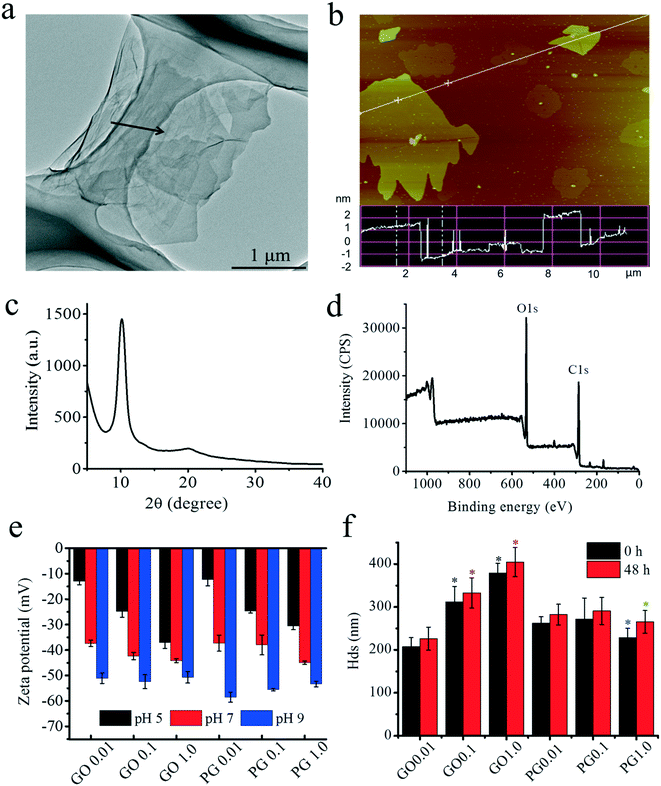 | ||
| Fig. 1 Characterization of GO. (a) TEM imaging; (b) AFM imaging; (c) X-ray photoelectron spectrum; (d) X-ray powder diffraction spectrum; (e) zeta potential of GO and PAHs with GO (PG). (f) Hds of GO and PAHs with GO (PG). Black and red “*” denote significant differences from GO 0.01 at 0 h and 48 h, respectively, at p < 0.05. Blue “*” represents significant differences between pristine GO and PG at 0 h. Green “*” represents significant differences between pristine GO and PG at 48 h. In Fig. 1e and f, the tested concentration of PAHs was 10 μg L−1, and the tested concentrations of GO ranged from 0.01–1.0 mg L−1 as labeled after GO or PG. | ||
GO regulates the uptake of PAHs
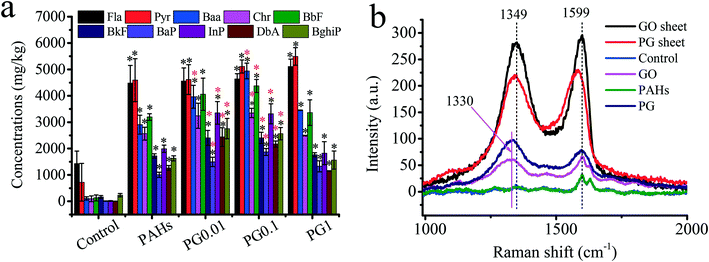
Roots are the primary route of exposure to pollutants in plants.18,45 Therefore, studying how the accumulation of PAHs in rice roots is affected by GO is crucial. To date, information regarding the effects of GO on the accumulation of PAHs in crops is lacking. As shown in Fig. 2a, compared with the PAH group, the contents of Fla and Pyr with relatively low molecular weights slightly increased by 0.5–19.5% in the rice roots as the concentration of GO increased in the PG group. In contrast, the concentrations of PAHs (Baa, Chr, BbF, BkF, BaP, InP, DbA and BghiP) with relatively high molecular weights in the PG groups clearly increased in the rice roots. In particular, when exposed to low doses (0.01 mg L−1 and 0.1 mg L−1) of GO, the concentrations of PAHs increased by 26.4–92.5% compared with the concentrations of PAHs when exposed to PAHs alone. Our recent work confirmed that GO exposure triggered the permeability of plant cell walls and had a carrier effect,11,30 which could promote the uptake of PAHs. As shown in Fig. 1f, the Hd increased as the concentrations of GO increased. In addition to uptake, GO covered the biological surface and PG with more negative charges at high GO concentrations (the plant cells exhibited negative charges),30 which explained the low uptake of PAHs at high concentrations (1.0 mg L−1) of GO. Moreover, to verify the carrier effects of GO, the Raman spectra of the rice roots were assessed. As shown in Fig. 2b, the peaks at approximatively 1349 cm−1 and 1599 cm−1 are ascribed to the typical D and G bands of GO, respectively. Compared with pristine GO sheets, PAHs adsorbed on GO exhibited a redshift from 9 cm−1 and 28 cm−1 at the D and G bands, respectively, due to the interconnected sp2 carbons; π-electrons were delocalized within the ribbon after PAH adsorption.46 The D and G bands were also detected in roots exposed to GO and PG. Our recent work also revealed the carrier effects of arsenic on GO.11 The Raman spectra results verified the uptake of GO and PG but did not provide quantitative information due to the inhomogeneous distribution of nanoparticles in biological tissues.47GO regulates oxidative stress of PAHs
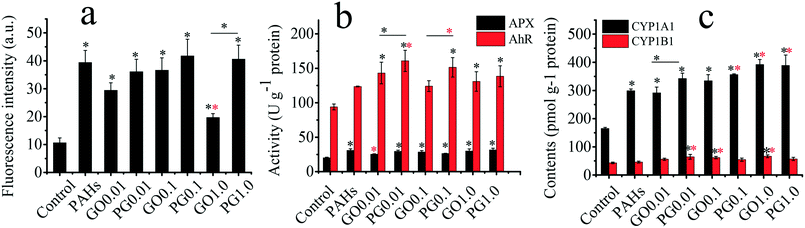
As shown in Fig. 3a, the levels of ROS in the GO, PAH and PG groups were significantly increased by 85–245%, 271% and 240–293%, respectively, compared with those in the control group. The ROS levels for treatment with both 0.01 and 0.1 mg L−1 GO were higher than the ROS level for treatment with 1.0 mg L−1 GO, suggesting complex interactions among plants, GO and free radicals. On the one hand, GO inhibited the hydroxyl radical and nitroxide free species signals.48 The hydroxyl radical and nitroxide free species are vital in plant oxidative systems. On the other hand, GO as a xenobiotic could trigger free radicals in the biological matrix. Ascorbate peroxidase (APX) plays a vital defensive role against ROS.49 As shown in Fig. 3b, the activity of APX in the GO, PAH and PG groups was significantly higher than that in the control group, demonstrating that the plants exhibited adaptive responses to oxidative stress. PAHs induced aryl hydrocarbon receptor (AhR) expression, leading to the induction of a battery of xenobiotic enzymes, including cytochrome P450 1A1 (CYP1A1) and cytochrome P450 1B1 (CY1B1).50–52 As shown in Fig. 3b, the activity of AhR in the GO, PAH and PG groups markedly increased by 32–52%, 31.6% and 47–71% compared with that in the control group, respectively. The AhR levels in the PG group were 12–30% higher than those in the PAH group. As shown in Fig. 3c, the CYP1A1 content in the GO, PAH and PG groups was significantly increased by 76–137% compared with the CYP1A1 content in the control group, and the CYP1A1 content in the PG groups was 15–30% higher than the content in the PAH groups. The CYP1B1 content in the GO and PG groups was also markedly higher than the content in the control group, and the CYP1B1 content was increased by 18–40% in the PG group compared with the CYP1B1 content in the PAH groups. Therefore, GO upregulated the expression of AhR, CYP1A1 and CYP1B1 induced by PAHs. Oxidative stress is closely linked to root development.53 As shown in Fig. S1,† the root tip widths in the GO, PAH and PG groups were significantly larger than those in the control group. Analysis of the gray intensity of the root tips revealed that the relative abundance of cytoplasm in the GO, PAH and PG groups was lower than that in the control group. The relative abundance of cytoplasm in the PG group decreased by 32.99% compared with that in the PAH group. Therefore, GO intensified the oxidative stress and the developmental disturbance induced by PAHs in the roots.Toxicological mechanisms
Proteomics is an effective tool for revealing the global responses of plants to biotic and abiotic stresses.54 Using LC-MS/MS, 426 differentially expressed proteins were screened from approximately 3000 proteins. As shown in Fig. S2,† using HCL analysis, the samples were divided into two clusters (i.e., GO and PAHs/PG/control). The PAHs/PG/control cluster was further divided into two sub-clusters (i.e., PG and PAHs/control). Similar results were achieved in the HCL analysis. The abovementioned cluster analysis indicated that GO intensified the protein alterations induced by PAHs. Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of differently expressed proteins was conducted, as shown in Fig. S3,† using OmicsBean. Downregulated KEGG pathways were observed in ribosomes, while exposure to GO upregulated the ribosome pathway compared with exposure to PAHs alone. The upregulated KEGG pathway was mainly involved in metabolic-related processes, such as carbon metabolism, amino acid biosynthesis and glycolysis/gluconeogenesis. Previous studies have proposed that redox imbalances (Fig. 3) in the cell redox state affect translation, influencing mRNA levels and stability, amino acid availability, carbon metabolism and energy balance.55,56As a multi-omics approach, the combination of proteomics and metabolomics represents a powerful tool to comprehensively evaluate the biological effects of nanomaterials.57 Therefore, the metabolites were studied. Approximately 200 peaks in each sample were analyzed using GC-MS with a derivatization procedure. As shown in Fig. S4a,† 82 metabolites were identified, including amino acids, carbohydrates, fatty acids and alkanes. As shown in Fig. S4b,† the following two clusters were revealed: the control group and another group; thus, exposure to both PAHs and GO disturbed the biological metabolism.
To clarify the alterations in metabolism, the upregulated and downregulated metabolic pathways were identified using MetaboAnalyst 3.0. The downregulated KEGG pathways were primarily related to aminoacyl-tRNA biosynthesis and metabolic processes, such as galactose and alanine/aspartate/glutamate metabolism. The availability of aminoacyl-tRNA affected protein translation,55 which was consistent with the ribosome results from the proteomic analysis. Amino acid metabolism and sugar metabolism are related to glycolysis and the TCA cycle, which are related to energy status maintenance.58 To resist the redox imbalances induced by GO and PAHs (Fig. 3), the regulated pathways according to both the metabolomic and proteomic analyses were involved in translation, amino acid metabolism, and sugar metabolism, which are related to energy status maintenance and carbon skeletons.
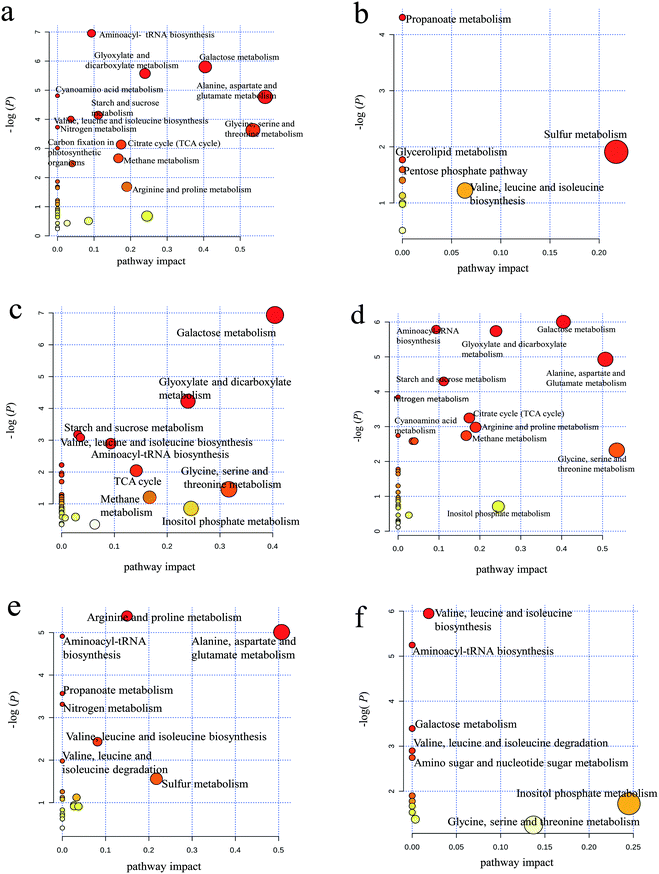
As shown in Fig. 4a, aminoacyl-tRNA biosynthesis, galactose metabolism, alanine/aspartate/glutamate metabolism and glycine/serine/threonine metabolism were clearly downregulated by PAHs, but PAHs upregulated the propanoate metabolic pathway (Fig. 4b). This result is consistent with the findings of previous studies demonstrating that the metabolic disruption by PAHs was mainly due to changes in amino acid, carbohydrate and energy metabolism.59–61 KEGG analysis of proteins showed that ribosomes and carbon metabolism were remarkably downregulated and upregulated (Fig. S3a and b†), respectively, which is consistent with the downregulation of aminoacyl-tRNA biosynthesis.
Similar downregulation of galactose metabolism was observed in the GO group (Fig. 4c), but upregulated pathways were not observed for GO. According to the KEGG analysis, ribosomes and the biosynthesis of amino acids were remarkably downregulated and upregulated (Fig. S3c and d†), respectively, which is not entirely consistent with the metabolism results shown in Fig. 4c, thus supporting the need for integrated proteomic and metabolomic analyses. The downregulated metabolic pathways in the PG groups were more similar to those in the PAH group than to those in the GO group (Fig. 4d), suggesting that PAHs played a dominant role in the metabolic disturbance. The abovementioned results were consistent with the results of the KEGG analysis of proteins (Fig. S3e and f†). Nitrogen metabolism (e.g., arginine/proline metabolism and alanine/aspartate/glutamate metabolism) was downregulated in the PG group compared with that in the PAH groups (Fig. 4e), and valine/leucine/isoleucine biosynthesis and inositol phosphate metabolism were upregulated in the PG group compared with that in the PAH groups (Fig. 4f). Nitrogen metabolism is related to energy metabolism,62 which is consistent with the remarkable inhibition of ATP synthase delta in the protein analysis, as shown in Fig. 3. Moreover, as shown in Fig. S3g and h,† the disturbance in the ribosomes contributed to the alterations in nitrogen metabolism.
The connections between protein alterations and metabolic pathway disturbance are further clarified in Fig. S5 and Table S1.† The metabolites which were significantly regulated by PAHs and PG exposure were mainly related to amino acids, sugars and fatty acids. The protein Q2R480 related to carbon metabolism was significantly upregulated upon PAH exposure. Glycolysis/gluconeogenesis related proteins (e.g., downregulated proteins Q8S3Q3, Q6H703 and Q0J8A4 and upregulated proteins Q5SNH5, Q40677, Q7FAH2, Q6Z9G0, Q69V57, Q2RAK2, Q2QM55 and Q6H798) and biosynthesis of amino acids related proteins (e.g., downregulated proteins Q6H703, Q0J8A4, Q9XEA8 and Q6K969 and upregulated proteins Q5SNH5, Q40677, Q7FAH2, Q6Z9G0, Q69V57, Q2RAK2, Q2QM55, Q6H798, Q93Y73 and Q6L4H5) were significantly altered by PAHs and PG exposure. Both carbon metabolism related protein Q2R480 and glycolysis/gluconeogenesis related protein Q8S3Q3 regulate the metabolic pathway from glucose-6-p to fructose-6-p. Importantly, many significantly altered proteins (e.g., downregulated proteins Q6H703 and Q0J8A4 and upregulated protein Q5SNH5, Q40677, Q7FAH2, Q6Z9G0, Q69V57, Q2RAK2 and Q2QM55) regulated glycolysis/gluconeogenesis and biosynthesis of amino acids together.
Proteomic exploration of transmembrane transport
Proteomics was employed to explore the effects of GO and PAHs on specific transmembrane transport proteins. Proteomics analysis revealed that significantly regulated transmembrane transport proteins in the treated groups were mainly related to ATP synthesis and aquaporins (Fig. 5). Previous studies also demonstrated that PAHs cross the plant cell membrane via aquaporins and by energy-consuming active pathways.63,64 Therefore, to further verify the roles of energy and aquaporins in the uptake of PAHs by GO, a PAH uptake experiment was performed through inhibiting active transfer by reducing temperature and inhibiting aquaporins with glycerol, respectively.65,66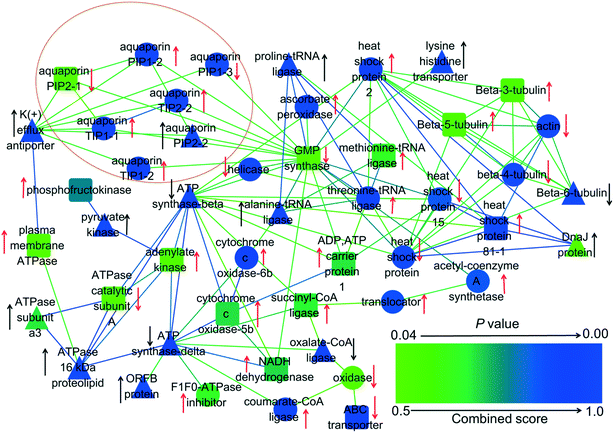 | ||
| Fig. 5 Interaction network of 48 differentially expressed proteins related to transmembrane transport in rice roots exposed to GO, PAHs and PG. The red arrows denote the proteins that were upregulated or downregulated by PAHs. The black arrows denote significant alterations in proteins induced by GO during exposure to PAHs. The up and down arrows represent upregulated and downregulated protein expression, respectively. The triangles show that GO significantly aggravates the expression of proteins in the PAH group. The circles indicate that GO significantly mitigated the deviant expression of proteins in the PAH group. The round rectangles indicate that GO had no significant effects on the differentially expressed proteins in the PAH group. p-Values represent the degree of deviation between the PG and PAH groups. The specific p-values are listed in Table S2 in the ESI.† | ||
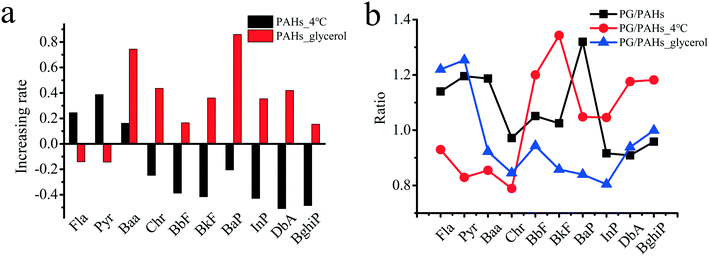
In addition to the inhibition of aquaporins for the PAH uptake pathway, experiments considering glycerol are environmentally relevant. Glycerol is the main by-product in the process of biodiesel production and is applied in various industries, such as in the production of cosmetics, food, tobacco products, and alkyd resin.67 As shown in Fig. 6a, the uptake of seven PAHs (i.e., BaP, Chr, BbF, BkF, InP, BghiP and DbA) at 4 °C significantly decreased by 20.5–50.9% compared with that at 25 °C, while the uptake of the other three PAHs (i.e., Fla, Pyr and Baa) increased by 16.2–38.7%. In general, low temperatures severely reduce the active transport of molecules.68,69 The uptake of PAHs (i.e., Chr, BbF, BkF, BaP, InP, DbA and BghiP) with higher molecular weights and hydrophobicity may depend on active transport more than those of the other three PAHs (i.e., Baa, Fla and Pyr) with relatively lower molecular weights and hydrophobicity. Due to glycerol exposure, the uptake of PAHs (i.e., Fla and Pyr) decreased by 14.1% and 14.4%, respectively, suggesting that Fla and Pyr with relatively smaller benzene rings and reduced hydrophobicity primarily entered the rice roots through aquaporins. However, the uptake of other PAHs (i.e., BghiP, BbF, InP, BkF, DbA, Chr, Baa and BaP) increased by 15.4–86.0% after exposure to glycerol, suggesting that the uptake of these PAHs did not depend on aquaporins. Glycerol may induce reorganization of the cytoskeleton,70 which contributes to the accumulation of PAHs with higher molecular weights and hydrophobicity. Interestingly, as shown in Fig. 6b and compared with Fig. 6a, GO played opposite roles in the uptake of several PAHs, except for Baa, Chr and BaP (Fig. 6a and b), at low temperature and in the presence of glycerol. For example, the uptake of BbF, BkF, InP, DbA and BghiP was upregulated, while the uptake of Fla and Pyr was downregulated by GO at a low temperature. Glycerol exposure induced results that were opposite to those observed under the low temperature conditions. The above results suggest that GO may carry small-ring PAHs into plants via a non-aquaporin pathway.
The passive process in the proteomics analysis mainly involved aquaporins, as denoted by the circle in Fig. 5. Aquaporins are channel proteins that facilitate the transport of water across plant cell membranes.71 PAHs with small rings cross plant cell membranes along with water absorption, which is facilitated via aquaporins.63,72 Four aquaporins (PIP1-2, TIP1-1, TIP1-2 and TIP2-2) were upregulated and two aquaporins (PIP1-3 and PIP2-1) were downregulated by PAHs. According to the results in Fig. 6a, the four upregulated aquaporins (i.e., PIP1-2, TIP1-1, TIP1-2 and TIP2-2) likely regulated the uptake of PAHs with low molecular weights and relatively high water solubility (i.e., Pyr and Baa). GO mitigated the differential expression of PIP1-2, TIP1-1, TIP1-2, TIP2-2 and PIP1-3 induced by PAHs. The above results indicate that the regulation of PAH uptake through aquaporin expression is altered by GO.
Except for aquaporins, PAHs upregulated 19 proteins (e.g., acetyl-coenzyme A synthetase, heat shock protein, cytochrome c oxidase-6b, threonine-tRNA ligase, dicarboxylate/tricarboxylate transporter, ADP, ATP carrier protein 1, tubulin, methionine-tRNA ligase, NADH dehydrogenase, and plasma membrane ATPase) and downregulated 9 proteins (e.g., actin, tubulin, heat shock protein, ABC transporter, and ATPase catalytic subunit A), which are related to transmembrane transport. Integrating the results shown in Fig. 6a, the 19 upregulated aquaporins (e.g., acetyl-coenzyme A synthetase, heat shock protein) likely regulated the uptake of PAHs with high molecular weights and relatively low water solubility (i.e., BaP, Chr, BbF, BkF, InP, BghiP and DbA).
Moreover, GO significantly enhanced the upregulated proteins (e.g., actin, tubulin, heat shock protein, ABC transporter, ATPase catalytic subunit A) and the downregulated proteins (e.g., actin, tubulin, heat shock protein, ABC transporter, ATPase catalytic subunit A) induced by PAHs. In contrast, GO mitigated the differential expression of acetyl-coenzyme A synthetase, actin, tubulin, cytochrome c oxidase-6b, dicarboxylate/tricarboxylate transporter, heat shock protein and threonine-tRNA ligase induced by PAHs. All these proteins are related to ATP synthesis and active transport. For example, acetyl-coenzyme A is a crucial metabolite in carbon and energy metabolism.73 Heat shock proteins are chaperones that assist in the proper folding, stability and transport of proteins across cellular membranes.74 The data described above revealed the differentially expressed proteins related to transmembrane transport in rice roots exposed to GO, PAHs and PG, explaining the underlying molecular mechanisms of PAH uptake by GO.
Additionally, testing PAH uptake in rice at 4 °C resulted in cold stress to the rice and subsequently caused adverse effects on plant growth, which was directly related to PAH uptake. Thus, the biomass data from rice grown at 25 °C and 4 °C and from rice grown in glycerol are provided in Fig. S6.† The significant differences in biomass between groups were analyzed using one-way ANOVA. No significant differences in the root biomass were observed between control and treated groups due to the short-term exposure at 25 °C, 4 °C, or to glycerol. The biomass in the leaves of rice co-exposed to PAHs, GO and glycerol was higher than that in rice co-exposed to PAHs and GO at 25 °C.
Oxidative stress and underlying molecular mechanisms
As shown in Fig. 3a–c, both PAHs and PAHs with GO promoted oxidative stress. GO did not significantly affect ROS levels when rice was exposed to PAHs. However, the expression levels of AhR, CYP1A1 and CYP1B1 enzymes in the GO group with PAHs were significantly higher than those in the PAH group. To investigate the mechanism, proteins related to oxidative stress were screened using a proteomic approach, as shown in Fig. 7. PAHs altered the expression of many oxidative stress-related proteins. There were many upregulated proteins, including aldehyde oxidase, ascorbate peroxidase 1, ascorbate peroxidase 3, peroxidase 5, sulfite reductase, 2-Cys peroxiredoxin, GSTU6, 6-phosphogluconate dehydrogenase, NADP-isocitrate dehydrogenase, fructose-bisphosphate aldolase, alcohol dehydrogenase and phosphofructokinase and a few downregulated proteins, including 1-Cys peroxiredoxin, cysteine proteinase inhibitor, GST, and peroxidase 2. Aldehyde oxidase serves as an important biological source of ROS,75 which is consistent with the increased ROS levels induced by PAHs. Correspondingly, ROS-scavenging proteins were also regulated by PAHs, such as ascorbate peroxidase 1, ascorbate peroxidase 3, and peroxidase. Previous studies have also verified that sulfite reductase and peroxiredoxin, which are upregulated by PAHs, can protect plants from oxidative stress.76,77 Upregulated 6-phosphogluconate dehydrogenase and NADP-isocitrate dehydrogenase can promote NADPH regeneration, and NADPH is an essential reductive coenzyme for antioxidative systems in the PAH group.78 The upregulated fructose-bisphosphate aldolase participates in carbohydrate metabolism pathways79,80 The downregulated cysteine proteinase inhibitor and GST also play important roles in the regulation of plant stress responses in the PAH group.81,82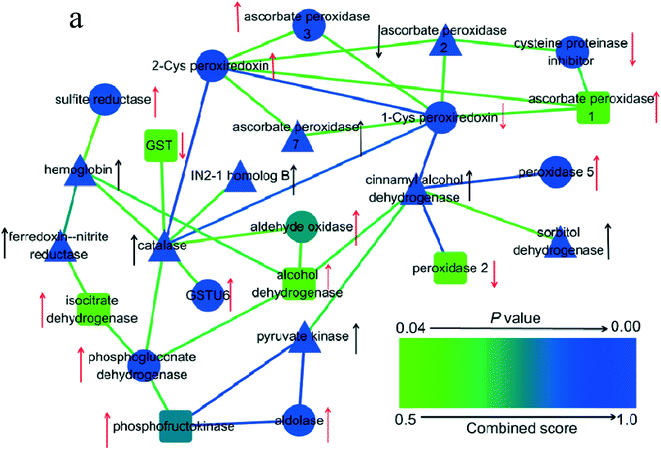 | ||
| Fig. 7 Interaction network of 25 differentially expressed proteins related to oxidative stress in plant roots exposed to PAHs and PG. The red arrows denote the proteins that were upregulated and downregulated by PAHs. The black arrows denote significant alterations in the proteins induced by GO during exposure to PAHs. The up- and down-directed arrows represent upregulated or downregulated protein expression, respectively. The triangles represent significant GO aggravation of the expression of proteins in the PAH group. The circles indicate that GO significantly mitigated the deviant expression of proteins in the PAHs group. The round rectangles indicate that GO had no significant effect on the differentially expressed proteins in the PAH group. p-Values represent the degree of deviation between the PG and PAH groups. The specific p-values are listed in Table S3 in the ESI.† | ||
The oxidative stress-related proteins (sorbitol dehydrogenase, hemoglobin, catalase isozyme B, pyruvate kinase, ferredoxin–nitrite reductase, cinnamyl alcohol dehydrogenase, ascorbate peroxidase 7, IN2-1 homolog B) in the PAH group were significantly upregulated by GO. In contrast, the ascorbate peroxidase 2 protein in the PAH group was downregulated by GO. GO also mitigated the downregulation of the cysteine proteinase inhibitor and 1-Cys peroxiredoxin and the upregulation of ascorbate peroxidase 3, GSTU 6, peroxidase 5, alcohol dehydrogenase and 2-Cys peroxiredoxin, which were induced by PAHs. The above results suggest the complex regulation of oxidative stress-relevant proteins by GO and PAHs at the omics level.
Moreover, protein–protein interactions between oxidative stress and transmembrane transport were established, as presented in Fig. S7.† The red and black colored proteins are related to oxidative stress and transmembrane transport, respectively. APX, phosphofructokinase and pyruvate kinase played dual roles in oxidative stress and transmembrane transport.
Metabolites related to the uptake of PAHs and oxidative stress
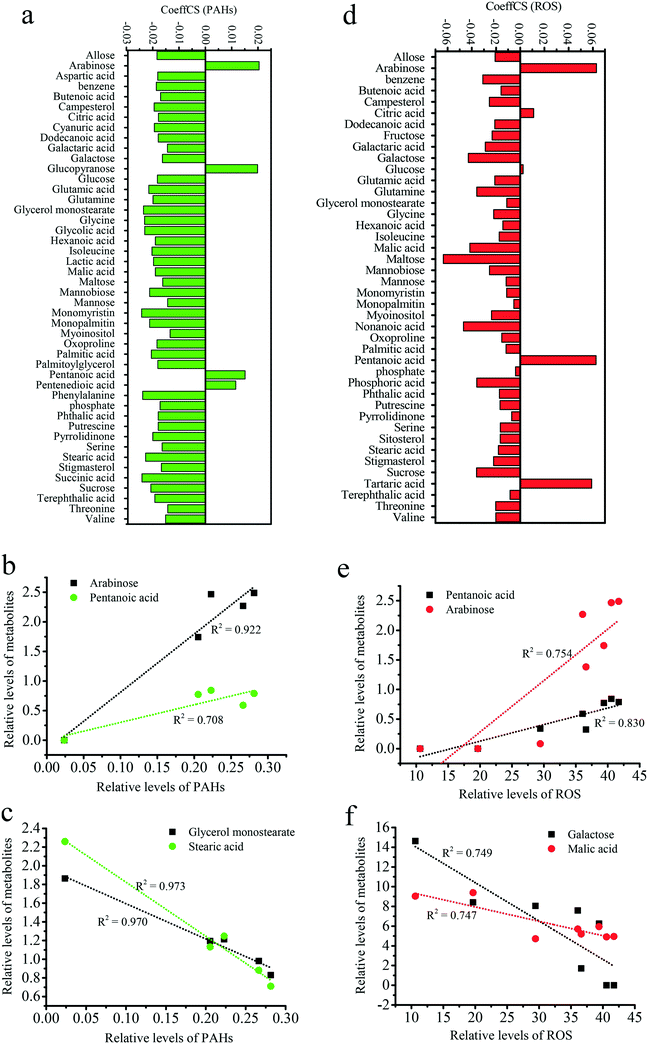
To determine the relationships among metabolites, PAH uptake and ROS levels, an OPLS-DA analysis was performed with the metabolites as X variables and PAH uptake or ROS as the Y variable. The metabolites with high VIP values greatly contributed to PAH uptake or ROS levels, as shown in Fig. S8.† The links between metabolism and PAH uptake or ROS levels are presented in Fig. 8. The positive and negative CoeffCS values reflect the positive and negative correlations between the metabolites and PAH uptake, respectively (Fig. 8a). Arabinose, pentanoic acid, pentanedioic acid and glucopyranose positively contributed to PAH uptake. As shown in Fig. 8b, robust, positive linear correlations were observed between arabinose (R2 = 0.922) and pentanoic acid (R2 = 0.708) metabolism and PAH uptake. The secretion of mucus (e.g., arabinose) increased when organisms were exposed to gold nanoparticles, which was considered a tolerance mechanism against nanoparticle uptake.83 Glycerol monostearate, stearic acid, phenylalanine, and monomyristin negatively contributed to the level of PAH uptake (Fig. 8a). As shown in Fig. 8c, robust, negative linear correlations were observed for glycerol monostearate (R2 = 0.973) and stearic acid (R2 = 0.970) metabolism with PAH uptake. Thus, increases in solid lipids suppress the uptake of PAHs by GO. The effects of glycerol on the uptake of PAHs are presented in Fig. 6a. pentanoic acid, arabinose and tartaric acid positively contributed to ROS levels (Fig. 8d). As shown in Fig. 8e, robust, positive linear correlations were observed for pentanoic acid (R2 = 0.830) and arabinose (R2 = 0.754) metabolism with ROS levels. The associations of pentanoic acid and arabinose with oxidative stress have not been previously reported. Galactose, malic acid and maltose negatively contributed to ROS levels (Fig. 8d). As shown in Fig. 8f, robust, negative linear correlations were observed for galactose (R2 = 0.794) and malic acid (R2 = 0.747) metabolism with ROS levels. Zinc oxide nanoparticles have been shown to induce oxidative stress through excessive galactose.84 Excessive malic acid triggers cell apoptosis and glucose oxidation.85 However, downregulation of galactose and malic acid are linked to greater oxidative stress upon GO and PAH exposure, supporting metabolic homeostasis during biological responses.Due to the emergence of GO and the similarity of its six-carbon rings with many PAHs, an understanding of the biological response to GO and PAH co-exposure is critical. However, information regarding GO and its mechanisms is lacking. We illustrated the effects of GO on the uptake pathways of PAHs and on the oxidative stress induced by PAHs. To explore the underlying detailed mechanisms in the abovementioned phenomena, proteomic screening was performed to elucidate the specific protein–protein interactions related to the uptake of PAHs and oxidative stress during co-exposure to GO and PAHs. A metabolomic approach was used together with OPLS-DA to screen for specific metabolites that contribute to the uptake of PAHs and the increase in ROS levels. The combination of a global protein and metabolic analysis and a targeted analysis of biological end points (e.g., uptake of PAHs and ROS) provided well-validated insight into the mechanisms of nanomaterial toxicity. In summary, this study illustrated the enhanced adverse effects of PAHs induced by GO in rice and explored the specific underlying molecular mechanisms, providing new implications for potential environmental risks of emergent GO.
Materials and methods
Characterization of GO
GO nanosheets (XF002-1) were purchased from Nanjing XFNANO Materials Tech Co., Ltd., China. The detailed methods used to characterize GO were described in our previous study.86 Briefly, the morphology of GO was examined using field emission transmission electron microscopy (TEM, JEM-2010 FEF, JEOL, Japan) and atomic force microscopy (AFM, Nanoscope IV, Veeco, USA). To identify the composition and phase of GO, the X-ray diffraction (XRD) pattern was examined using a Bruker D8 Advance X-ray diffractometer (Bruker, Germany) at 40 kV and 40 mA with Cu Kα radiation (k = 1.5406 Å). The 2θ angle ranged from 5° to 40° at a scanning rate of 4 min−1. X-ray photoelectron spectroscopy (XPS) was conducted using a PHI5000 Versa Probe with a monochromatic Al Kα X-ray source (1486.6 eV). The spectra were analyzed using Casa-XPS V2.3.13 software. The hydrodynamic diameters (Hds) and zeta potentials were determined by performing dynamic light scattering using a ZetaPALS instrument equipped with a 30 mW and 635 nm laser (BI-200SM, Brookhaven, USA).Cultivation of rice seedlings
Rice seeds (14ZJ02, Oryza sativa japonica) were obtained from the Tianjin Academy of Agricultural Sciences, Tianjin, China. PAHs (Z-014G) were purchased from AccuStandard Inc., USA. The rice seeds were sterilized using 10% H2O2 for 30 min. Subsequently, the seeds were completely rinsed with ultrapure water and then evenly sprinkled on trays with double-layer gauze. The culture trays were placed in an illumination incubator (LRH-250-Gb, China) at 25 °C in the light (566 μmol m−2 s−1) for 14 h and at 20 °C in the dark for 10 h. After a five day culture, the rice seedlings were transplanted into 50 mL glass flasks containing a quarter strength culture solution from the International Rice Research Institute formula and cultured for 3 days, and then, the rice seedlings were cultured with a half-strength culture solution for another 3 days to allow for adaptation to the growth environment. The components of the culture solution from the International Rice Research Institute formula are listed in Table S4.† Subsequently, the rice seedlings were exposed to GO or GO with 10 μg L−1 PAHs (PG) for 7 days. This duration (7 days) is frequently used in rice seedling experiments.87Graphene production is estimated to reach 1200 tons per annum by 2019, and GO plays an important role in the manufacture and applications of graphene-based nanomaterials due to its advantageous properties.88 According to the life-cycle of nanomaterials, GO is likely to have a similar fate as carbon nanotubes in which more than 80% of manufactured GO will be potentially released into the environment.89 Furthermore, there is great potential for GO exposure in environments where GO is manufactured and applied.90 GO concentrations of 0.01 mg L−1 to 100 mg L−1, or even higher have been used in previous studies that have explored its toxicological mechanisms.91–94 To analyze the environmental risks of GO centered exposure during its manufacture and applications and compared with the results of previous studies, GO concentrations of 0.01, 0.1 and 1.0 mg L−1 were tested in the present work.
As is well known, PAH pollution is prevalent in the environment owing to emissions from industrial activities and incomplete combustion of fuels. According to PAH concentrations in contaminated surface water,95 10 μg L−1 PAH was used in the present work. To explore the uptake pathways and given the detection limit of GC-MS, each PAH at 100 μg L−1 in water was used in the PAH uptake experiment. PAHs with large molecular weights and four or more benzene rings have been widely assessed in toxicological studies due to their high stability, high hydrophobicity and low biodegradability.96 Therefore, the present study investigated PAHs with four or more rings namely fluoranthene (Fla), pyrene (Pyr), benz(a)anthracene (Baa), chrysene (Chr), benzo(b)fluoranthene (BbF), benzo(k)fluoranthene (BkF), benzo(a)pyrene (BaP), indeno(1,2,3-cd)pyrene (InP), dibenz(a,h)anthracene (DbA) and benzo(g,h,i)perylene (BghiP).
Plant growth and oxidative stress
Plant roots are the main tissues exposed to nanoparticles in soil and water.97,98 The fresh roots and leaves of eight plants were accurately weighed with an electronic balance. Morphological alterations in the rice root tips were then observed under a light microscope (Olympus IX71, Japan). The rice seedling roots were washed with distilled water, and approximately 2 μm-long root tips were cut using a sharp stainless-steel knife. The root tips were placed on glass slides with a coverslip, and images were obtained under white light. The root width and color difference (representing the relative abundance of cytoplasm) in the root tips were analyzed using ImageJ 1.43. APX/AhR activities and CYP1A1/CYP1B1 contents were spectrophotometrically determined using BYE97079, BYE97168, BYE97170 and BYE97171 assay kits, respectively, according to the manufacturer's instructions (Shanghai Bangyi Biotechnology Co., Ltd., China). 2′,7′-Dichlorodihydrofluorescein diacetate (DCFH-DA) was used as a fluorescence probe to measure intracellular ROS. The seedling roots were washed ten times with phosphate-buffered saline (PBS), incubated with 10 μM DCFH-DA in the dark at 25 °C for 30 min and then immediately cut approximately 2 μm from the root tips. The root tips were placed on glass slides with coverslips and imaged under a microscope with an excitation wavelength of 485 nm and an emission wavelength of 535 nm (Olympus IX71, Japan).GO analysis and PAH detection
The rice seedling roots were washed with distilled water and cross-sectioned at 1.5 cm from the root tips using a sharp stainless-steel knife. The cross-sections were placed on glass slides, and the typical D and G bands of GO were analyzed using a Raman spectrometer (DXR Microscope, USA) with an excitation wavelength of 780 nm from a diode-pumped solid state (DPSS) laser. The seedling roots were rinsed repeatedly with methanol and then accurately weighed. The samples were homogenized by grinding and mixing with anhydrous Na2SO4 (mass ratio, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10) and then placed into stainless tubes with an accelerated solvent (E-916, BUCHI, Switzerland). Subsequently, phenanthrene-d10, chrysene-d12, and perylene-d12 were added as internal standards to account for possible losses during the extraction, and the samples were covered with a glass fiber membrane. The samples were extracted using dichloromethane/acetone (1
10) and then placed into stainless tubes with an accelerated solvent (E-916, BUCHI, Switzerland). Subsequently, phenanthrene-d10, chrysene-d12, and perylene-d12 were added as internal standards to account for possible losses during the extraction, and the samples were covered with a glass fiber membrane. The samples were extracted using dichloromethane/acetone (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) at 120 °C and 1500 psi for 15 min, followed by preheating for 5 min. The flush volume was 80%, and the purge time was 100 s. The samples were collected with matched glass bottles, and the extraction was repeated 3 times.99 The samples were concentrated to 2–3 mL using a rotary evaporator at 30 °C, purified with a silica gel column, eluted with 10 mL dichloromethane–hexane (volume ratio, 3
1, v/v) at 120 °C and 1500 psi for 15 min, followed by preheating for 5 min. The flush volume was 80%, and the purge time was 100 s. The samples were collected with matched glass bottles, and the extraction was repeated 3 times.99 The samples were concentrated to 2–3 mL using a rotary evaporator at 30 °C, purified with a silica gel column, eluted with 10 mL dichloromethane–hexane (volume ratio, 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7), dried via nitrogen blow-off and dissolved in 300 μL dichloromethane. The samples were analyzed using gas chromatography-mass spectrometry (GC-MS; 6890 N/5973, Agilent, USA) in selective ion monitoring (SIM) mode.100 The oven program was as follows: 70 °C for 1 min, 10 °C min−1 up to 260 °C and maintenance for 4 min, and 5 °C min−1 up to 300 °C and maintenance for 5 min.
7), dried via nitrogen blow-off and dissolved in 300 μL dichloromethane. The samples were analyzed using gas chromatography-mass spectrometry (GC-MS; 6890 N/5973, Agilent, USA) in selective ion monitoring (SIM) mode.100 The oven program was as follows: 70 °C for 1 min, 10 °C min−1 up to 260 °C and maintenance for 4 min, and 5 °C min−1 up to 300 °C and maintenance for 5 min.
Proteomic analysis
Proteins were extracted from the rice roots using 8 M urea, and 100 μg protein was reduced by adding 5 mM dithiothreitol for 0.5 h at 56 °C and alkylated by adding 20 mM iodoacetamide for 0.5 h at room temperature in the dark. Then, 5 mM dithiothreitol was added, and the samples were kept in the dark for 15 min. The protein samples were finally transferred to a 10 kDa filter unit, washed with 8 M urea and 50 mM ammonium bicarbonate twice, and digested with trypsin at a mass ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 enzyme/protein overnight at 37 °C. The reaction was stopped by adding 1% formic acid. Liquid chromatography-tandem mass spectrometry (LC-MS/MS) was performed using a Thermo Scientific Easy-nLC 1000 liquid chromatography system coupled to a Q-Exactive Plus via a nanoelectrospray ion source (Thermo, USA). The spectral data were searched against the rice RefSeq database in Proteome Discoverer 1.4.1.14 using Mascot (version 2.3.01, Matrix Science) to achieve a false discovery rate <1%.
50 enzyme/protein overnight at 37 °C. The reaction was stopped by adding 1% formic acid. Liquid chromatography-tandem mass spectrometry (LC-MS/MS) was performed using a Thermo Scientific Easy-nLC 1000 liquid chromatography system coupled to a Q-Exactive Plus via a nanoelectrospray ion source (Thermo, USA). The spectral data were searched against the rice RefSeq database in Proteome Discoverer 1.4.1.14 using Mascot (version 2.3.01, Matrix Science) to achieve a false discovery rate <1%.
Metabolic analysis
Plant roots are considered the main route of plant exposure to nanoparticles in soil or water matrices and may harbor a major amount of accumulated nanoparticles, leading to physical or chemical toxicity to the plants.101 To analyze the metabolism in the rice roots, the rice seedling culture was terminated on the seventh day using liquid nitrogen, and the seedling roots were collected using a sharp stainless-steel knife. The samples were homogenized by grinding with 4.5 mL of cold methanol–chloroform–ultrapure water (volume ratio, 2.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) solution. The homogenate was extracted via ultrasound (300 W, 1 h) in an ice-water bath, followed by centrifugation at 9000g for 10 min to collect the supernatant. Subsequently, the pellets were resuspended in 4 mL of cold methanol–chloroform–ultrapure water (volume ratio, 2.5
1) solution. The homogenate was extracted via ultrasound (300 W, 1 h) in an ice-water bath, followed by centrifugation at 9000g for 10 min to collect the supernatant. Subsequently, the pellets were resuspended in 4 mL of cold methanol–chloroform–ultrapure water (volume ratio, 2.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) solution and extracted using ultrasound for 30 min at 300 W. The suspension was centrifuged at 9000g for 10 min again and mixed with the first supernatant, and then, ultrapure water (500 μL) was added, followed by centrifugation at 9000g for 5 min. Methanol and water were removed from the upper phase via nitrogen blow-off and vacuum freeze-drying, respectively. The lower phase of chloroform was also removed via nitrogen blow-off. O-Methyl amine hydrochloride (20 mg mL−1, 50 μL) and N-methyl-N-(trimethylsilyl) trifluoroacetamide (80 μL) were used as derivatives. The samples were analyzed using GC-MS (6890A/5977A, Agilent, USA). The oven program was as follows: 80 °C for 2 min, 15 °C min−1 up to 325 °C and then maintenance for 6 min. The mass spectrometer was operated in full scan mode from 80 to 800 amu.
1) solution and extracted using ultrasound for 30 min at 300 W. The suspension was centrifuged at 9000g for 10 min again and mixed with the first supernatant, and then, ultrapure water (500 μL) was added, followed by centrifugation at 9000g for 5 min. Methanol and water were removed from the upper phase via nitrogen blow-off and vacuum freeze-drying, respectively. The lower phase of chloroform was also removed via nitrogen blow-off. O-Methyl amine hydrochloride (20 mg mL−1, 50 μL) and N-methyl-N-(trimethylsilyl) trifluoroacetamide (80 μL) were used as derivatives. The samples were analyzed using GC-MS (6890A/5977A, Agilent, USA). The oven program was as follows: 80 °C for 2 min, 15 °C min−1 up to 325 °C and then maintenance for 6 min. The mass spectrometer was operated in full scan mode from 80 to 800 amu.
Statistical analysis
All treatments included at least three replicates, and the error bars represent the standard deviation (mean ± SD). The data were analyzed using one-way ANOVA and compared using post hoc Tukey's test with SPSS 21.0. A p-value < 0.05 indicated statistical significance. The proteomic data were clustered using DAVID bioinformatics resources 6.8, and the interaction network was achieved using STRING and Cytoscape 3.3.0.Conflicts of interest
The authors declare no competing financial interests.Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (grant nos. 21407085, 21577070, 21722703 and 31770550), the Tianjin Natural Science Foundation (grant no. 16JCQNJC08400), the Central Public Research Institutes Basic Funds for Research and Development (Agro-Environmental Protection Institute, Ministry of Agriculture), the Fundamental Research Funds for the Central Universities, the Hundred Young Academic Leaders Training Program and 111 program, Ministry of Education, China (T2017002).References
- I. Iavicoli, V. Leso, D. H. Beezhold and A. A. Shvedova, Toxicol. Appl. Pharmacol., 2017, 329, 96–111 CrossRef CAS PubMed.
- A. Naumov, F. Grote, M. Overgaard, A. Roth, C. E. Halbig, K. Nørgaard, D. M. Guldi and S. Eigler, J. Am. Chem. Soc., 2016, 138, 11445–11448 CrossRef CAS PubMed.
- R. Spitz Steinberg, M. Cruz, N. G. Mahfouz, Y. Qiu and R. H. Hurt, ACS Nano, 2017, 11, 5670–5679 CrossRef CAS PubMed.
- H. Y. Mao, S. Laurent, W. Chen, O. Akhavan, M. Imani, A. A. Ashkarran and M. Mahmoudi, Chem. Rev., 2013, 113, 3407–3424 CrossRef CAS PubMed.
- M. Zhang, B. Gao, J. Chen, Y. Li, A. E. Creamer and H. Chen, Chem. Eng. J., 2014, 255, 107–113 CrossRef CAS.
- S. Sharma, S. Singh, A. K. Ganguli and V. Shanmugam, Carbon, 2017, 115, 781–790 CrossRef CAS.
- Y. Zhao, L. Zhi, Q. Wu, Y. Yu, Q. Sun and D. Wang, Nanotoxicology, 2016, 10, 1469–1479 CrossRef CAS PubMed.
- L. Zhi, M. Qu, M. Ren, L. Zhao, Y. Li and D. Wang, Carbon, 2017, 113, 122–131 CrossRef CAS.
- C. M. Park, K. H. Chu, J. Heo, N. Her, M. Jang, A. Son and Y. Yoon, J. Hazard. Mater., 2016, 309, 133–150 CrossRef CAS PubMed.
- B. Glomstad, D. Altin, L. Sorensen, J. F. Liu, B. M. Jenssen and A. M. Booth, Environ. Sci. Technol., 2016, 50, 2660–2668 CrossRef CAS PubMed.
- X. Hu, J. Kang, K. Lu, R. Zhou, L. Mu and Q. Zhou, Sci. Rep., 2014, 4, 6122 CrossRef CAS PubMed.
- Y. M. Chen, X. G. Hu, J. Sun and Q. X. Zhou, Nanotoxicology, 2016, 10, 42–52 CAS.
- O. Akhavan, E. Ghaderi, H. Emamy and F. Akhavan, Carbon, 2013, 54, 419–431 CrossRef CAS.
- P. Begum, R. Ikhtiari and B. Fugetsu, Carbon, 2011, 49, 3907–3919 CrossRef CAS.
- M. Qu, Y. Li, Q. Wu, Y. Xia and D. Wang, Nanotoxicology, 2017, 11, 520–533 CrossRef CAS PubMed.
- O. Akhavan, E. Ghaderi, S. A. Shirazian and R. Rahighi, Carbon, 2016, 97, 71–77 CrossRef CAS.
- Y. Wang, C. H. Chang, Z. Ji, D. C. Bouchard, R. M. Nisbet, J. P. Schimel, J. L. Gardea-Torresdey and P. A. Holden, ACS Nano, 2017, 11, 5753–5765 CrossRef CAS PubMed.
- N. Zuverza-Mena, D. Martinez-Fernandez, W. Du, J. A. Hernandez-Viezcas, N. Bonilla-Bird, M. L. Lopez-Moreno, M. Komarek, J. R. Peralta-Videa and J. L. Gardea-Torresdey, Plant Physiol. Biochem., 2017, 110, 236–264 CrossRef CAS PubMed.
- A. Cox, P. Venkatachalam, S. Sahi and N. Sharma, Plant Physiol. Biochem., 2016, 107, 147–163 CrossRef CAS PubMed.
- J. I. Kwak and Y.-J. An, Environ. Res., 2016, 151, 368–382 CrossRef CAS PubMed.
- X. G. Hu, D. D. Li, Y. Gao, L. Mu and Q. X. Zhou, Environ. Int., 2016, 94, 8–23 CrossRef CAS PubMed.
- A. Bianco, Angew. Chem., Int. Ed., 2013, 52, 4986–4997 CrossRef CAS PubMed.
- R. De La Torre-Roche, J. Hawthorne, Y. Q. Deng, B. S. Xing, W. J. Cai, L. A. Newman, C. Wang, X. M. Ma and J. C. White, Environ. Sci. Technol., 2012, 46, 9315–9323 CrossRef CAS PubMed.
- R. De La Torre-Roche, J. Hawthorne, Y. Deng, B. Xing, W. Cai, L. A. Newman, Q. Wang, X. Ma, H. Hamdi and J. C. White, Environ. Sci. Technol., 2013, 47, 12539–12547 CrossRef CAS PubMed.
- F. Schwab, T. D. Bucheli, L. Camenzuli, A. Magrez, K. Knauer, L. Sigg and B. Nowack, Environ. Sci. Technol., 2013, 47, 7012–7019 CrossRef CAS PubMed.
- W. Zou, X. Li, Z. Lai, X. Zhang, X. Hu and Q. Zhou, ACS Appl. Mater. Interfaces, 2016, 8, 33165–33174 CAS.
- I. L. Gunsolus and C. L. Haynes, Anal. Chem., 2015, 88, 451–479 CrossRef PubMed.
- B. Dalzon, C. Aude-Garcia, V. Collin-Faure, H. Diemer, D. Béal, F. Dussert, D. Fenel, G. Schoehn, S. Cianférani and M. Carrière, Nanoscale, 2017, 9, 9641–9658 RSC.
- L. Chupani, E. Zusková, H. Niksirat, A. Panáček, V. Lünsmann, S.-B. Haange, M. von Bergen and N. Jehmlich, Sci. Total Environ., 2017, 579, 1504–1511 CrossRef CAS PubMed.
- X. Hu, K. Lu, L. Mu, J. Kang and Q. Zhou, Carbon, 2014, 80, 665–676 CrossRef CAS.
- V. R. Patel, K. Eckel-Mahan, P. Sassone-Corsi and P. Baldi, Nat. Methods, 2012, 9, 772–773 CrossRef CAS PubMed.
- G. E. Bragin, T. F. Parkerton, A. D. Redman, D. J. Letinksi, J. D. Butler, M. L. Paumen, C. A. Sutherland, T. M. Knarr, M. Comber and K. den Haan, Environ. Toxicol. Chem., 2016, 35, 2948–2957 CrossRef CAS PubMed.
- F. A. Bezza and E. M. N. Chirwa, Chem. Eng. J., 2017, 309, 563–576 CrossRef CAS.
- J. Feng, X. Li, J. Zhao and J. Sun, Environ. Sci. Pollut. Res., 2017, 24, 18195–18203 CrossRef CAS PubMed.
- M. X. Liu, Y. Y. Yang, X. Y. Yun, M. M. Zhang and J. Wang, J. Soil Water Conserv., 2016, 71, 327–334 CrossRef.
- G. Liu, W. Guo, J. Niu, X. An and L. Zhao, J. Soils Sediments, 2017, 17, 229–239 CrossRef CAS.
- J. Church, X. Wang, J. Calderon, W. H. Lee, H. J. Cho and L. Zhai, J. Nanosci. Nanotechnol., 2016, 16, 1620–1623 CrossRef CAS PubMed.
- S. Mahpishanian, H. Sereshti and M. Ahmadvand, J. Environ. Sci., 2017, 55, 164–173 CrossRef PubMed.
- Z. Zahra, N. Waseem, R. Zahra, H. Lee, M. A. Badshah, A. Mehmood, H.-K. Choi and M. Arshad, J. Agric. Food Chem., 2017, 65, 5598–5606 CrossRef CAS PubMed.
- C. M. Rico, S. Majumdar, M. Duarte-Gardea, J. R. Peralta-Videa and J. L. Gardea-Torresdey, J. Agric. Food Chem., 2011, 59, 3485–3498 CrossRef CAS PubMed.
- Y. Li, Q. Wu, Y. Zhao, Y. Bai, P. Chen, T. Xia and D. Wang, ACS Nano, 2014, 8, 2100–2110 CrossRef CAS PubMed.
- E. Hashemi, O. Akhavan, M. Shamsara, M. Daliri, M. Dashtizad and A. Farmany, Colloids Surf., B, 2016, 146, 770–776 CrossRef CAS PubMed.
- L. Hu, P. Jiang, P. Zhang, G. Bian, S. Sheng, M. Huang, Y. Bao and J. Xia, J. Mater. Sci., 2016, 51, 8296–8309 CrossRef CAS.
- M. Kryuchkova, A. Danilushkina, Y. Lvov and R. Fakhrullin, Environ. Sci.: Nano, 2016, 3, 442–452 RSC.
- A. D. Servin and J. C. White, NanoImpact, 2016, 1, 9–12 CrossRef.
- K. N. Kudin, B. Ozbas, H. C. Schniepp, R. K. Prud'Homme, I. A. Aksay and R. Car, Nano Lett., 2008, 8, 36–41 CrossRef CAS PubMed.
- M. H. Lahiani, E. Dervishi, I. Ivanov, J. Chen and M. Khodakovskaya, Nanotechnology, 2016, 27, 265102 CrossRef PubMed.
- X. Hu, W. Kang and L. Mu, Environ. Sci. Technol., 2017, 51, 5425–5433 CrossRef CAS PubMed.
- S. H. Habib, H. Kausar and H. M. Saud, BioMed Res. Int., 2016, 2016, 1–10 CrossRef PubMed.
- R. J. Wall, A. Fernandes, M. Rose, D. R. Bell and I. R. Mellor, Environ. Int., 2015, 76, 49–56 CrossRef CAS PubMed.
- S. Tabrez and M. Ahmad, Environ. Monit. Assess., 2013, 185, 2977–2987 CrossRef CAS PubMed.
- Y. Landkocz, F. Ledoux, V. André, F. Cazier, P. Genevray, D. Dewaele, P. J. Martin, C. Lepers, A. Verdin and L. Courcot, Environ. Pollut., 2017, 221, 130–140 CrossRef CAS PubMed.
- A. E. Pradas del Real, V. Vidal, M. Carrière, H. Castillo-Michel, C. M. Levard, P. Chaurand and G. Sarret, Environ. Sci. Technol., 2017, 51, 5774–5782 CrossRef CAS PubMed.
- C. Hu, B. K. Ham, H. M. El-shabrawi, D. Alexander, D. Zhang, J. Ryals and W. J. Lucas, Plant J., 2016, 87, 442–454 CrossRef CAS PubMed.
- M. Moore, N. Gossmann and K.-J. Dietz, Trends Plant Sci., 2016, 21, 388–397 CrossRef CAS PubMed.
- N. Suzuki, S. Koussevitzky, R. Mittler and G. Miller, Plant, Cell Environ., 2012, 35, 259–270 CrossRef CAS PubMed.
- S. Gioria, J. L. Vicente, P. Barboro, R. La Spina, G. Tomasi, P. Urban, A. Kinsner-Ovaskainen, R. Francois and H. Chassaigne, Nanotoxicology, 2016, 10, 736–748 CrossRef CAS PubMed.
- L. Wang, J. Fu, M. Li, L. Fragner, W. Weckwerth and P. Yang, Front. Plant Sci., 2016, 7, 750 Search PubMed.
- S. A. Brown, J. R. McKelvie, A. J. Simpson and M. J. Simpson, Environ. Pollut., 2010, 158, 2117–2123 CrossRef CAS PubMed.
- S. A. Brown, A. J. Simpson and M. J. Simpson, Environ. Chem., 2009, 6, 432–440 CrossRef CAS.
- K. Hardonnière, E. Saunier, A. Lemarié, M. Fernier, I. Gallais, C. Helies-Toussaint, B. Mograbi, S. Antonio, P. Bénit and P. Rustin, Sci. Rep., 2016, 6, 30776 CrossRef PubMed.
- C. Filippi, A. Pryde, P. Cowan, T. Lee, P. Hayes, K. Donaldson, J. Plevris and V. Stone, Nanotoxicology, 2015, 9, 126–134 CrossRef CAS PubMed.
- Z.-X. Chen, H.-G. Ni, X. Jing, W.-J. Chang, J.-L. Sun and H. Zeng, Chemosphere, 2015, 131, 192–200 CrossRef CAS PubMed.
- X. Zhan, X. Zhang, X. Yin, H. Ma, J. Liang, L. Zhou, T. Jiang and G. Xu, J. Environ. Qual., 2012, 41, 188–196 CrossRef CAS PubMed.
- Y. Liang, J. Si and V. Römheld, New Phytol., 2005, 167, 797–804 CrossRef CAS PubMed.
- F. J. Zhao, J. F. Ma, A. A. Meharg and S. P. McGrath, New Phytol., 2009, 181, 777–794 CrossRef CAS PubMed.
- C. A. G. Quispe, C. J. R. Coronado and J. A. Carvalho Jr, Renewable Sustainable Energy Rev., 2013, 27, 475–493 CrossRef CAS.
- A. C. Barnes, C. Benning and R. L. Roston, Plant Physiol., 2016, 171, 2140–2149 CrossRef CAS PubMed.
- S. Deng, J. Sun, R. Zhao, M. Ding, Y. Zhang, Y. Sun, W. Wang, Y. Tan, D. Liu, X. Ma, P. Hou, M. Wang, C. Lu, X. Shen and S. Chen, Plant Physiol., 2015, 169, 530–548 CrossRef CAS PubMed.
- A. Bellettre, J. P. Couillerot, A. S. Blervacq, S. Aubert, E. Gout, J. L. Hilbert and J. Vasseur, Plant Physiol. Biochem., 2001, 39, 503–511 CrossRef CAS.
- O. Postaire, C. Tournaire-Roux, A. Grondin, Y. Boursiac, R. Morillon, A. R. Schaffner and C. Maurel, Plant Physiol., 2010, 152, 1418–1430 CrossRef CAS PubMed.
- F. Chaumont and S. D. Tyerman, Plant Physiol., 2014, 164, 1600–1618 CrossRef CAS PubMed.
- M. J. Hynes and S. L. Murray, Eukaryotic Cell, 2010, 9, 1039–1048 CrossRef CAS PubMed.
- H. Hosseinzadeh, S. Mehri, A. Heshmati, M. Ramezani, A. Sahebkar and K. Abnous, J. Pharm. Sci., 2014, 22, 5 Search PubMed.
- T. K. Kundu, M. Velayutham and J. L. Zweier, Biochemistry, 2012, 51, 2930–2939 CrossRef CAS PubMed.
- M. Wang, Y. Jia, Z. Xu and Z. Xia, Front. Plant Sci., 2016, 71843 Search PubMed.
- A. Perkins, K. J. Nelson, D. Parsonage, L. B. Poole and P. A. Karplus, Trends Biochem. Sci., 2015, 40, 435–445 CrossRef CAS PubMed.
- F. J. Corpas and J. B. Barroso, Front. Environ. Sci., 2014, 2, 55 Search PubMed.
- L. Zhou, H. Xu, S. Mischke, L. W. Meinhardt, D. Zhang, X. Zhu, X. Li and W. Fang, Hortic. Res., 2014, 1, 14029 CrossRef PubMed.
- M. Chen, S. Mishra, S. A. Heckathorn, J. M. Frantz and C. Krause, J. Plant Physiol., 2014, 171, 235–242 CrossRef CAS PubMed.
- R. Li, W. Wang, W. Wang, F. Li, Q. Wang, Y. Xu and S. Wang, Electron. J. Biotechnol., 2015, 18, 368–375 CrossRef CAS.
- I. Cummins, D. J. Wortley, F. Sabbadin, Z. He, C. R. Coxon, H. E. Straker, J. D. Sellars, K. Knight, L. Edwards, D. Hughes, S. S. Kaundun, S.-J. Hutchings, P. G. Steel and R. Edwards, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 5812–5817 CrossRef CAS PubMed.
- A. Patwa, A. Thiéry, F. Lombard, M. K. Lilley, C. Boisset, J.-F. Bramard, J.-Y. Bottero and P. Barthélémy, Sci. Rep., 2015, 5, 11387 CrossRef PubMed.
- Y. Wei, Y. Li, J. Jia, Y. Jiang, B. Zhao, Q. Zhang and B. Yan, NanoImpact, 2016, 3, 1–11 CrossRef.
- Y.-P. Hsiao, W.-W. Lai, S.-B. Wu, C.-H. Tsai, S.-C. Tang, J.-G. Chung and J.-H. Yang, Toxins, 2015, 7, 81–96 CrossRef PubMed.
- X. Hu, Y. Gao and Z. Fang, Carbon, 2016, 109, 65–73 CrossRef CAS.
- G. Dixit, A. P. Singh, A. Kumar, S. Dwivedi, F. Deeba, S. Kumar, S. Suman, B. Adhikari, Y. Shukla, P. K. Trivedi, V. Pandey and R. D. Tripathi, Sci. Rep., 2015, 5, 16205 CrossRef CAS PubMed.
- A. Zurutuza and C. Marinelli, Nat. Nanotechnol., 2014, 9, 730–734 CrossRef CAS PubMed.
- J. D. Lanphere, B. Rogers, C. Luth, C. H. Bolster and S. L. Walker, Environ. Eng. Sci., 2014, 31, 350–359 CrossRef CAS PubMed.
- J. Zhao, Z. Wang, J. C. White and B. Xing, Environ. Sci. Technol., 2014, 48, 9995–10009 CrossRef CAS PubMed.
- L. Chen, C. Wang, H. Li, X. Qu, S.-T. Yang and X.-L. Chang, Environ. Sci. Technol., 2017, 51, 10146–10153 CrossRef CAS PubMed.
- C. Hu, Q. Wang, H. Zhao, L. Wang, S. Guo and X. Li, Chemosphere, 2015, 128, 184–190 CrossRef CAS PubMed.
- P. F. M. Nogueira, D. Nakabayashi and V. Zucolotto, Aquat. Toxicol., 2015, 166, 29–35 CrossRef CAS PubMed.
- S. Ouyang, X. Hu and Q. Zhou, ACS Appl. Mater. Interfaces, 2015, 7, 18104–18112 CAS.
- P. U. Singare, Environ. Monit. Assess., 2016, 188, 599 CrossRef PubMed.
- I. Shirdel, M. R. Kalbassi, M. Shokri, R. Olyaei and I. Sharifpour, Ecotoxicol. Environ. Saf., 2016, 130, 207–213 CrossRef CAS PubMed.
- M. Ashfaq, N. Verma and S. Khan, Environ. Sci.: Nano, 2017, 4, 138–148 RSC.
- I. Jośko, P. Oleszczuk and E. Skwarek, J. Hazard. Mater., 2017, 331, 200–209 CrossRef PubMed.
- H. Yin, Q. Tan, Y. Chen, G. B. Lv and X. D. Hou, Microchem. J., 2011, 97, 138–143 CrossRef CAS.
- K. E. Ugwu and P. O. Ukoha, Environ. Geochem. Health, 2017, 1–15 Search PubMed.
- N. A. Anjum, S. S. Gill, A. C. Duarte, E. Pereira and I. Ahmad, J. Nanopart. Res., 2013, 15, 1896 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available: Tables S1–S4 and Fig. S1–S8. See DOI: 10.1039/c7en00803a |
| This journal is © The Royal Society of Chemistry 2018 |