Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
QM/MM calculations reveal a bridging hydroxo group in a vanadium nitrogenase crystal structure†
Bardi
Benediktsson
 a,
Albert Th.
Thorhallsson
a and
Ragnar
Bjornsson
a,
Albert Th.
Thorhallsson
a and
Ragnar
Bjornsson
 *ab
*ab
aScience Institute, University of Iceland, Dunhagi 3, 107 Reykjavik, Iceland
bDepartment of Inorganic Spectroscopy, Max-Planck-Institut für Chemische Energiekonversion, Stiftstrasse 34-36, 45470 Mülheim an der Ruhr, Germany. E-mail: ragnar.bjornsson@cec.mpg.de
First published on 23rd May 2018
Abstract
A new 1.2 Å crystal structure of vanadium nitrogenase, isolated under turnover conditions, recently revealed a light atom ligand (OH or NH) replacing the bridging S2B sulfide of the FeV cofactor. QM/MM calculations on the new structure now reveal the light-atom ligand to be a bridging hydroxo group, probably derived from water binding to the cofactor.
Nitrogen is present in the atmosphere in the form of dinitrogen and the element is essential for life. Only diazotrophs, however, possess the capability of activating and breaking the triple bond of dinitrogen, a reaction performed by the nitrogenase enzymes.1,2 Nitrogenases are metalloenzymes that catalyse the reduction of dinitrogen to two molecules of ammonia for each molecule of dinitrogen. Different forms exist that can be distinguished by the nature (or absence) of the heterometal that is present in the active site. Molybdenum nitrogenase contains an iron–molybdenum cofactor (FeMoco), vanadium nitrogenase contains an iron–vanadium cofactor (FeVco) and the iron nitrogenase contains an all-iron cofactor (FeFeco).
The Mo nitrogenase is the most active and has been studied most extensively while the V nitrogenase has in recent years gained more attention following the discovery of its unique catalytic properties3–5 and recently the first crystal structure became available.6 Despite intense research efforts, the mechanism of biological nitrogen reduction is far from understood and fundamental information such as the substrate binding site is lacking for all nitrogenase forms.
The chemistry of the nitrogenases differ, with V nitrogenase requiring more electrons and protons for N2 reduction and more H2 is produced per N2 reacted,7 as compared to Mo nitrogenase. This is possibly a consequence of the heterometal but the crystal structure recently revealed that FeVco differs from FeMoco with respect to a bridging ligand where either a carbonate or nitrate ligand was found in place of sulfide S3A.6 Additionally, the Fe sites are more reduced in FeVco than FeMoco according to Fe XAS experiments and theoretical calculations, a consequence of the different heterometal.8
Recently, Sippel et al.9 described the isolation of a turnover state of V nitrogenase that they were able to crystallize. The 1.2 Å crystal structure revealed the same unusual CO32−/NO3− ligand but additionally, the S2B bridging sulfide was missing and a light-atom ligand had replaced it (Fig. 1). At this resolution it was not possible to distinguish between C, N and O. A carbon ligand can be plausibly ruled out and only NH and OH were considered as likely candidates for the light-atom ligand, as there is a strong hydrogen bond between the ligand atom and the carbonyl group of nearby residue Gln176. Electron density analysis also found evidence of a protonated ligand fitting better. Interestingly the S2B sulfide atom that the ligand had replaced could be found, likely in the form of an SH− ion, 7 Å away. These results led the authors to propose a mechanism for nitrogen reduction, with the light atom ligand interpreted as an NH group. As the crystallographic study was not able to conclusively identify the nature of the ligand, it is of utmost importance to clarify the identity of this ligand conclusively. In this study, we carefully examine the OH and NH possibilities via quantum mechanics/molecular mechanics (QM/MM) calculations.
 | ||
| Fig. 1 The newly discovered bridging ligand, shown in purple, as it appears in the crystal structure of the turnover state of VFe nitrogenase by Sippel et al.9 The ligand atom has replaced a bridging sulfide that now appears close to the cofactor, probably in the form of SH−. | ||
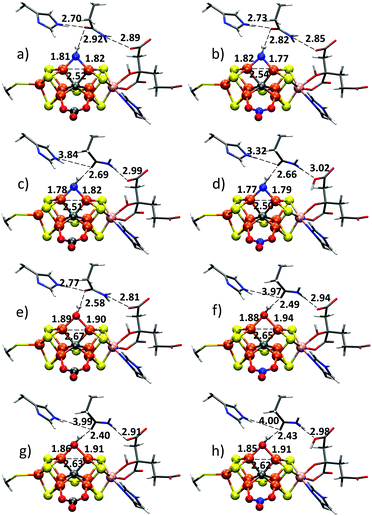
As shown in Fig. 2, the light atom X bridges Fe atoms no. 2 and 6. It makes a strong hydrogen bond to the carbonyl group of the nearby Gln176 residue that appears in a different conformation compared to the VFe resting state structure (or the analogous Gln191 in MoFe protein). This strong hydrogen bond between the carbonyl oxygen and atom X allows one to deduce that the ligand must be protonated, likely by one proton.
The hydrogen bond between X and OGln176 is strong: 2.51 Å in one instance of the active site pocket and 2.39 Å in the other. In this context it is worth noting that O(H)⋯O hydrogen bonds found in crystal structures of small molecules are in the range of 2.54–2.82 Å (heavy atom distances) while N(H)⋯O hydrogen bonds are longer, in the range of 2.75–2.98 Å.10 The stronger hydrogen bonding of OH donor compared to NH donors would thus suggest OH as a more likely candidate for the XH ligand, but due to the nature of a metal-bridging NH/OH this is not so clear-cut as such a ligand could have unusual electronic properties, particularly seeing as the metal–sulfur clusters of these enzymes are unique. There are few examples of Fe-bridging NH ligands in the literature and the oxidation state of such a ligand is not clear (likely either a nitrene, HN0 or nitrido, HN2−). A structural argument for distinguishing between OH or NH, in our view, necessitates quantum chemical calculations that are capable of both describing the unusual electronic structure accurately as well as accounting for the protein environment.
In a recent study we described a QM/MM model of the resting state of the MoFe protein of nitrogenase.11 By combining broken-symmetry DFT calculations with the TPSSh functional12,13 with a classical forcefield (CHARMM3614) we demonstrated that our QM/MM approach allowed direct determination of the charge of the cofactor, that the cofactor is populating a specific electronic state and that the alkoxide group is protonated. This analysis was possible via structural comparison alone and indicates the accuracy of our theoretical approach as well as the high resolution (1.0 Å) of the experimental crystal structure15 of the resting state.
Here we apply the same QM/MM approach to the VFe protein, using the 1.2 Å crystal structure of the ligand-bound state as a starting point (PDB ID: 6FEA). A 38![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 917 atom spherical QM/MM model was prepared as detailed in the ESI.† All heavy atoms were kept fixed at crystallographic positions during the preparation phase. QM/MM geometry optimizations (using TPSSh and CHARMM36 for QM and MM descriptions respectively; see ESI† for further information) were performed for both NH and OH ligands. As the cofactor redox state is unknown, but the spin-state was found to be S = 3/2, we considered two possibilities: same redox level as the resting state, labelled E0 and a state oxidized by two electrons, labelled Eox. We note that resting state FeVco has a more reduced Fe oxidation state than FeMoco according to Fe XAS8 and the cofactor charge could be defined as either [VFe7S8C(CO3)]2− or [VFe7S8C(NO3)]−. The S2B-ligand substitution (at the Fe2–Fe6 site) will affect the charge, as will the identity of the ligand at the Fe4–Fe5 site. We considered both CO32− and NO3− as reasonable possibilities as discussed in the original crystallographic study of the resting state.6 CO32− and NO3− are isoelectronic and likely to behave similarly but the difference in charge will certainly affect the electrostatic environment in the active site.
917 atom spherical QM/MM model was prepared as detailed in the ESI.† All heavy atoms were kept fixed at crystallographic positions during the preparation phase. QM/MM geometry optimizations (using TPSSh and CHARMM36 for QM and MM descriptions respectively; see ESI† for further information) were performed for both NH and OH ligands. As the cofactor redox state is unknown, but the spin-state was found to be S = 3/2, we considered two possibilities: same redox level as the resting state, labelled E0 and a state oxidized by two electrons, labelled Eox. We note that resting state FeVco has a more reduced Fe oxidation state than FeMoco according to Fe XAS8 and the cofactor charge could be defined as either [VFe7S8C(CO3)]2− or [VFe7S8C(NO3)]−. The S2B-ligand substitution (at the Fe2–Fe6 site) will affect the charge, as will the identity of the ligand at the Fe4–Fe5 site. We considered both CO32− and NO3− as reasonable possibilities as discussed in the original crystallographic study of the resting state.6 CO32− and NO3− are isoelectronic and likely to behave similarly but the difference in charge will certainly affect the electrostatic environment in the active site.
Due to the already high negative charge of FeVco compared to FeMoco, a more negative charge than 2− or redox state beyond E0 is highly unlikely. In fact, due to the high negative charge of the E0 models, the NH group, when interpreted as a nitrido NH2− group and including a CO32− ligand, resulted in an electronic structure with unbound electrons (positive orbital energies), an obvious sign of something wrong with the model. This did not occur with models with an OH− ligand, likely due to the lower charge. Finally we note that calculating an oxidized state can also be interpreted as assuming a nitrene NH0 group replacing S2− instead of nitrido NH2−.
The QM/MM optimized geometries for all models are shown in Fig. 3 and structural parameters are tabulated in Table 1 (details of the QM region are present in table as well as ESI†). We start by discussing the effect of an NH or OH ligand on the hydrogen-bonding in the active site. An NH model with E0 charge and a CO32− ligand (Fig. 3a) is found to result in a long N(H)⋯O hydrogen bond between NH and Gln176 oxygen (2.92 Å; close to what would be expected for N(H)⋯O hydrogen bonds), notably longer than the N(H)⋯O hydrogen bonds between His180 and Gln176 (2.70 Å), and Gln176 and homocitrate (2.89 Å) which is opposite to what is found in the experimental structure (Fig. 2). This is far from the crystallographic distances of 2.51 and 2.39 Å. This model also resulted in positive orbital energies and hence unbound electrons. This is a clear sign of too high negative charge and we thus additionally explored a model where the homocitrate carboxylic acid groups were protonated. Protonation of homocitrate results in all-negative orbital energies but this does not result in a structure that is considerably closer to the crystal structure (see ESI†), the N(H)⋯OGln176 distance is 2.88 Å. These calculated hydrogen bond distances are in fact similar to the experimental N(H)⋯O hydrogen bonds between His180 and Gln176, and Gln176 and homocitrate, in the crystal structure.
| Fe2–Fe6 lig. | NH | NH | NH | NH | OH | OH | OH | OH | Crystal |
|---|---|---|---|---|---|---|---|---|---|
| Fe4–Fe5 lig. | CO32− | NO3− | CO32− | NO3− | CO32− | NO3− | CO32− | NO3− | |
| Redox state | E 0 | E 0 | E ox | E ox | E 0 | E 0 | E ox | E ox | ABC,DEF |
| Q176–X | 2.92 | 2.82 | 2.69 | 2.66 | 2.58 | 2.49 | 2.40 | 2.43 | 2.51, 2.39 |
| Q176–H180 | 2.70 | 2.73 | 3.84 | 3.32 | 2.77 | 3.97 | 3.99 | 4.00 | 2.85, 2.83 |
| Q176–HC | 2.89 | 2.85 | 2.99 | 3.02 | 2.81 | 2.94 | 2.91 | 2.98 | 2.87, 2.91 |
| Fe2–X | 1.81 | 1.82 | 1.78 | 1.77 | 1.89 | 1.88 | 1.86 | 1.85 | 2.01, 2.08 |
| Fe6–X | 1.82 | 1.77 | 1.82 | 1.79 | 1.90 | 1.94 | 1.91 | 1.91 | 2.01, 2.05 |
| Fe2–Fe6 | 2.52 | 2.54 | 2.51 | 2.50 | 2.67 | 2.65 | 2.63 | 2.62 | 2.64, 2.65 |
| ∠Fe2–Fe6–Q176–X | 6.01 | 3.03 | −6.98 | −0.66 | 5.68 | −11.39 | −7.79 | −10.18 | 4.30, 6.11 |
| C–X | 2.91 | 2.91 | 2.91 | 2.90 | 2.90 | 2.93 | 2.92 | 2.93 | 3.05, 3.15 |
| V–Fe5 | 2.73 | 2.70 | 2.70 | 2.75 | 2.74 | 2.71 | 2.73 | 2.76 | 2.71, 2.70 |
| V–Fe6 | 2.88 | 3.04 | 2.93 | 2.88 | 2.81 | 2.87 | 2.91 | 2.94 | 2.79, 2.78 |
| V–Fe7 | 2.71 | 2.66 | 2.77 | 2.81 | 2.71 | 2.74 | 2.73 | 2.96 | 2.77, 2.76 |
| Fe5–Fe6 | 2.65 | 2.64 | 2.68 | 2.62 | 2.63 | 2.66 | 2.70 | 2.64 | 2.58, 2.59 |
| Fe5–Fe7 | 2.67 | 2.60 | 2.66 | 2.63 | 2.66 | 2.60 | 2.66 | 2.62 | 2.64, 2.58 |
| Fe6–Fe7 | 2.53 | 2.53 | 2.50 | 2.72 | 2.51 | 2.56 | 2.73 | 2.58 | 2.58, 2.64 |
| Fe4–Fe5 | 2.77 | 2.79 | 2.74 | 2.65 | 2.74 | 2.65 | 2.61 | 2.67 | 2.79, 2.78 |
| Fe3–Fe7 | 2.60 | 2.58 | 2.56 | 2.58 | 2.58 | 2.59 | 2.56 | 2.58 | 2.60, 2.60 |
| Fe1–Fe2 | 2.68 | 2.67 | 2.70 | 2.71 | 2.69 | 2.71 | 2.73 | 2.73 | 2.69, 2.68 |
| Fe1–Fe3 | 2.67 | 2.65 | 2.65 | 2.66 | 2.64 | 2.63 | 2.66 | 2.67 | 2.67, 2.67 |
| Fe1–Fe4 | 2.59 | 2.60 | 2.59 | 2.56 | 2.62 | 2.57 | 2.62 | 2.57 | 2.59, 2.59 |
| Fe2–Fe3 | 2.60 | 2.62 | 2.81 | 2.79 | 2.58 | 2.65 | 2.80 | 2.85 | 2.63, 2.64 |
| Fe2–Fe4 | 2.64 | 2.68 | 2.67 | 2.67 | 2.65 | 2.63 | 2.70 | 2.66 | 2.64, 2.63 |
| Fe3–Fe4 | 2.69 | 2.63 | 2.68 | 2.63 | 2.66 | 2.62 | 2.68 | 2.62 | 2.64, 2.65 |
| Tot. charge | 3− | 2− | 1− | 0 | 2− | 1− | 0 | 1+ | |
| [VFe7S8C(YO3)X]n | 2− | 1− | 0 | 1+ | 1− | 0 | 1+ | 2+ |
Furthermore, modelling the ligand as an NH group results in a very short Fe2–Fe6 distance of 2.52 Å (0.12–0.13 Å shorter than the crystal structure) and gives short Fe–N bonds of 1.82 and 1.81 Å (compared to 2.01–2.08 Å in the crystal structure). We note in this context that distances involving light atoms bonded to heavy atoms have higher uncertainties than heavy–heavy atom distances due to Fourier ripple effects.16 This will also affect the hydrogen-bonding distance. The metal–metal distances, however, in contrast will be better resolved. The 0.12–0.13 Å Fe2–Fe6 contraction due to an NH ligand that is predicted by our calculations, is uncomfortably large and should have been resolved in the 1.2 Å crystal structure, were it present. Previous MoFe crystal structures with resolution between 1.0–2.0 Å have shown that these Fe–Fe distances have an average deviation of only 0.03 Å and maximum deviation of 0.05 Å (see ESI†).
An NH model with NO3− instead of CO32− as the other bridging ligand (Fig. 3b) gives a shorter N(H)⋯O distance (2.82 Å), an improvement, but also appears inconsistent with the crystal structure. When oxidized NH models (Fig. 3c and d) are calculated with CO32− and NO3−, the N(H)⋯O distance is reduced to 2.69 and 2.66 Å, respectively, which is closer to the experimental distance but still far off. This stronger hydrogen bond, however, also results in a weakening of the hydrogen bonding between His180 and Gln176 that does not fit well with the experimental structure (off by 0.5–1.0 Å). Furthermore, the Fe2–Fe6 distance is still always too short and as seen in Table 1 the more oxidized charge results in an increased Fe2–Fe3 distance of 2.81 Å (CO32−) or 2.79 Å (NO3−) that seems incompatible with the experimental structure. The V–Fe6 distances are also too long for all NH models. As there is very little observed change in the Fe–Fe and V–Fe distances when going from the resting state V–Fe crystal structure to the turnover V–Fe crystal structure, these changes are hard to accept. All things considered, ligand X in the crystal structure is highly unlikely to be an NH.
In sharp contrast to the NH models, the model with an OH ligand and E0 charge (Fig. 3e and Table 1), results in a 2.58 Å O(H)⋯OGln176 distance that is in much better agreement with the crystallographic hydrogen bond distance. The Fe2–Fe6 distance (2.67 Å calc., 2.64–2.65 Å in crystal) is also in good agreement with the crystal structure and the Fe–O bond lengths (∼1.9 Å) are closer to the experimental Fe–X bond lengths (2.03–2.05 Å). Substituting CO32− for NO3− (Fig. 3f) gives a shorter O(H)⋯OGln176 hydrogen bond but this disrupts the hydrogen bonding between His180 and Gln176. Seeing as CO32− and NO3− are isoelectronic ligands, this reveals a sensitivity of the hydrogen-bonding network to the electrostatic environment in the active site. The more oxidized OH models (Fig. 3g and h) result in even stronger O(H)⋯OGln176 hydrogen bonding (in better agreement with the experimental distances) but this also disrupts the His180–Gln176 hydrogen bonding.
As with NH, the removal of 2 electrons results in too long metal–metal distances: Fe2–Fe3, V–Fe6 and V–Fe7, making an oxidized state unlikely. These changes likely occur due to removal of electrons from metal pairs with delocalized electron character. The ∠Fe2–Fe6–Q176–X dihedral angle in Table 1 also reveals a slightly different orientation of the OH group in the oxidized models as well as the E0-NO3− model, compared to the E0-CO32− model and the crystal structure.
We additionally performed QM cluster calculations where a continuum solvation model replaced the explicit protein MM environment and constraints were used to keep an approximate active site geometry. These results, shown in the ESI,† reveal that the cluster models are not capable of describing the local active site geometry well enough as the hydrogen bond between O(H)⋯OGln176 is predicted to be considerably weaker than with the QM/MM models. Seeing as the QM/MM results revealed a sensitivity to the electrostatic environment, this is perhaps not surprising. The OH models still gave Fe2–Fe6 distances closer to experiment than NH models. This demonstrates the utility of our QM/MM models for accurately describing molecular structure of the active site in nitrogenases.
In conclusion, our study clearly resolves the identity of the unknown ligand species in the new VFe crystal structure and reveals it to be a hydroxo group instead of an NH. While there is always uncertainty associated with distances in crystal structures, particularly involving light atoms we feel the data safely rules out all NH models based on deviations of light–light atom distances as large as 0.4–0.5 Å and in particular based on deviations of metal–metal distances of 0.12–0.20 Å. In addition to the OH assignment, a CO32− ligand is preferred over NO3− in our calculations and only an E0 charge state fits well, i.e. the model shown in Fig. 3e. The identity of the ligand species as an OH instead of NH perhaps reduces the mechanistic significance of the new crystal structure. On the other hand, the discovery of a hydroxo group bound to the cofactor (likely derived from a water molecule) hints at a previously unknown role of water in the redox states or mechanism of nitrogenases. Alternatively, the structure might correspond to a reversible side-reaction, perhaps analogous to the Ni-A and Ni-B states of [NiFe] hydrogenase where solvent-derived hydroxo bridges between Ni and Fe have been suggested.17,18 More importantly, this new structure further establishes the importance of Fe2 and Fe6 in ligand binding (a previous MoFe crystal structure19 showed a bridging CO between Fe2 and Fe6) as well as further showing the lability of the bridging sulfides in these intriguing cofactors.
R. B. acknowledges support from the Icelandic Research Fund, grants 141218051 and 162880051, and the University of Iceland Research Fund. R. B. thanks Oliver Einsle for useful discussions. Open Access funding provided by the Max Planck Society.
Conflicts of interest
There are no conflicts to declare.References
- B. K. Burgess, Chem. Rev., 1990, 90, 1377–1406 CrossRef.
- L. C. Seefeldt, B. M. Hoffman and D. R. Dean, Annu. Rev. Biochem., 2009, 78, 701–722 CrossRef PubMed.
- C. C. Lee, Y. Hu and M. W. Ribbe, Science, 2010, 329, 642 CrossRef PubMed.
- J. Hu, C. C. Lee and M. W. Ribbe, Science, 2011, 333, 753–755 CrossRef PubMed.
- J. G. Rebelein, C. C. Lee, Y. Hu and M. W. Ribbe, Nat. Commun., 2016, 7, 13641 CrossRef PubMed.
- D. Sippel and O. Einsle, Nat. Chem. Biol., 2017, 13, 956–960 CrossRef PubMed.
- D. Rehder, J. Inorg. Biochem., 2000, 80, 133–136 CrossRef PubMed.
- J. A. Rees, R. Bjornsson, J. K. Kowalska, F. A. Lima, J. Schlesier, D. Sippel, T. Weyhermüller, O. Einsle, J. A. Kovacs and S. DeBeer, Dalton Trans., 2017, 46, 2445–2455 RSC.
- D. Sippel, M. Rohde, J. Netzer, C. Trncik, J. Gies, K. Grunau, I. Djurdjevic, L. Decamps, S. L. A. Andrade and O. Einsle, Science, 2018, 359, 1484–1489 CrossRef PubMed.
- A. Langkilde, S. M. Kristensen, L. Lo Leggio, A. Mølgaard, J. H. Jensen, A. R. Houk, J. C. Navarro Poulsen, S. Kauppinen and S. Larsen, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2008, 64, 851–863 CrossRef PubMed.
- B. Benediktsson and R. Bjornsson, Inorg. Chem., 2017, 56, 13417–13429 CrossRef PubMed.
- J. Tao, J. Perdew, V. Staroverov and G. Scuseria, Phys. Rev. Lett., 2003, 91, 146401 CrossRef PubMed.
- V. N. Staroverov, G. E. Scuseria, J. Tao and J. P. Perdew, J. Chem. Phys., 2003, 119, 12129–12137 CrossRef.
- R. B. Best, X. Zhu, J. Shim, P. E. Lopes, J. Mittal, M. Feig and A. D. Mackerell, J. Chem. Theory Comput., 2012, 8, 3257–3273 CrossRef PubMed.
- T. Spatzal, M. Aksoyoglu, L. Zhang, S. L. A. Andrade, E. Schleicher, S. Weber, D. C. Rees and O. Einsle, Science, 2011, 334, 940 CrossRef PubMed.
- R. W. Strange, M. Ellis and S. S. Hasnain, Coord. Chem. Rev., 2005, 249, 197 CrossRef.
- M. Carepo, D. L. Tierney, C. D. Brondino, T. C. Yang, A. Pamplona, J. Telser, I. Moura, J. J. Moura and B. M. Hoffman, J. Am. Chem. Soc., 2002, 124, 281–286 CrossRef PubMed.
- W. Lubitz, H. Ogata, O. Rüdiger and E. Reijerse, Chem. Rev., 2014, 114, 4081–4148 CrossRef PubMed.
- T. Spatzal, K. A. Perez, O. Einsle, J. B. Howard and D. C. Rees, Science, 2014, 345, 1620–1623 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Details about QM/MM calculations, additional data on alternative models and cluster models and Cartesian coordinates for all structures. See DOI: 10.1039/c8cc03793k |
| This journal is © The Royal Society of Chemistry 2018 |