Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Tunable plasmonic colorimetric assay with inverse sensitivity for extracellular DNA quantification†
Roger M.
Pallares
 ab,
Nguyen T. K.
Thanh
ab,
Nguyen T. K.
Thanh
 *a and
Xiaodi
Su
*bc
*a and
Xiaodi
Su
*bc
aBiophysics Group, Department of Physics and Astronomy, University College London, Gower Street, London W1C 6BT, UK. E-mail: ntk.thanh@ucl.ac.uk
bInstitute of Materials Research and Engineering, A*STAR (Agency for Science, Technology and Research), 2 Fusionopolis Way, Innovis, #8-03, Singapore, 138634, Singapore. E-mail: xd-su@imre.a-star.edu.sg
cDepartment of Chemistry, National University of Singapore, Block S8, Level 3, 3 Science Drive 3, Singapore, 117543, Singapore
First published on 14th August 2018
Abstract
Extracellular DNA (eDNA) is a biomolecule commonly used to characterize microorganism communities in soil and aqueous environments. In this work we developed a gold nanorod (AuNR)-based colorimetric assay with inverse sensitivity and tunable dynamic range for eDNA. The effects of three key parameters, such as AuNR aspect ratio, DNA length and structure, have been identified allowing the assay to reach the detection levels necessary for the quantification of environmental eDNA.
The degradation of organic matter results in the constant release of eDNA into the ecosystem.1,2 The DNA is initially broken down by cellular nucleases inside the cell and by microorganisms once released into the environment.1 Because the degradation of eDNA occurs very fast,3 the fragments that remain in the environment vary in size but are predominantly below 100 base pairs (bp).4 Those short fragments, however, can survive for thousands of years under specific conditions,5 reaching concentrations as high as 2 μg g−1 in terrestrial soils and 0.5 g m−2 in superficial sea sediments.6 Since eDNA is a fundamental component of bacterial biofilms, participates in horizontal gene transfer, and is a nutritional source for microbes,6 eDNA quantification has been used to obtain information of the microbial communities in soils and their movement through aqueous soil phases.4,6 eDNA is mostly double stranded DNA (dsDNA) because single stranded oligonucleotides (ssDNA) do not exist for long periods of time in the environment.7 Nevertheless, ssDNA plays a short but important role in the initial formation of some bacterial biofilms.7 Therefore, quantification of both dsDNA and ssDNA is important for eDNA characterization.
DNA is generally quantified by UV spectroscopy, fluorescent intercalating dyes or quantitative real-time polymerase chain reaction (qPCR).8,9 The eDNA concentrations in environmental samples, however, are close to the limit of detection (LOD) of the first two techniques, resulting in low reliability. qPCR, on the other hand, is based on time consuming enzymatic amplification reactions.9 Gold nanoparticle based colorimetric assays have been developed as fast and straightforward alternatives for DNA detection and quantification.10,11 Those assays take advantage of the unique optical properties of gold nanoparticles, such as strong absorption coefficients and size-dependent optical responses,12,13 and measure the color change in solution caused by DNA-induced nanoparticle aggregation and/or dispersion. Based on the aggregation principle, gold nanoparticle assays for DNA can be classified into two big groups: crosslinking assays,10,11 where the hybridization between a target and ssDNA probe functionalized gold nanoparticles causes the aggregation of the latter; and non-crosslinking assays,14 where the interaction between DNA and spherical gold nanoparticles is purely electrostatic. On one hand, the crosslinking assays take advantage of the higher stability of the ssDNA–nanoparticle conjugates that give less false positive results. Furthermore, since the early designs, several important parameters, such as nanoparticle size and concentration, have been optimized to maximize the performance of the crosslinking assays.15 On the other hand, the non-crosslinking assays are also appealing because they are faster and do not require any biofunctionalization.
We recently reported a AuNR-based colorimetric assay16 for the quantification of cell-free DNA (cfDNA): a double stranded oligonucleotide of typically 180 bp, originated from apoptotic cells and used as a blood circulating cancer biomarker. This non-crosslinking assay was based on the electrostatic interactions between dsDNA molecules and cetyltrimethylammonium bromide (CTAB)-coated AuNRs. The DNA concentration dependent AuNR aggregation profile led to an inverse sensitivity (i.e. higher analytical response at lower DNA concentrations) (Scheme 1a). This was a unique analytical concept that enhanced the signal-to-noise ratio at concentrations close to the LOD of conventional assays, such as proportional (Scheme 1b) and inversely proportional sensors (Scheme 1c). Although this analytical method could efficiently detect the concentrations of circulating cfDNA associated with several types of cancers, it was unclear whether DNA structure and length as well as the nanorod aspect ratio (AR) could affect and improve the analytical performance, such as signal-to-noise ratio and LOD. In addition, it was not known if the assay could provide an inverse sensitivity response when quantifying short DNA molecules within the scale lengths of eDNA (<100 bp) or ssDNA.
Here we address the above concerns by characterizing the effect of three key parameters, i.e. the AR of the AuNRs, the DNA length and the DNA structure, on the inverse sensitivity assay performance. We show that high aspect ratio nanoparticles display better signal-to-noise ratios in the quantification of dsDNA molecules shorter than 100 bp. Larger rods (AR of 3.3) are more robust against DNA length variations, while shorter nanoparticles (AR of 2.0) are more sensitive to oligonucleotide size. Moreover, the sensor capabilities have been further expanded to the quantification of ssDNA. All these results indicate that several parameters are interconnected in the colorimetric response, and thorough optimization must be done for optimal analytical performance against eDNA.
Scheme 1d illustrates the colorimetric assay mechanism with inverse sensitivity, where the positively charged AuNRs display different aggregation profiles when they electrostatically interact with different concentrations of negatively charged dsDNA.16,17 In the absence of dsDNA, AuNRs are dispersed in solution, whereas in the presence of a small amount of dsDNA, the oligonucleotides neutralize the positive charge of the rods, causing a large degree of aggregation. As the concentration of dsDNA is increased, the oligonucleotides start covering the AuNR surface, providing electrostatic repulsion between the rod particles, and in turn they re-disperse. These changes can be tracked by the drop and recovery of the localized surface plasmon (LSP) resonance peak, respectively.
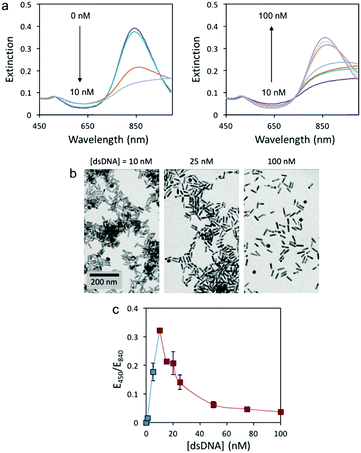
In this study, we initially tested the colorimetric assay with 3.8 AR rods and 59 bp dsDNA (sequence in Table S1, ESI†) because oligonucleotides around 60 bp have been found to be efficient extracellular vectors for bacterial transformation.4 Since environmental samples are biologically complex and full of potential interfering species, conventional analytical protocols require eDNA extraction and isolation before quantification. Hence, Tris buffer (8 mM, pH 7.4) was used in these experiments since it is the most common buffer used in DNA extraction procedures. AuNRs were synthesized following a previously published protocol.18 The UV-Vis spectra of the AuNRs in the presence of different amounts of 59 bp dsDNA display two different trends (Fig. 1a). At low dsDNA concentrations (0 to 10 nM) the LSP band (840 nm) red-shifts with increasing oligonucleotide concentration. At higher concentrations (10 to 100 nM), however, the LSP band blue-shifts with increasing dsDNA concentration. Transmission electron microscopy (TEM) images (Fig. 1b) confirmed that the changes in the solution color were caused by the aggregation of AuNRs at low dsDNA concentration (10 nM) and re-dispersion at higher dsDNA concentrations (25 and 100 nM). Our previous work with cfDNA (oligonucleotides around 180 bp) proved that aggregation and re-dispersion of AuNRs at low and high dsDNA concentrations, respectively, were caused by the change in the overall charge of CTAB-coated AuNRs from positive to neutral to negative with increasing dsDNA concentration.16 Similar behavior has also been reported when dsDNA interacts with conjugated polymers.19
For the current 59 bp dsDNA, the response curve of the colorimetric assay is plotted as the extinction ratio at 450 and 840 nm (E450/E840) as a function of dsDNA concentration (Fig. 1c). Two concentration regimes are observed: a narrow regime (0 to 10 nM dsDNA) with directly proportional sensitivity, and a wide regime (10 to 100 nM dsDNA) with inverse sensitivity. The inverse sensitive limit (ISL) was calculated as the lowest concentration that begun to manifest the inverse sensitivity response. For the 59 bp dsDNA with AuNRs of 3.8 AR, the ISL was identified at 10 nM and displayed a signal-to-noise ratio of 68, 23-fold higher than conventional LOD.20 It is worth mentioning that the concentration regime with inverse sensitivity was 90% of the full response curve. Therefore, the response of this colorimetric assay was very close to the performance of an ideal inverse sensitivity assay. In order to confirm if the concentration of a sample is within the inverse sensitivity part of the response curve, a second test must be performed where the sample is diluted. Only under an inverse sensitivity regime, the dilution displays a higher intensity response than the original sample.
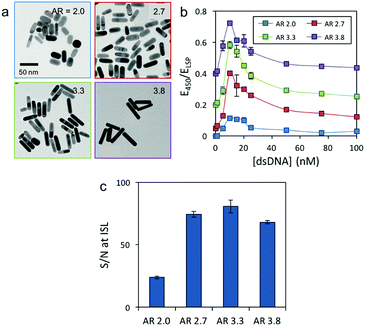
To study whether AuNR morphology affects the colorimetric assay response, we synthesized AuNRs with different dimensions (Table S3, ESI†) and AR (2.0, 2.7 and 3.3) in addition to 3.8 (Fig. 2a). The concentration of Au was kept constant in all four solutions for better comparison (14 mg L−1). All AuNR solutions displayed two distinct responses to the 59 bp dsDNA, i.e. at low oligonucleotide concentrations the LSP band red-shifted, and at higher concentrations it blue-shifted (Fig. S1, ESI†). The sensing response of the different AuNRs is plotted using the extinction ratio between 450 nm and their respective LSP maximum (E450/ELSP) versus the DNA concentration (Fig. 2b). Interestingly, all the AuNRs displayed the same ISL (10 nM) but different maximum intensities at the ISL. The AuNRs with an AR of 3.3 displayed the highest signal-to-noise ratio at the ISL (81), closely followed by 2.7 (74) and 3.8 (68) AR rods (Fig. 2c). The shortest AuNRs, however, showed a significantly lower signal-to-noise ratio of 24. These findings suggest that high AR nanocrystals are better candidates for constructing this inverse sensitivity colorimetric assay for eDNA, because they provide better signal-to-noise ratios at low analyte concentrations. One of the main challenges when quantifying eDNA is the concentration variability, which depends on several factors, such as the origin of the sample and the extraction protocol. Therefore, an analytical assay with either a large or tunable dynamic range is required. The following experiments proved that our colorimetric assay could address the challenge and provide a tunable dynamic range and thus an adjustable ISL by controlling the concentration of nanoparticles in solution. Fig. S2 (ESI†) depicts the response curve of the assay as a function of AuNR (AR 3.8) dilution. When the number of nanoparticles in solution was decreased, the dynamic range and the ISL shifted to smaller dsDNA concentrations. By combining the assay response curves of four solutions with different concentrations of AuNRs (AR 3.8), we were able to cover several ranges of concentrations associated with eDNA in aquatic environments (see Table S2 for the dynamic ranges and ISLs of different AuNR solutions compared with the literature values of eDNA from different waterbodies, ESI†).
To further confirm whether this colorimetric assay could display inverse sensitivity with various short oligonucleotides within the scale lengths of eDNA (<100 bp) and to determine the effect of DNA length on the assay ISL, we tested three different short oligonucleotides (44, 59 and 74 bp long) using AR 3.3 and 2.0 AuNRs. The UV-Vis spectra of the AuNRs with an AR of 3.3 were firstly recorded in the presence of different concentrations of dsDNA (Fig. S3, ESI†), and the response curves were plotted as E450/ELSP as a function of DNA concentration (Fig. 3a). The three response curves showed similar E450/ELSP variations with an ISL of 10 nM for the longer dsDNA (59 and 74 bp) and 15 nM for the shorter 44 bp oligonucleotide. Interestingly, when shorter AuNRs with an AR of 2.0 were used, higher E450/ELSP sensitivity toward dsDNA length was observed. The ISL significantly decreased when longer DNA molecules were quantified (i.e. ISL of 25, 10 and 5 nM for 44, 59 and 74 bp DNA, respectively) (Fig. 3b). We hypothesize that the lower ISL for longer oligonucleotides is because the larger the dsDNA molecules, the more efficient they can interact with several nanoparticles at the same time, and fewer oligonucleotides are required to cause nanoparticle aggregation. These results also suggested that the AR of the AuNRs and the dsDNA length are interconnected, and shorter rods are more sensitive to oligonucleotide size, while longer rods are more robust against DNA length variations. The tolerance against size variation makes larger AR rods better candidates for the quantification of environmental samples, since eDNA presents size variability.
 | ||
| Fig. 3 Comparison between the response curves of different sized dsDNA and AuNRs with an AR of (a) 3.3 and (b) 2.0. All response curves have been offset for easier comprehension. | ||
Finally, we investigated whether the colorimetric assay could quantify ssDNA in an inverse sensitivity regime, and how was its performance compared to dsDNA. AuNRs with an AR of 3.3 were mixed with different concentrations of 59 mer ssDNA or 59 bp dsDNA, and the extinction spectra of the solutions were recorded (Fig. S4, ESI†). The response curves showed that the sensor displayed inverse sensitivity for ssDNA. Nevertheless, the ISL was higher (25 nM) than that for dsDNA (10 nM) (Fig. 4). These observations indicated that higher amounts of ssDNA were required to neutralize the positive charge of CTAB coated AuNRs and cause their aggregation. We attributed the distinct behavior between ssDNA and dsDNA to their structural differences that led to the different interaction with the AuNRs. First, compared to dsDNA, the ssDNA backbone has half phosphate units, which leads to weaker electrostatic interaction with the nanoparticles than dsDNA. Second, dsDNA has a more rigid structure with more exposed phosphates,21 which further facilitates the interaction with the surroundings. Not only those parameters affected the ISL, but also contributed to the higher sensitivity in terms of the E450/ELSP response to DNA concentration changes. For instance, for the 59 bp dsDNA, an increase of the concentration from 10 nM (ISL) to 25 nM caused a E450/ELSP drop of 48.3%, whereas 5-fold higher ssDNA variation was necessary to cause similar intensity changes (i.e. increasing from 25 nM (ISL) to 100 nM ssDNA caused a decrease of intensity of 45.6%).
 | ||
| Fig. 4 Comparison between the response curves of single and double stranded DNA. AuNRs with an AR of 3.3 were used. All response curves have been offset for easier comprehension. | ||
In summary, we identified the AR of AuNRs, DNA length, and DNA structure (single and double-stranded DNA) as important factors that affect the sensing performance of the colorimetric assay with inverse sensitivity for eDNA. Although AuNR morphology did not affect the ISL of the assays, AuNRs with a higher AR displayed better signal-to-noise ratios and were more robust against oligonucleotide size variation. AuNRs with a smaller AR (i.e. 2.0), however, were more sensitive to DNA length variations. By controlling the concentration of AuNRs, the assay could cover a very wide range of concentrations associated with eDNA in several waterbody environments (from 19.2 to 3835 ng mL−1). Moreover, ssDNA could be quantified under an inverse sensitivity response but with a higher ISL in comparison to dsDNA. All these observations highlight the interconnection between several parameters, which define the assay analytical performance. Based on these results, one can choose the appropriate sensing set up for environmental DNA samples according to the ISL requirement.
RMP thanks UCL and A*STAR for his PhD fellowship. NTKT thanks EPSRC for funding. XS thanks A*STAR JCO funding 14302FG096.
Conflicts of interest
There are no conflicts to declare.Notes and references
- K. M. Nielsen, P. J. Johnsen, D. Bensasson and D. Daffonchio, Environ. Biosaf. Res., 2007, 6, 37–53 CrossRef PubMed.
- G. F. Ficetola, C. Miaud, F. Pompanon and P. Taberlet, Biol. Lett., 2008, 4, 423–425 CrossRef PubMed.
- A. Torti, M. A. Lever and B. B. Jørgensen, Mar. Genomics, 2015, 24, 185–196 CrossRef PubMed.
- S. Overballe-Petersen, K. Harms, L. A. A. Orlando, J. V. M. Mayar, S. Rasmussen, T. W. Dahl, M. T. Rosing, A. M. Poole, T. Sicheritz-Ponten, S. Brunak, S. Inselmann, J. de Vries, W. Wackernagel, O. G. Pybus, R. Nielsen, P. J. Johnsen, K. M. Nielsen and E. Willerslev, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 19860–19865 CrossRef PubMed.
- E. M. Golenberg, Philos. Trans. R. Soc. London, Ser. B, 1991, 333, 419–427 CrossRef PubMed.
- D. Vorkapic, K. Pressler and S. Schild, Curr. Genet., 2016, 62, 71–79 CrossRef PubMed.
- M. Zweig, S. Schork, A. Koerdt, K. Siewering, C. Sternberg, K. Thormann, S. V. Albers, S. Molin and C. Van der Does, Environ. Microbiol., 2014, 16, 1040–1052 CrossRef PubMed.
- K. Nielsen, H. S. Mogensen, J. Hedman, H. Niederstätter, W. Parson and N. Morling, Forensic Sci. Int.: Genet., 2008, 2, 226–230 CrossRef PubMed.
- S. Shokralla, J. L. Spall, J. F. Gibson and M. Hajibabaei, Mol. Ecol., 2012, 21, 1794–1805 CrossRef PubMed.
- C. a Mirkin, R. L. Letsinger, R. C. Mucic and J. J. Storhoff, Nature, 1996, 382, 607–609 CrossRef PubMed.
- R. Elghanian, J. J. Storhoff, R. C. Mucic, R. L. Letsinger and C. A. Mirkin, Science, 2010, 1078, 1078–1081 Search PubMed.
- S. Eustis and M. A. El-Sayed, Chem. Soc. Rev., 2006, 35, 209–217 RSC.
- R. M. Pallares, Y. Wang, S. H. Lim, N. T. K. Thanh and X. Su, Nanomedicine, 2016, 11, 2845–2860 Search PubMed.
- R. Kanjanawarut and X. Su, Anal. Chem., 2009, 81, 6122–6129 CrossRef PubMed.
- V. S. Godakhindi, P. Kang, M. Serre, N. A. Revuru, J. M. Zou, M. R. Roner, R. Levitz, J. S. Kahn, J. Randrianalisoa and Z. Qin, ACS Sens., 2017, 2, 1627–1636 CrossRef PubMed.
- R. M. Pallares, S. L. Kong, T. H. Ru, N. T. K. Thanh, Y. Lu and X. Su, Chem. Commun., 2015, 51, 14524–14527 RSC.
- R. M. Pallares, M. Bosman, N. T. K. Thanh and X. Su, Nanoscale, 2016, 8, 19973–19977 RSC.
- R. M. Pallares, X. Su, S. H. Lim and N. T. K. Thanh, J. Mater. Chem. C, 2016, 4, 53–61 RSC.
- R. M. Pallares, L. Sutarlie, N. T. K. Thanh and X. Su, Sens. Actuators, B, 2018, 271, 97–103 CrossRef.
- G. L. Long and J. D. Winefordner, Anal. Chem., 1983, 55, 712A–724A CrossRef.
- J. Ambia-Garrido, A. Vainrub and B. M. Pettitt, Comput. Phys. Commun., 2010, 181, 2001–2007 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8cc05465g |
| This journal is © The Royal Society of Chemistry 2018 |