Open Access Article
Open Access ArticleGeneration of a functional, semisynthetic [FeFe]-hydrogenase in a photosynthetic microorganism†
Adam
Wegelius‡
a,
Namita
Khanna‡§
a,
Charlène
Esmieu
b,
Giovanni Davide
Barone
a,
Filipe
Pinto¶
cd,
Paula
Tamagnini
cd,
Gustav
Berggren
 *b and
Peter
Lindblad
*b and
Peter
Lindblad
 *a
*a
aMicrobial Chemistry, Department of Chemistry-Ångström, Uppsala University, Box 523, SE-751 20 Uppsala, Sweden. E-mail: Peter.Lindblad@kemi.uu.se
bMolecular Biomimetics, Department of Chemistry-Ångström, Uppsala University, Box 523, SE-751 20 Uppsala, Sweden. E-mail: Gustav.Berggren@kemi.uu.se
ci3S – Instituto de Investigação e Inovação em Saúde, IBMC – Instituto de Biologia Molecular e Celular, Universidade do Porto, 4200-135 Porto, Portugal
dFaculdade de Ciências, Departamento de Biologia, Universidade do Porto, 4169-007 Porto, Portugal
First published on 25th September 2018
Abstract
[FeFe]-Hydrogenases are hydrogen producing metalloenzymes with excellent catalytic capacities, highly relevant in the context of a future hydrogen economy. Here we demonstrate the synthetic activation of a heterologously expressed [FeFe]-hydrogenase in living cells of Synechocystis PCC 6803, a photoautotrophic microbial chassis with high potential for biotechnological energy applications. H2-Evolution assays clearly show that the non-native, semi-synthetic enzyme links to the native metabolism in living cells.
Broader context[FeFe]-Hydrogenases are metalloenzymes known for their spectacular performance in hydrogen production. In natural systems the synthesis of the active site containing a unique [2Fe]-subcluster is dependent on a complicated enzymatic maturation machinery. Recently, a synthetic maturation procedure was successfully employed for the first time in vivo in the heterotrophic bacteria Escherichia coli. Here we demonstrate the synthetic in vivo activation of a heterologously expressed [FeFe]-hydrogenase in a unicellular cyanobacterium and describe how the non-native, semi-synthetic enzyme links to the native metabolism in a living photosynthetic cell. Our model organism, Synechocystis PCC 6803, represents a photoautotrophic microbial chassis with high potential for biotechnological energy applications. The activated hydrogenase evolves hydrogen both in light and in darkness with an activity directly linked to the metabolic status of the cell. The procedure reported in the present work opens up unique possibilities to investigate not only [FeFe]-hydrogenases but also other metalloenzymes in a photosynthetic microbial cell environment, completely bypassing the many challenges of e.g. biological maturation and regulations. |
Cyanobacteria, photosynthetic microbial chassis with high potential for biotechnological applications, are considered strong candidates for photobiological fuel and chemical production.1 Almost all cyanobacteria contain an inherent capacity to produce hydrogen gas catalyzed by a nitrogenase and/or a bi-directional hydrogenase.2 Wild type (WT) cells of Synechocystis harbor a single, bidirectional, oxygen sensitive [NiFe]-hydrogenase encoded by the hox-genes. Hydrogen production occurs both under dark, fermentative conditions and, in a short burst, during the transition from darkness to light.3 In darkness, the hydrogen production is dependent on external or internal fermentation substrate(s).4 In light, the native hydrogenase can utilize electrons from PSII-catalyzed water splitting and/or fermentative sources, although it is rapidly inactivated by oxygen generated by PSII.3,4 However, neither nitrogenase nor [NiFe]-hydrogenase can be considered Nature's premier H2 production catalysts. Indeed, the [FeFe]-hydrogenases (HydA), found in some eukaryotes and in prokaryotes outside the cyanobacteria phylum, are known to have catalytic activities that vastly outperform any hydrogen producing enzyme naturally occurring in cyanobacteria.2 Moreover, the [FeFe]-hydrogenases are also much less prone to catalyse the oxidation of H2 to protons and electrons.2 Thus, the introduction of such an [FeFe]-hydrogenase is expected to result in significant enhancement in H2 production from the cyanobacterial host.
The heterologous expression of HydA is complicated by the requirement for efficient co-expression of its dedicated maturation machinery.5 At the core of the enzyme sits the unique H-cluster, responsible for the remarkable catalytic activity. This cofactor consists of a [4Fe–4S] cluster connected to a [2Fe] subcluster via a cysteine residue.6 In nature, biosynthesis of the [2Fe] subcluster requires three hydrogenase specific maturases, HydE, HydF and HydG.7 HydEFG assemble the subcluster and transfer it to the unmatured HydA protein, already containing the [4Fe–4S] cluster inserted by the universal iron–sulfur cluster synthesis machinery.8
Co-expression of HydA and the three maturases in Escherichia coli (E. coli) results in an active enzyme and subsequent hydrogen production.7
In recent years, it was discovered that maturation of HydA can be readily accomplished even without the involvement of the HydEFG machinery. This was first demonstrated for purified, unmatured HydA which was shown to form an active enzyme by addition of synthetic analogues of the [2Fe] subcluster.9,10 For a few years, this technique was limited to in vitro conditions with purified HydA. However, recently artificial maturation of HydA was demonstrated in vivo in living cells of E. coli.11–13 In the present work, we employ, for the first time, this technique beyond E. coli, and synthetically activate heterologously expressed HydA in living cells of the unicellular cyanobacterium Synechocystis PCC 6803. Moreover, we demonstrate that this artificially maturated, “semi-synthetic”, HydA links to the native metabolism of the cells and is active both in light conditions and in darkness. To our knowledge, this is the first example of synthetic activation of a metalloenzyme within any photosynthetic microorganism. The ability to artificially mature hydrogenases in living cyanobacterial cells is an important step forward in hydrogen based bioenergy research. It will facilitate screening photosynthetic hosts and enable straightforward investigations of enzymes with non-native [2Fe]-subclusters and/or amino acid sequences in a photosynthetic environment. This will be an important tool in the search for efficient catalysts for photo-biohydrogen production and future renewable energy systems.
We expressed the [FeFe]-hydrogenase HydA1 from the green algae Chlamydomonas reinhardtii (CrHydA1) in WT and a hydrogenase deficient mutant (ΔhoxYH) of Synechocystis PCC 6803 (Synechocystis WT and Synechocystis Δhox, respectively) using a broad host range shuttle vector. To reliably express the hydA1-gene in the cyanobacterial host strains, we used the Ptrccore promoter together with a bicistronic design adapter (supplementary Note 1, ESI†). The employment of the engineered, hydrogenase free, host strain Synechocystis Δhox offers a unique opportunity to elucidate the activity of the heterologous hydrogenase without interference from any native hydrogen metabolism.
When an anaerobic culture of Synechocystis Δhox CrHydA1 was incubated with 100 μg (156 nmol) of synthetic [2Fe] subcluster mimic [Fe2(adt)(CN)2(CO)4]2− (complex 1, adt = –SCH2NHCH2S–)14 significant hydrogen accumulation was observed after 48 h incubation in darkness with glucose-supplemented growth medium (Fig. 1). Cells of Synechocystis Δhox CrHydA1 not receiving complex 1, but otherwise handled identically, did not reveal any detectable hydrogen production. Likewise, Synechocystis Δhox without the unmatured HydA did not exhibit hydrogen production when treated with complex 1 (data not shown). Together this demonstrates the necessity of having both the heterologously expressed unmatured HydA and the synthetic cofactor present for proton reduction to occur. Under the same conditions, hydrogen production from Synechocystis WT CrHydA1 increased upon addition of complex 1 (Fig. 1). It is thus apparent that the native hydrogen production can be increased by addition of a second hydrogen producing enzyme to the biological system. Moreover, it is noteworthy that hydrogen production was significantly higher in Synechocystis Δhox cells containing an active CrHydA1 than in WT cells, underscoring the possibility to increase Synechocystis hydrogen productivity through enzyme optimization. However, the combined hydrogen production from the native NiFe-hydrogenase and the activated HydA did not reach a higher level than observed in cells of Synechocystis Δhox CrHydA1 with activated HydA which may indicate an underlying limitation of substrate under the experimental conditions employed. Time profiles of the hydrogen production (Fig. S1, ESI†) revealed that all the observed hydrogen was produced already within 24 h of incubation, further strengthening this remark.
We further assayed the hydrogen production from the semi-synthetic CrHydA1 enzyme expressed in Synechocystis Δhox cells under altered growth conditions. After treatment with complex 1 under anaerobic conditions, cells were incubated in darkness or in light (100 μmol of photons m−2 s−1 with DCMU (3-(3,4-dichlorophenyl)-1,1-dimethylurea)) in media with or without nitrate (BG11 and BG110, respectively) and/or glucose, giving rise to 8 different environmental conditions. After 48 h, accumulated hydrogen was measured (Fig. 2a and c). Activations in BG110 were done with cells already deprived of nitrate for 12 h, showing clear signs of on-going chlorosis. Vast differences in hydrogen production became apparent in the different environmental conditions examined, and most notable was the difference between light with DCMU (light + DCMU) (Fig. 2a) and darkness (Fig. 2c). The highest recorded accumulation of hydrogen in darkness (194 ± 15 nmol per OD per mL, obtained without glucose and nitrate) was almost 20 times higher than the highest accumulation in light (10 ± 2 nmol per OD per mL, obtained in nitrate free media supplemented with glucose). Presence of DCMU in the growth medium had no effect on the hydrogen accumulation under dark conditions (data not shown). Cells incubated in light + DCMU and BG11 without glucose showed low, but detectable (below 1 nmol per OD per mL), levels of hydrogen accumulation, while it was undetectable in darkness. The considerable differences in accumulated hydrogen, depending on medium composition and light regime, clearly indicate that the activated hydrogenase is operating in vivo and is coupled to the metabolic status of the living cells. Exactly how the complex enters the cell to activate the hydrogenase is not known. The anionic nature of the subcluster mimic may suggest an involvement of transmembrane transporters.
Nitrate depleted media consistently gave higher hydrogen production than corresponding condition with nitrate (Fig. 2a and c). This is in line with what has previously been reported for native hydrogen production in Synechocystis WT3 and is a consequence of slowing down the reductant-demanding process of nitrate assimilation, thereby making the reduction capacity available for other cellular processes.15 It is interesting to note how the heterologous, synthetically activated [FeFe]-hydrogenase is able to connect with the cell metabolism and respond to cellular changes in the redox balance similarly to the [NiFe]-hydrogenase in Synechocystis WT. Considering the effect of glucose, it is apparent that the hydrogen production is glucose dependent when nitrate is present in the media. The effect of glucose under nitrogen deprived conditions is more complicated. While clearly showing a positive effect on hydrogen accumulation in light + DCMU (Fig. 2a), the addition of glucose in darkness had a negative effect (Fig. 2c), decreasing the accumulated hydrogen to about one fourth. Synechocystis is known to accumulate storage compounds, like PHB (poly[(R)-3-hydroxybutyrate]) upon nitrogen starvation,16,17 and addition of external carbon sources can enhance this accumulation.18,19 We speculate that the decrease in hydrogen production upon addition of glucose results from the enabling of anabolic cell processes, making the cell environment less reducing compared to when both nitrate and glucose are absent. Under such conditions the cells have to survive exclusively by catabolism of internal compounds. The exact nature of the response to glucose in our highly engineered, semi-synthetic system under these particular conditions clearly needs further investigations.
Time profiles for the hydrogen production of nitrate deprived cells (Fig. 2b and d) shows no significant hydrogen production after 24 h in BG110 light + DCMU. This is also the case for cells in BG110 in darkness when glucose is provided and in BG11 with glucose in both light and darkness (Fig. S2, ESI†). In contrast, when neither nitrate nor glucose is present in darkness, hydrogen production does not plateau but seemingly continues for the duration of the experiment (Fig. 2d), the only condition in this study where significant hydrogen production could be detected beyond 24 h. Compared to the reported activity from semi-synthetic CrHydA1 in E. coli11 where hydrogen production ceased after 3–4 h, the lifetime of the hydrogen production by the synthetically activated, heterologously expressed CrHydA1 in Synechocystis is remarkable. Hydrogen production beyond 24 h is statistically verified, and this again signifies the interdependence of and direct link to cellular metabolism as well as the long-term supply of intrinsic reductant power to the activated non-native hydrogenase.
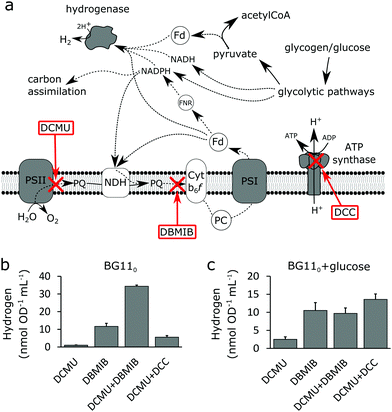
To further experimentally address the electron flow resulting in hydrogen production under the environmental conditions examined, specific inhibitors (schematically outlined in Fig. 3a) were used. The low hydrogen accumulation in light + DCMU, compared to dark conditions, may indicate a competitive relationship with the cyclic electron flow around PSI. Indeed, when DBMIB (2,5-dibromo-3-methyl-6-isopropyl-1,4-benzoquinone), known to inhibit the electron transfer from PQ to cytochrome b6f, was added instead of, or in addition to, DCMU in nitrogen deprived light conditions without glucose, a substantial increase in accumulated hydrogen after 48 h was observed (Fig. 3b). This was 34 times higher when DBMIB was added in addition to DCMU and 11 times higher when only DBMIB was used. Limited oxygen evolution could be detected in the latter case and the lower hydrogen production is probably a result of oxygen inactivation due to the absence of PSII inhibition. As expected, no detectable hydrogen was accumulated in light when neither DCMU nor DBMIB was present. The considerable increase in hydrogen production upon DBMIB inhibition rules out any plastoquinone (PQ) dependent pathway as the main contributor of reductants to the activated CrHydA1.
A reduced net electron influx due to the PSII inhibition, together with an operating cyclic electron transport around PSI, may rapidly lead to high levels of ATP being accumulated in the cells. ATP inhibition of key enzymes in glycolytic pathways is well studied and has been reported for pyruvate kinase in cyanobacteria.20 When ATP-synthase was inhibited with DCC (N,N′-dicyclohexylmethanediimine),21 in addition to the PSII inhibition, a limited positive effect on hydrogen production (5-fold increase) could be observed in nitrogen and glucose deprived cells (Fig. 3b), suggested to be a consequence of decreased ATP inhibition of the glycolytic pathways. Interestingly, ATPase inhibition by DCC has been reported to increase the NADPH/NADP+-ratio in nitrogen starved cells of Synechocystis WT under aerobic conditions15 which may also explain the observed increased hydrogen production.
Addition of DBMIB to cells with glucose dependent hydrogen production in BG11 light (Fig. 3c) had a distinct positive effect on hydrogen accumulation, although not as drastic as under nitrogen deprivation. DBMIB inhibition alone resulted in the same production of hydrogen as the combined DCMU + DBMIB inhibition, indicating that hydrogen production had already ceased for other reasons when the oxygen levels became high enough to inactivate the activated hydrogenase. DCC inhibition resulted in a 6-fold increase, similar to the result obtained without nitrate and glucose.
The results from the inhibition studies, together with the high production in darkness, strongly imply that the hydrogen production from the activated CrHydA1 is driven mainly by a fermentative, PQ independent source of electrons. This result is surprising since the electron donor to CrHydA1 in the native Chlamydomonas host is known to be reduced ferredoxin from the photosynthetic electron transport chain.22,23 Based on our results, we suggest that either a fermentatively reduced ferredoxin or NADH/NADPH from the glycolytic pathways is the electron donor to the semi-synthetic HydA in the Synechocystis cells.
Conclusions
With this report we demonstrate, for the first time, how the concept of in vivo synthetic activation of [FeFe]-hydrogenases can be taken beyond E. coli and employed to a living photosynthetic microorganism, the unicellular cyanobacterium Synechocystis PCC 6803. The in vivo generation of functional semi-synthetic, or artificial, metalloenzymes is a very young research field, and the interplay between such non-native enzymes and their hosts remain to be established.11,12,24 We show that the semi-synthetic heterologous hydrogenase is evidently linked to cellular metabolism, both in light and darkness, resulting in significant and sustained hydrogen production. Indeed, the heterologous [FeFe]-hydrogenase significantly outperforms the activity of the native [NiFe]-hydrogenase, underscoring the possibility to improve H2 production from cyanobacteria via enzyme optimization.The reported procedure enables easy and robust acquirement of active [FeFe]-hydrogenases operating in a living cyanobacterial cell environment, avoiding e.g. the challenges of any biological maturation system co-expression. This opens up for unique studies of photobiohydrogen production from modified [FeFe]-hydrogenases and other biotechnical applications in a photosynthetic cell environment where molecular tools are well-studied and available. In addition, it will aid in the search for more oxygen tolerant variants of [FeFe]-hydrogenases as well as provide a platform for efficient screening of modified cofactors and/or enzymes in photosynthetic cells. In a wider context, the successful preparation of an active semi-synthetic hydrogenase in Synechocystis PCC 6803 highlights how synthetic chemistry can be used as a tool to manipulate these cellular factories via in vivo preparation of artificial enzymes with novel reactivates.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The European Research Council (ERC, G. B. contract no. 714102), the Swedish Energy Agency (contract no. 11674-5), the KA Wallenberg Foundation (contract no. 2011.0067), the Wenner-Gren foundations (C. E. and G. B.), and the NordForsk NCoE program NordAqua (project no. 82845) are gratefully acknowledged for funding. Professor Alfonso Jaramillo, University of Warwick, UK is gratefully acknowledged for providing the [FeFe] hydrogenase gene (hydA1).References
- R. Miao, A. Wegelius, C. Durall, F. Liang, N. Khanna and P. Lindblad, in Modern Topics in the Phototrophic Prokaryotes: Environmental and Applied Aspects, ed. P. Hallenbeck, 2017, vol. 11, pp. 351–393 Search PubMed.
- N. Khanna, P. Raleiras and P. Lindblad, in The Physiology of Microalgae, Developments in Applied Phycology 6, ed. M. Borowitzka, J. Raven and J. Beardall, 2016, vol. 5, pp. 101–127 Search PubMed.
- F. Gutthann, M. Egert, A. Marques and J. Appel, Biochim. Biophys. Acta, Bioenerg., 2007, 1767, 161–169 CrossRef CAS PubMed.
- L. Cournac, G. Guedeney, G. Peltier and P. M. Vignais, J. Bacteriol., 2004, 186, 1737–1746 CrossRef CAS PubMed.
- J. B. Broderick, A. S. Byer, K. S. Duschene, B. R. Duffus, J. N. Betz, E. M. Shepard and J. W. Peters, J. Biol. Inorg. Chem., 2014, 19, 747–757 CrossRef CAS PubMed.
- Y. Nicolet, C. Piras, P. Legrand, C. E. Hatchikian and J. C. Fontecilla-Camps, Structure, 1999, 7, 13–23 CrossRef CAS PubMed.
- P. W. King, M. C. Posewitz, M. L. Ghirardi and M. Seibert, J. Bacteriol., 2006, 188, 2163–2172 CrossRef CAS PubMed.
- D. W. Mulder, E. S. Boyd, R. Sarma, R. K. Lange, J. A. Endrizzi, J. B. Broderick and J. W. Peters, Nature, 2010, 465, 248–251 CrossRef CAS PubMed.
- G. Berggren, A. Adamska, C. Lambertz, T. R. Simmons, J. Esselborn, M. Atta, S. Gambarelli, J. M. Mouesca, E. Reijse and W. Lubitz, Nature, 2013, 499, 66–69 CrossRef CAS PubMed.
- J. Esselborn, C. Adamska-Vekates, T. Simmons, G. Berggren, J. Noth, J. Siebel, A. Heimschemeier, V. Artero, E. Reijerse, M. Fontecave, W. Lubitz and T. Happe, Nat. Chem. Biol., 2013, 9, 607–609 CrossRef CAS PubMed.
- N. Khanna, C. Esmieu, L. S. Meszaros, P. Lindblad and G. Berggren, Energy Environ. Sci., 2017, 10, 1563–1567 RSC.
- J. A. Birrell, O. Rüdiger, E. J. Reijse and W. Lubitz, Joule, 2017, 1, 61–76 CrossRef CAS.
- L. S. Mészáros, B. Németh, C. Esmieu, P. Ceccaldi and G. Berggren, Angew. Chem., Int. Ed., 2018, 57, 2596–2599 CrossRef PubMed.
- H. Li and T. B. Rauchfuss, J. Am. Chem. Soc., 2002, 124, 726–727 CrossRef CAS PubMed.
- W. Hauf, M. Schlebusch, J. Hüge, J. Kopka, M. Hagemann and K. Forchhammer, Metabolites, 2013, 3, 101–118 CrossRef CAS PubMed.
- R. Schwarz and K. Forchhammer, Microbiology, 2005, 151, 2503–2514 CrossRef CAS PubMed.
- B. Panda, P. Jain, L. Sharma and N. Mallick, Bioresour. Technol., 2006, 97, 1296–1301 CrossRef CAS PubMed.
- M. Miyake, K. Kataoka, M. Shirai and Y. Asada, J. Bacteriol., 1997, 179, 5009–5013 CrossRef CAS PubMed.
- K. Sudesh, K. Taguchi and Y. Doi, Int. J. Biol. Macromol., 2002, 30, 97–104 CrossRef CAS PubMed.
- V. L. Knowles, C. S. Smith, C. R. Smith and W. C. Plaxton, J. Biol. Chem., 2001, 276, 20966–20972 CrossRef CAS PubMed.
- H. Zürrer, M. Snozzi and R. Bachofen, FEBS Lett., 1983, 153, 151–155 CrossRef.
- A. Melis and T. Happe, Photosynth. Res., 2004, 80, 401–409 CrossRef CAS PubMed.
- M. L. Ghirardi, M. C. Posewitz, P. C. Maness, A. Dubini, J. Yu and M. Seibert, Annu. Rev. Plant Biol., 2007, 58, 71–91 CrossRef CAS PubMed.
- M. Jeschek, R. Reuter, T. Heinisch, C. Trindler, J. Klehr, S. Panke and T. R. Ward, Nature, 2016, 537, 661–677 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Including: methods & suppl figures, table, note. See DOI: 10.1039/c8ee01975d |
| ‡ Equal contribution. |
| § Current address: Department of Biotechnology, Birla Institute of Technology & Science, Pilani Dubai Campus, Dubai International Academic City, P.O. Box 345055, Dubai, United Arab Emirates. |
| ¶ Current address: School of Biological Sciences & Centre for Synthetic and Systems Biology, University of Edinburgh, Edinburgh EH9 3FF, UK. |
| This journal is © The Royal Society of Chemistry 2018 |