Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A novel EI-GC/MS method for the accurate quantification of anti-aging compound oleoylethanolamine in C. elegans†
Andrea
Annibal
 a,
Özlem
Karalay
a,
Christian
Latza
a and
Adam
Antebi
*ab
a,
Özlem
Karalay
a,
Christian
Latza
a and
Adam
Antebi
*ab
aMax Planck Institute for Biology of Ageing, Cologne, Germany. E-mail: aantebi@age.mpg.de
bCologne Excellence Cluster on Cellular Stress Responses in Aging-Associated Diseases (CECAD), University of Cologne, Cologne, Germany
First published on 30th April 2018
Abstract
Endocannabinoids and related N-acyl ethanolamine-derived lipids affect a diverse array of physiological processes and pathological conditions. In the roundworm C. elegans, several endocannabinoid-like molecules have been implicated in regulating axon regeneration, energy balance and food intake as well as aging. One such molecule oleoylethanolamine (OEA) has been shown to promote life extension through nuclear receptor signal transduction, and its accurate quantitation therefore is of high interest. Using a combination of electron impact ionization (EI) and collision induced dissociation coupled to gas chromatography (GC), we found unique fragmentation ions of OEA and designed a specific MRM method for its accurate quantification. Our method should provide a reproducible and robust way to measure OEA dynamics under different genetic, pharmacologic and environmental perturbations.
Introduction
N-Acylethanolamines (NAEs) are lipid molecules that exhibit diverse bioactivities and regulate many aspects of animal behavior. This class of compounds includes arachidonylethanolamine (anandamide),1,2 an endogenous cannabinoid that binds to cannabinoid receptors to influence behavior, lipid metabolism, inflammation and cancer.3–6 They also include cannabinoid-related bioactive compounds such as palmitoylethanolamine (PEA), oleoylethanolamine (OEA) and linoleoylethanolamine (LEA),7 which regulate nociception, inflammation, as well as complex behaviors governing appetite and feeding via G-proteins-couple receptors or via PPARα.8–11 NAE signaling relies on a variety of proteins for synthesis and activity, including enzymes, lipases and receptors, many of which are conserved from worms to mammals.12Caenorhabditis elegans (C. elegans) is a facile model organism to study endocannabinoids and related molecules because of its ease of genetic manipulation and well described physiology. Importantly the nematode harbors enzymes involved in their production and breakdown, as well as receptors that mediate their activity. For example, N-arachidonylethanolamine works through several G protein coupled endocannabinoid receptors, to regulate axon regeneration, nociception, and feeding.8,13 Another compound, eicosapentaenoyl ethanolamine (EPEA) is among the most abundant bioactive molecules and has pro-aging properties. EPEA levels drop upon dietary restriction and its supplementation abolishes longevity induced by dietary restriction or S6 kinase reduction. In addition it inhibits formation of dauer larvae, a long-lived larval stage.14 Nevertheless, the downstream targets of EPEA remain elusive. By contrast, oleoylethanolamine (OEA) stands out as a bioactive NAE that has anti-aging properties.15 The C. elegans lysosomal lipase, LIPL-4 functions in the lipolysis of fats used in the production of OEA. LIPL-4 overexpressing transgenic lines or by OEA and chemical analogues supplementation increase worm life span, showing a key link between lysosomal function and longevity.16,17 Apparently OEA binds to the nuclear hormone receptor, NHR-80, and activates both NHR-80 and its binding partner NHR-49 to facilitate life span extension. Interestingly, this mechanism closely resembles the manner in which OEA activates mammalian PPARα nuclear receptor to regulate metabolism.18
Despite these elegant studies, the interplay between various NEAs, longevity and dietary response remains unclear, due in part to lack of information on the variety of NAE species and their bioactivities in the nematode. One of the main limitations is that robust methods for the detection, measurement and quantification of these species are lacking. Notably, NAEs are present in low abundance in biological samples, thus many technical issues and problems arise during their extraction, enrichment, and analysis.
Mass spectrometry (MS) is the first choice as an analytical technique to study endocannabinoids. Lipidomic techniques have been developed for the quantification of NEAs mostly using gas chromatography (GC)-MS, or more recently liquid chromatography (LC)-MS analyses. Mechoulam et al. discovered AEA using GC-MS with chemical ionization (CI) with isobutane and further optimized using methane as a reagent.19 Kempe and coworkers used negative chemical ionization mass spectrometry for the profiling of NEAs,20 whereas Kasai et al. used fast atom bombardment ionization coupled with a double-focusing sector mass spectrometry to retrieve spectra and fragmentation patterns of endocannabinoids.21 NEAs were also detected using UHPLC/HPLC coupled with high-resolution based mass spectrometry.18
Although LC-MS and GC-MS are generally similar systems with comparable detection limits, they are often in disagreement for the analysis of the endocannabinoids, especially for OEA quantification.22–24 Despite LC-MS methods providing high sensitivity, the accurate measurement of OEA can be influenced by the presence of OEA isomers (18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), such as, vaccinoyl ethanolamide (VEA), elaidoyl ethanolamide (EEA), and trans-VEA, derived from vaccenic acid (n-7), elaidic acid (n-9), and trans-vaccenic acid (n-7), which co-elute with OEA and decrease sensitivity.25–27 Because of the specific biological activity of OEA, it is crucial to obtain diagnostic fragments and the corresponding mass spectrometry pattern, which will help to design more precise MS-based approaches for quantitative analysis and resolve structural isomers.
1), such as, vaccinoyl ethanolamide (VEA), elaidoyl ethanolamide (EEA), and trans-VEA, derived from vaccenic acid (n-7), elaidic acid (n-9), and trans-vaccenic acid (n-7), which co-elute with OEA and decrease sensitivity.25–27 Because of the specific biological activity of OEA, it is crucial to obtain diagnostic fragments and the corresponding mass spectrometry pattern, which will help to design more precise MS-based approaches for quantitative analysis and resolve structural isomers.
Therefore in this study we compiled a comprehensive electron impact ionization (EI) gas chromatography-mass spectrometry (GC-MS) analysis of OEA, which we used to develop a sensitive MRM method. We then measured the OEA concentration in several worm strains including lipl-4(tm4417), the long lived LIPL-4 overexpressor, the insulin/IGF mutated receptor strain (daf-2) and in the genetic dietary restriction model eat-2. Our methods should greatly facilitate the analysis of OEA and other NEAs in metabolomic studies.
Material and methods
Chemicals
The following chemicals were purchased by Sigma-Aldrich, GmbH: methanol, N-methyl-N-trimethylsilyl-trifluoroacetamide (MSTFA) and n-hexane (and cyclo-hexane). Chloroform was purchase by Merck KGaA (Darmstadt, Germany) and AM3102 (N-(1R)-2-hydroxy-1-methylethyl-9Z-octadecenamide) and oleoylethanolamine by Cayman, Biomol, GmbH. VEA (11-vaccenic acid ethanolamine) and PeEA (6-petroselinic acid ethanolamine) were synthetized by Waldemar Röhrig.28Strains
All strains were grown and maintained on NGM agar seeded with E. coli (OP50) at 20 °C. Standard procedures for culturing and maintaining strains were used.29 Strains used: N2 (wild-type), daf-2(e1370)III, eat2(ad465)II, lipl-4(tm4417)V and the worm strain raxls3(ges-1p::lipl-4::sl2gfp), named as LIPL-4(oe) was kindly provided by Meng Wang.OEA extraction and derivatization
Synchronized young adult worms were collected in four independent replicates and homogenized with a Qiagen tissue lyser for 30 min at 4 °C. The homogenate volume, which corresponds to 200 μg protein, was subjected to Bligh and Dyer extraction (chloroform![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol, 2
methanol, 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) for 1 hour at 4 °C (ref. 30) and dried using a speed-vac. Pure OEA, AM3102, or worm lipid extracts were derivatized with 20 μL of MSTFA for 30 minutes at 70 °C. Samples were dried using a constant argon flow to remove the excess of the derivatizing agent and reconstituted in 10 μL of cyclohexane prior to injection into the GC/MS.
1) for 1 hour at 4 °C (ref. 30) and dried using a speed-vac. Pure OEA, AM3102, or worm lipid extracts were derivatized with 20 μL of MSTFA for 30 minutes at 70 °C. Samples were dried using a constant argon flow to remove the excess of the derivatizing agent and reconstituted in 10 μL of cyclohexane prior to injection into the GC/MS.
GC-QQQ measurement
GC-MS analyses were carried out on a GC/MS (Triple Quadrupole GC 7890A + 7000 QQQ, Agilent, Waldbronn, Germany) interfaced with a robotic auto sampler PAL 1 system (PAL LHX-xt). One microliter of the derivatized worm sample was injected into the liner (4 mm splitless, single taper liner with glass wool) by pulsed-splitted mode at the temperature of 200 °C. Analytes were separated using a HP5-ms Ultra Inert column (15 m × 250 μm × 0.25 μm) (Agilent) using a modified thermal gradient proposed by Hardison S. et al.31 Briefly, initial oven temperature was set to 180 °C and held for 1 min, then the temperature was increased by 30 °C min−1 to 300 °C at 6 min and held for 1 min. At end the temperature was increased up to 325 °C by 30 °C min−1 in 1.83 min. The temperature of the transfer line was set to 200 °C and helium was used as the carrier gas at a flow rate of 3 mL min−1 in constant flow mode. Ions were generated using an electron impact ion source (−70 eV, 300 °C) and analyzed by a triple quadrupole. Product ion scan was acquired from 50 to 400 m/z using CE of 0, 5, 10 and 20. Multiple reaction monitoring measurement was achieved using the transition for OEA (397.3 ≥ 188) and AM3102 (411.3 ≥ 202.2) using wide resolution.Data were analyzed using MassHunter Work station Software, Qualitative Analysis, Version B.06.00. Relative response for OEA was calculated by dividing the peak area of the analyte to the AM3102 peak area and further normalized to protein concentration.
ESI-MS analysis of OEA
AM3102 (methyl-OEA) and OEA were analyzed using robotic nano-flow ion source (TriVersa Nanomate, Advion Inc.) coupled with a high resolution accurate mass (HRAM) mass spectrometer (Q-Exactive Plus, Thermo Fischer Scientific GmbH, Bremen, Germany). The robotic source was equipped with a nano electrospray chip with 1.4 K ionization voltage and 0.3 psi back pressure. The S-lenses RF levels were set to 65. Mass spectra were acquired from 200 to 600 m/z at resolution 70.000 at m/z 400 in the positive ion mode using an isolation window of 0.4 amu. Tandem mass spectra were obtained by CID using 10, 20, 25 normalized collision energy (NCE). Data were analyzed using Xcalibur software version 4.0.Pharyngeal pumping rate assay
Pharyngeal pumping was assessed by observing the number of pharyngeal contractions during a 30 s interval using twenty synchronized young adult worms in three biological replicates as previously described.32Statistical analysis
Statistical analysis were performed using GraphPad Prism software 5.04. The p-values were calculated with one-way ANOVA test and Dunnet's test multiple comparisons.Results
EI and collision-induced dissociation (CID) behavior of OEA
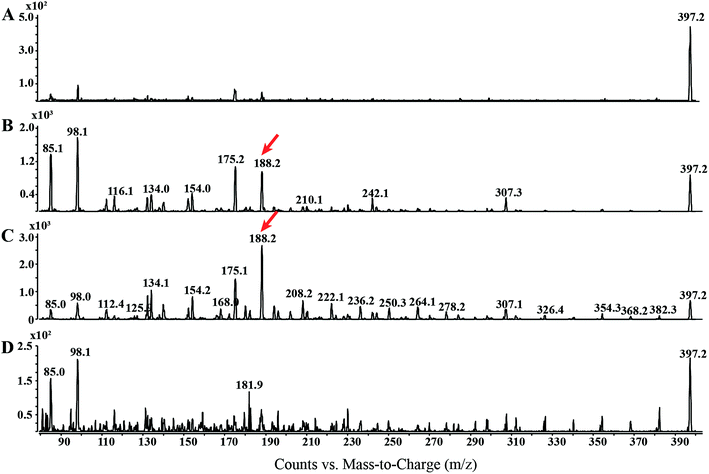
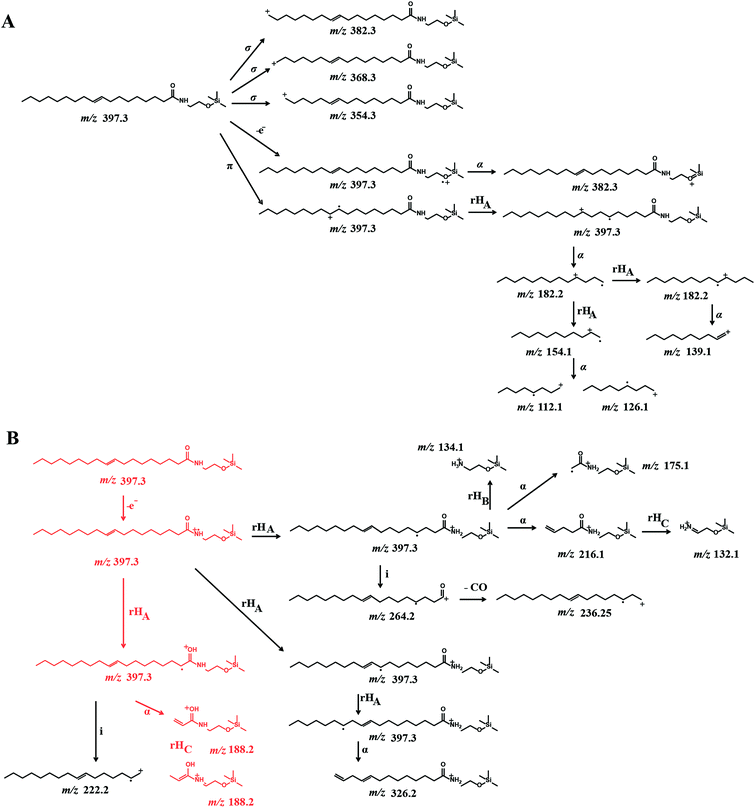
In order to garner useful diagnostic structural information, we chose to use EI combined with CID to analyze OEA fragmentation patterns. The EI-CID fragmentation behavior of OEA was investigated by performing a product ion scan (PIS) at different collision energies (CE), namely 0, 5, 10, 20 (Fig. 1). Mass spectra of OEA were characterized by a molecular ion [M˙+] base peak at m/z 397.3. OEA did not undergo in-source fragmentation if no collision gas was applied, and only few fragments were detected in the tandem mass spectrum as shown in Fig. 1A. Additionally high collision energy (>20 CE) resulted in loss of diagnostic ions (Fig. 1D). Optimal tandem mass spectra were thus retrieved by application of low collision energy (CE 10) to obtain informative structural fragments (Fig. 1C).EI-MS generated characteristic ions for trimethylsilyl (TMS) derivatives, such as a product ion at m/z 382.3, which corresponds to the loss of one methyl group [M-15]+, and loss of trimethylsilanol group [M-90]+ at m/z 307.1.33,34 The tandem mass spectrum was also characterized by the presence of different ions, which are presumably formed by hydrogen elimination (rH), alpha (α), sigma (σ) and allylic cleavage35 (Fig. 2). Sigma cleavage at the allyl fatty acid position led to the formation of ions at 382.3 m/z (C22H44NO2Si+), at m/z 368.3 (C21H42NO2Si+) and at m/z 354.3 (C20H40NO2Si+). Product ion at m/z 382.3 can also be generated by alpha cleavage at the trimethylsilyl group with the formation of oxonium ion (Fig. 2A). Allylic cleavage is the most significant fragmentation mode due to resonance stabilization.36 Cleavages occurred at the double bond with hydrogen elimination leading to product ion at m/z 154.2 (C11H22˙+), and a further fragmentation by alpha cleavage may generate two daughter ions at m/z 112.1 (C8H16˙+) and at m/z 126.1 (C9H18˙+). Fragmentation ion at m/z 139.9 (C10H19+) can be formed by alpha cleavage from the ion at m/z 182.2 (C13H26˙+) (Fig. 2A). The ion at m/z 85 (C6H13+) is already reported to be formed by McLafferty rearrangement.37 The OEA precursor ion at m/z 397.3 may also undergo even electron ion cleavage hydrogen elimination (rHB) leading to the formation of product ions at m/z 134.1 (C5H16NOSi+) (Fig. 2B), and the daughter ion at m/z 132.2 (C5H14NOSi+) can be generated by gamma hydrogen elimination (rHC). Additionally the ion at m/z 175.1 (C7H17NO2Si˙+) could be formed by cleavage at the alpha carbon from the precursor ion.
Further specific fragments are formed by induction cleavage at the nitrogen position, which leads to the formation of ion at m/z 264.1 (C18H32O˙+), which upon loss of –CO forms the ion at m/z 236.2 (C17H32˙+).38 The characteristic ion of oleic acid fragment was detected at m/z 222.1 (C16H30˙+) and is formed by hydrogen elimination and subsequent induction cleavage.39,40
Finally the ion at m/z 188.2 (C8H18NO2Si˙+) from the OEA precursor is proposed to be formed by an initial cleavage at the carbon alpha and a possible structural rearrangement obtained via gamma hydrogen elimination (rHC). Under these EI-CID condition, no product ion at m/z 267, which correspond to the loss of [M-130]+ was observed.41
We additionally compared the formation of similar product ions using soft ionization techniques, such as electrospray ionization (ESI) in combination with lower collision energy using a Q exactive platform. ESI-CID-MS analysis of OEA, in positive ion mode, led to the formation of three major fragments, the precursor ion at m/z 326.30518 (C20H40O2N), the ion [M-H2O]+ and the ion at m/z 62.06011, which was assigned to the ethanolamine moiety (C2H8ON) (ESI S1†). Although we retrieved the ethanolamine moiety product ion from the ESI-CID-MS analysis, we obtained more structural information using the EI-CID-GC/MS methodology.
Method validation
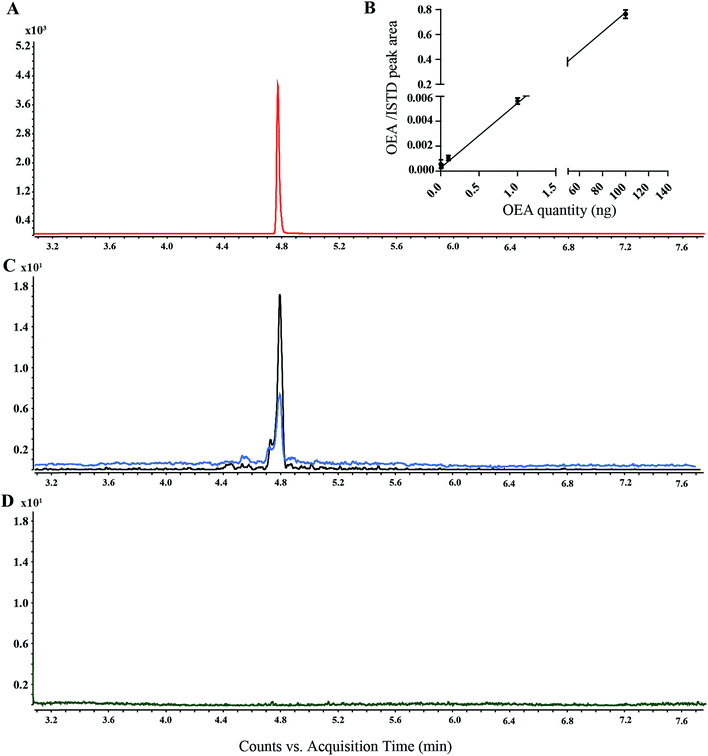
OEA and the corresponding methylated analogue (AM3102) were separated using a low bleed GC column with a stationary phase composed by phenyl-methylpolysiloxane.42 Oleoylethanolamine was detected at 4.8 min, whereas AM3102 at 4.74 min. For characterization of the instrumental parameters, the ion at m/z 188.2 and the ion at m/z 202.2 were selected for the MRM analysis of OEA (397.3 ≥ 188.2) and for AM3102 (411.3 ≥ 202.2) respectively, which led to the detection of a single chromatographic peak for pure standards (Fig. 3A).For the assessment of the method sensitivity and linearity parameters, OEA was dissolved in ethanol and was serially diluted from 100 ng to 1 pg. Each dilution step was derivatized and injected (Fig. 3B). The limits of detection (LOD) and quantification (LOQ) as well as the linear dynamic range (LDR) are summarized in the Table 1. The GC-MS/MS method exhibited high interday and intraday accuracy. The intraday error for methyl OEA and OEA were 6.67 and 8.89 (RSD%), respectively, whereas 9.69 and 9.31 were the error for the interday measurement over 5 days (Table 2). This method has an LOQ, 0.085 ng (0.2 pmol), and a precision <10%, which is below the range of previously reported methods (Table S1†). Although some publications have not reported these parameters, we could compare our method with works from Fontana et al., which used a similar instrument setup and obtained higher LOQ using a different derivatization step.41 ESI based methods showed a higher LOQ compared to the GC-MS approaches, nevertheless they have a precision between 8 and 14% (Table S1†).
| Compound | m/z | t R | LOD (pmol) | LOQ (pmol) | LDR | Slope | Intercept | R 2 | Recovery (%) |
|---|---|---|---|---|---|---|---|---|---|
| OEA | 397.3 | 4.77 | 0.07 | 0.2 | 0.54 × 102 | 6.5 × 103 | 1.3 × 103 | 0.99 | 83.63 |
| Compound | Intraday precision (n = 5) | Inter-day precision n = 5 per day | ||
|---|---|---|---|---|
| t R (min) ± SD (RSD%) | Peak area ± SD (RSD%) | t R (min) ± SD (RSD%) | Peak area ± SD (RSD%) | |
| AM3102 | 4.72 ± 0.005 (0.01) | 4443 ± 296.71 (6.67) | 4.73 ± 0.001 (0.026) | 4327 ± 410 (9.69) |
| OEA | 4.77 ± 0.0005 (0.01) | 4405 ± 391.76 (8.89) | 4.77 ± 0.005 (0.11) | 4199 ± 370.79 (9.31) |
For the analysis of recovery, we spiked in AM3102 (100 ng) into the lipid matrix of C. elegans extracts (n = 4). The calculated recovery was 83%, which is similar to previously published data.22,43 We additionally investigated the GC/MS behavior of two isomers of OEA, namely VEA (11-vaccenic acid ethanolamine) and PeEA (6-petroselinic acid ethanolamine). Our new method succeeded in the separation of the three isomers, which showed different retention times (ESI S2†). However, when we performed semi-quantitative analysis on transgenic worm lines, we only detected a single peak corresponding to OEA (Fig. 3C), presumably because OEA is known to be synthesized in vivo, whereas VAE and PeEA are up taken from different food sources, such as, milk and plant products.44,45
Optimization of OEA derivatization conditions
In this study, we initially used Fluka III, which consist of a trimethylsilylating mixture comprised of BSTFA, MSTFA and trimethylsilyl imidazole. Using Fluka III it was possible to detect the signal corresponding to pure OEA as well as for AM3102 (data not shown). However, it proved to be necessary to regularly clean the ion source and change the liner frequently in order to obtain reproducible results. For this reason we only used the N,O-bis(trimethylsilyl)trifluoroacetamide (MSTFA) containing reagents for the sylation experiment. Although the difference in silyl donor ability between BSTFA, trimethylsilyl imidazole and MSTFA may be assumed to be small, the use of the single MSTFA agent did not require constant cleaning and led to more reproducible results. The time of derivatization was set to 30 min, although derivatized sample was detectable after 10 min, in accordance with previous works.21Derivatization is used to increase the thermal stability and the volatility of the compounds, to avoid the analyte being absorbed into the injector.46,47 Our findings also suggest that the silylation derivatization was required for the detection of OEA, since other conditions failed to provide a detectable peak (Fig. 3D). OEA was reported by many researchers as TMS derivatives, which led to an improvement of the chromatographic performance by increased GC separation.31,48,49 However, Devane et al., and Maccarone et al., also reported non-derivatized anandamide from various biological samples.50–52
OEA quantification in worms
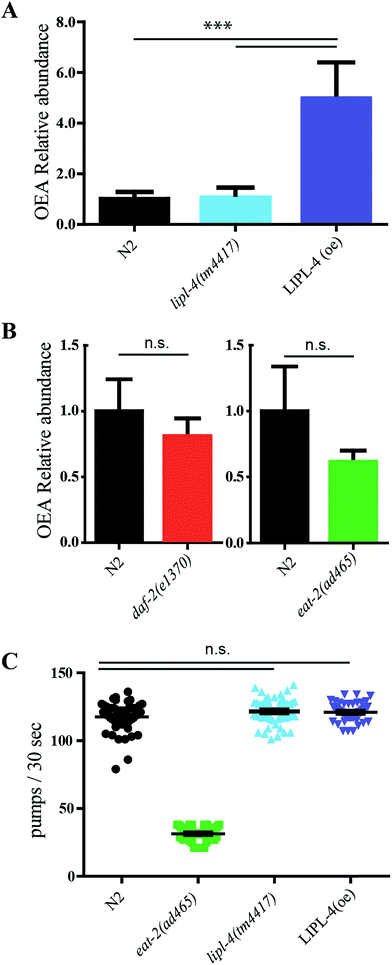
The novel MRM method was used to quantify OEA in four different worm strains. The relative level of OEA in the diverse genotypes was normalized to the wild type (N2) and plotted in Fig. 4A and B.We first focused on lipl-4 since its overexpression has been previously associated with increased production of OEA and extended life. As expected, the LIPL-4 overexpressor strain, raxls3(ges-1p::lipl-4::sl2gfp), showed a significant increase of 5-fold compared to the control sample. However, no significance changes of OEA levels were observed in the lipl-4(tm4417)V loss of function mutant control compared to the wild type control, suggesting that other activities in the nematode may also contribute basal levels of OEA.
Low levels of EPEA are associated with the genetic model of dietary restriction, eat-2, and this NAE has been considered to be pro-aging.18,53 Moreover lipl-4 expression is elevated in the long lived daf-2/InsR mutant.14,54 We therefore wondered whether either of these longevity models exhibited elevated levels of the anti-aging OEA. Surprisingly, no significant changes in OEA levels were observed for eat-2 or daf-2 (Fig. 4B), and if anything, OEA levels tended to be decrease in these genotypes.
Worm feeding behavior
OEA has been previously shown to be anorexigenic in mammals.55,56 We wondered whether elevated OEA could affect worm feeding behavior, which was assessed by monitoring the pharyngeal contraction rates in the transgenic genotypes. As previously shown, the control eat-2 decreased pumping rate.57 However, neither lipl-4 mutant nor LIPL-4 overexpressor, had any influence on pharyngeal pumping rates compared to wild type (Fig. 4C).Discussion
The present study illustrates a useful new methodology to accurately quantify OEA in C. elegans. OEA and other NAEs have been characterized by GC-MS coupled with chemical ionization, fast atom bombardment and electron impact coupled with often single/triple quadrupole and ion traps (Table S1†). In this work we revised and developed an EI-CID GC-MS approach to retrieve new diagnostic ion fragments for OEA that are simple, accurate, and sensitive. This new methodology was initially used to investigate OEA in C. elegans, but it can be translated also to other species.As expected, EI in combination with CID provided mass spectra rich in daughter ions. Using this approach we were able to unambiguously detect OEA using the novel fragment ion (C8H18NO2Si˙+). Although this ion source is commonly coupled with GC, few working groups have reported the use of electron impact ionization for the analysis of endocannabinoids. For example, Lucanic and coworkers used a GC system coupled to an ion trap with higher sensitivity, but this method requires significantly higher amounts of the starting material, (≈200 mg) for each single run. Furthermore endocannabinoids were quantified using diagnostic ions, such as, [M-15]+ (loss of one methyl group), and [M-90]+ (loss of trimethylsilylanol group), which might lead to non-specific signals.14,58,59
Additionally, most current methods require the extraction of endocannabinoids and further enrichment using solid phase extraction (SPE) or analytical fractionation.43 Pre-separation steps are often required to remove high abundance analytes, such as phospholipids and triacylglycerides, which represent in many cases the 60–80% of the total lipid extract. Recent works of Hardison S. et al. reported an indispensable purification step from the lipid crude extract using C18 SPE for the detection of AEA from the brain tissue.31 These purification methodologies rely on silica gel chromatography or homemade silica/CHCl3 containing Pasteur pipette.60 Furthermore NEAs from human tissues have been also fractionated by normal phase and reverse phase HPLC using silica column and C18 respectively.22
In contrast with the previous methods, we devised an easy and fast approach without any separation or fractionation, giving rise to more accurate and facile quantitation.
C. elegans possesses a smaller genome, proteome and metabolome than those of mammals.61 In comparison with the analysis of a complex tissue, which may lose proteins and metabolites during surgical removal, the worm lysate contains all 959 somatic cells. Note that because of the simplicity of the nematode biological matrix, the use of Bligh and Dyer in our investigations was sufficient to extract OEA without a pre-separation step. However, alternative methods, such as modified Folch extraction or basic and acid hydrolysis are required for plasma and other mammalian tissues.22,41
Previous studies suggest that lysosomal LIPL-4 helps produce OEA which works through nuclear hormone receptor NHR-80/NHR-49 complexes to increase life span.15,18 Using our new method we demonstrate that LIPL-4 is partially responsible for the production of OEA. On the one hand, we found that OEA production is highly correlated with the overexpression level of the LIPL-4 transgene. On the other hand, we could not detect a significant reduction in the lipl-4(tm4417)V mutant, suggesting that OEA might also be produced by other lipases. In this regard, it was previously suggested that lipl-4(tm4417)V is not a complete loss of function but a partial reduction of activity.18 In C. elegans at least eight lipases have been found, namely LIPL-1 to LIPL-8, which suggests that other lipases might be promiscuously involved in OEA production.53 Lysosomal lipases have also been implicated in affecting life span in other models.62 Homozygote knockout mice for lysosomal acid lipase (LIPA) exhibit massive triglyceride storage and shortened life span.63,64
Conceivably, OEA might not be the only product of LIPL-4 and other N-acylethanolamines-like molecules involved in ageing processes. Different NEAs have opposite effects on regulation of life span and how their specific activities interact remain unresolved. Folick et al. have shown that OEA is anti-aging: lipl-4 overexpression increases its production and supplementation with OEA and analogues extends life span. Works of Seah et al. indicate an increase expression of lysosomal lipase upon silencing of the nutrient sensor LET-363/mTOR.53 In contrast, Lucanic et al. found that the endocannabinoid, EPEA, is pro-aging. EPEA levels are down under dietary restriction, and its supplementation abolishes longevity due to dietary restriction and S6 kinase reduction.14 Along these lines, overexpression of nape-1 (N acyl phosphatidyl ethanolamines specific phospholipase D), the enzyme that carries out the last steps of biosynthesis, diminishes longevity.12,14 Conversely, overexpression of FAAH fatty acid amide hydrolase, the enzyme that degrades NAEs, enhances longevity. Based on other species, FAAH would be expected to degrade not only EPEA, but OEA as well. If so, EPEA depletion might be epistatic to OEA depletion for effects on life span. Future studies on the relative levels of various NAEs in these genotypes may shed light on the matter.
When we investigated levels of OEA in two long-lived models, namely daf-2 and the DR model eat-2, we found surprisingly that neither mutant affected levels of OEA, despite evidence that suggests these strains upregulate various lipases. Thus OEA triggered longevity might be specific to LIPL-4 overexpression. Alternately other NAEs may contribute to life span control.
OEA has been shown to affect feeding behavior via PPARα receptor to induce anorexia action in rodents.55 However, in the simple worm model, OEA might be not directly involved in food behavior or control of food intake,65 since we saw no obvious effect on feeding behavior in LIPL-4 overexpressing worms. Recent work from Oakes et al. indicated that 2-AG and AEA are the effectors of many cannabinoid-dependent behaviors, which include feeding as well as nociception and locomotion.66 Taken together, these observations may indicate that OEA is more specifically involved in the aging process rather than canonical cannabinoid and noradrenergic signaling in C. elegans.
Conclusion
We introduced a new methodology for the quantification in vivo of OEA in the nematode C. elegans. The detailed structural information in this study can be used to understand acyl-ethanolamines fragmentation mechanism and can be expanded to other endocannabinoids. To our knowledge this is the first report of the relative abundance of OEA in LIPL-4 over expressor, daf-2 and eat-2 strains. Moreover our studies reveal that LIPL-4 does not influence the feeding behavior in the nematode. These findings provide the bases for future mechanistic studies to investigate NAEs and the cannabinoid system in C. elegans.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors are thankful to Dr Meng Wang for kindly providing the LIPL-4 overexpressor strain. VEA and PeEA standards were kindly provided by Waldemar Röhrig. This work was supported by the Max Planck Society.References
- R. I. Wilson and R. A. Nicoll, Science, 2002, 296, 678–682 CrossRef CAS PubMed.
- A. Villanueva, S. M. Yilmaz, W. R. Millington, R. A. Cutrera, D. G. Stouffer, L. H. Parsons, J. F. Cheer and C. Feleder, Shock, 2009, 32, 614–620 CrossRef CAS PubMed.
- M. H. Bracey, M. A. Hanson, K. R. Masuda, R. C. Stevens and B. F. Cravatt, Science, 2002, 298, 1793–1796 CrossRef CAS PubMed.
- E. Dainese, G. De Fabritiis, A. Sabatucci, S. Oddi, C. B. Angelucci, C. Di Pancrazio, T. Giorgino, N. Stanley, M. Del Carlo, B. F. Cravatt and M. Maccarrone, Biochem. J., 2014, 457, 463–472 CrossRef CAS PubMed.
- M. Kaczocha, S. T. Glaser, J. Chae, D. A. Brown and D. G. Deutsch, J. Biol. Chem., 2010, 285, 2796–2806 CrossRef CAS PubMed.
- H. R. Freitas, A. R. Isaac, R. Malcher-Lopes, B. L. Diaz, I. H. Trevenzoli and R. A. De Melo Reis, Nutr. Neurosci., 2017, 1–20, DOI:10.1080/1028415x.2017.1347373.
- I. Ivanov, P. Borchert and B. Hinz, Anal. Bioanal. Chem., 2015, 407, 1781–1787 CrossRef CAS PubMed.
- S. I. Pastuhov, K. Fujiki, P. Nix, S. Kanao, M. Bastiani, K. Matsumoto and N. Hisamoto, Nat. Commun., 2012, 3, 1136 CrossRef PubMed.
- M. Gabrielli, N. Battista, L. Riganti, I. Prada, F. Antonucci, L. Cantone, M. Matteoli, M. Maccarrone and C. Verderio, EMBO Rep., 2015, 16, 213–220 CrossRef CAS PubMed.
- X. Xu, H. Guo, Z. Jing, L. Yang, C. Chen, L. Peng, X. Wang, L. Yan, R. Ye, X. Jin and Y. Wang, J. Cardiovasc. Pharmacol., 2016, 68, 280–291 CrossRef CAS PubMed.
- M. Koch, Front. Neurosci., 2017, 11, 293 CrossRef PubMed.
- N. Harrison, M. A. Lone, T. K. Kaul, P. Reis Rodrigues, I. V. Ogungbe and M. S. Gill, PLoS One, 2014, 9, e113007 Search PubMed.
- S. I. Pastuhov, K. Matsumoto and N. Hisamoto, Genes Cells, 2016, 21, 696–705 CrossRef CAS PubMed.
- M. Lucanic, J. M. Held, M. C. Vantipalli, I. M. Klang, J. B. Graham, B. W. Gibson, G. J. Lithgow and M. S. Gill, Nature, 2011, 473, 226–229 CrossRef CAS PubMed.
- L. R. Lapierre, A. Melendez and M. Hansen, Autophagy, 2012, 8, 144–146 CrossRef CAS PubMed.
- L. R. Lapierre, S. Gelino, A. Melendez and M. Hansen, Curr. Biol., 2011, 21, 1507–1514 CrossRef CAS PubMed.
- M. C. Wang, E. J. O'Rourke and G. Ruvkun, Science, 2008, 322, 957–960 CrossRef CAS PubMed.
- A. Folick, H. D. Oakley, Y. Yu, E. H. Armstrong, M. Kumari, L. Sanor, D. D. Moore, E. A. Ortlund, R. Zechner and M. C. Wang, Science, 2015, 347, 83–86 CrossRef CAS PubMed.
- R. Mechoulam, S. Ben-Shabat, L. Hanus, M. Ligumsky, N. E. Kaminski, A. R. Schatz, A. Gopher, S. Almog, B. R. Martin and D. R. Compton, et al. , Biochem. Pharmacol., 1995, 50, 83–90 CrossRef CAS PubMed.
- K. Kempe, F. F. Hsu, A. Bohrer and J. Turk, J. Biol. Chem., 1996, 271, 17287–17295 CrossRef CAS PubMed.
- H. F. Kasai, M. Tsubuki, K. Takahashi, T. Honda and H. Ueda, Anal. Sci., 2003, 19, 1593–1598 CrossRef CAS PubMed.
- J. Palandra, J. Prusakiewicz, J. S. Ozer, Y. Zhang and T. G. Heath, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2009, 877, 2052–2060 CrossRef CAS PubMed.
- U. Yapa, J. J. Prusakiewicz, A. D. Wrightstone, L. J. Christine, J. Palandra, E. Groeber and A. J. Wittwer, Anal. Biochem., 2012, 421, 556–565 CrossRef CAS PubMed.
- L. Walter, A. Franklin, A. Witting, T. Moller and N. Stella, J. Biol. Chem., 2002, 277, 20869–20876 CrossRef CAS PubMed.
- M. Igarashi, N. V. DiPatrizio, V. Narayanaswami and D. Piomelli, Biochim. Biophys. Acta, Mol. Cell Biol. Lipids, 2015, 1851, 1218–1226 CrossRef CAS PubMed.
- G. Appendino, A. Ligresti, A. Minassi, M. G. Cascio, M. Allara, O. Taglialatela-Scafati, R. G. Pertwee, L. De Petrocellis and V. Di Marzo, J. Med. Chem., 2009, 52, 3001–3009 CrossRef CAS PubMed.
- R. Vitale, G. Ottonello, R. Petracca, S. M. Bertozzi, S. Ponzano, A. Armirotti, A. Berteotti, M. Dionisi, A. Cavalli, D. Piomelli, T. Bandiera and F. Bertozzi, ChemMedChem, 2014, 9, 323–336 CrossRef CAS PubMed.
- W. Röhrig, R. Waibel, C. Perlwitz, M. Pischetsrieder and T. Hoch, Anal. Bioanal. Chem., 2016, 408, 6141–6151 CrossRef PubMed.
- S. Brenner, Genetics, 1974, 77, 71–94 CAS.
- D. B. Magner, J. Wollam, Y. Shen, C. Hoppe, D. Li, C. Latza, V. Rottiers, H. Hutter and A. Antebi, Cell Metab., 2013, 18, 212–224 CrossRef CAS PubMed.
- S. Hardison, S. T. Weintraub and A. Giuffrida, Prostaglandins Other Lipid Mediators, 2006, 81, 106–112 CrossRef CAS PubMed.
- C. Huang, C. Xiong and K. Kornfeld, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 8084–8089 CrossRef CAS PubMed.
- J. F. Rontani and C. Aubert, J. Am. Soc. Mass Spectrom., 2008, 19, 66–75 CrossRef CAS PubMed.
- J. J. Myher, L. Marai and A. Kuksis, Anal. Biochem., 1974, 62, 188–203 CrossRef CAS PubMed.
- J. F. Rontani, S. T. Belt, T. A. Brown and C. Aubert, Rapid Commun. Mass Spectrom., 2014, 28, 1937–1947 CrossRef CAS PubMed.
- V. Spitzer, Prog. Lipid Res., 1996, 35, 387–408 CrossRef CAS PubMed.
- C. E. Kientz and A. Verweij, J. Chromatogr. A, 1986, 355, 229–240 CrossRef CAS.
- A. Loidl-Stahlhofen, W. Kern and G. Spiteller, J. Chromatogr. B: Biomed. Sci. Appl., 1995, 673, 1–14 CrossRef CAS PubMed.
- Y. J. Xu and J. Zhang, Bioanalysis, 2013, 5, 1527–1543 CrossRef CAS PubMed.
- L. R. Alexander, J. B. Justice Jr and J. Madden, J. Chromatogr., 1985, 342, 1–12 CrossRef CAS PubMed.
- A. Fontana, V. Di Marzo, H. Cadas and D. Piomelli, Prostaglandins, Leukotrienes Essent. Fatty Acids, 1995, 53, 301–308 CrossRef CAS.
- X. Huang, J. Xue, Y. Wang, X. Wu and H. Tong, Anal. Methods, 2012, 4, 1132–1141 RSC.
- G. G. Muccioli and N. Stella, Anal. Biochem., 2008, 373, 220–228 CrossRef CAS PubMed.
- L. Ding, Y. Wang, M. Kreuzer, X. Guo, J. Mi, Y. Gou, Z. Shang, Y. Zhang, J. Zhou, H. Wang and R. Long, J. Dairy Res., 2013, 80, 410–417 CrossRef CAS PubMed.
- F. Ellenbracht, W. Barz and H. K. Mangold, Planta, 1980, 150, 114–119 CrossRef CAS PubMed.
- U. Shafique, S. Schulze, C. Slawik, S. Kunz, A. Paschke and G. Schuurmann, Anal. Chim. Acta, 2017, 949, 8–22 CrossRef CAS PubMed.
- Y. Y. Lee, J. M. Galano, C. Oger, C. Vigor, R. Guillaume, J. Roy, J. Y. Le Guennec, T. Durand and J. C. Lee, Lipids, 2016, 51, 1217–1229 CrossRef CAS PubMed.
- S. B. Sachs and F. Woo, J. Forensic Sci., 2007, 52, 308–319 CrossRef CAS PubMed.
- T. C. Kirkham, C. M. Williams, F. Fezza and V. Di Marzo, Br. J. Pharmacol., 2002, 136, 550–557 CrossRef CAS PubMed.
- W. A. Devane, L. Hanus, A. Breuer, R. G. Pertwee, L. A. Stevenson, G. Griffin, D. Gibson, A. Mandelbaum, A. Etinger and R. Mechoulam, Science, 1992, 258, 1946–1949 CAS.
- W. A. Devane, F. A. Dysarz III, M. R. Johnson, L. S. Melvin and A. C. Howlett, Mol. Pharmacol., 1988, 34, 605–613 CAS.
- M. Maccarrone, M. Attina, A. Cartoni, M. Bari and A. Finazzi-Agro, J. Neurochem., 2001, 76, 594–601 CrossRef CAS PubMed.
- N. E. Seah, C. D. de Magalhaes Filho, A. P. Petrashen, H. R. Henderson, J. Laguer, J. Gonzalez, A. Dillin, M. Hansen and L. R. Lapierre, Autophagy, 2016, 12, 261–272 CrossRef CAS PubMed.
- J. L. Watts, J. Clin. Med., 2016, 5, 19 CrossRef PubMed.
- A. Romano, R. Coccurello, G. Giacovazzo, G. Bedse, A. Moles and S. Gaetani, BioMed Res. Int., 2014, 2014, 203425 Search PubMed.
- P. D. Cani, M. L. Montoya, A. M. Neyrinck, N. M. Delzenne and D. M. Lambert, Br. J. Nutr., 2004, 92, 757–761 CrossRef CAS PubMed.
- N. F. Trojanowski, D. M. Raizen and C. Fang-Yen, Sci. Rep., 2016, 6, 22940 CrossRef CAS PubMed.
- T. Yoshida, J. Chromatogr. Sci., 2017, 1–9, DOI:10.1093/chromsci/bmx048.
- L. Ruiz-Aceituno, C. Carrero-Carralero, A. I. Ruiz-Matute, L. Ramos, M. L. Sanz and I. Martinez-Castro, J. Chromatogr. A, 2017, 1484, 58–64 CrossRef CAS PubMed.
- S. Gonzalez, J. Manzanares, F. Berrendero, T. Wenger, J. Corchero, T. Bisogno, J. Romero, J. A. Fuentes, V. Di Marzo, J. A. Ramos and J. Fernandez-Ruiz, Neuroendocrinology, 1999, 70, 137–145 CrossRef CAS PubMed.
- J. J. Moresco, P. C. Carvalho and J. R. Yates III, J. Proteomics, 2010, 73, 2198–2204 CrossRef CAS PubMed.
- H. Du, D. P. Witte and G. A. Grabowski, J. Lipid Res., 1996, 37, 937–949 CAS.
- H. Du, M. Heur, M. Duanmu, G. A. Grabowski, D. Y. Hui, D. P. Witte and J. Mishra, J. Lipid Res., 2001, 42, 489–500 CAS.
- H. Du, M. Duanmu, D. Witte and G. A. Grabowski, Hum. Mol. Genet., 1998, 7, 1347–1354 CrossRef CAS PubMed.
- R. Shen, Protein Cell, 2011, 2, 689–690 CrossRef PubMed.
- M. D. Oakes, W. J. Law, T. Clark, B. A. Bamber and R. Komuniecki, J. Neurosci., 2017, 37, 2859–2869 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ay00396c |
| This journal is © The Royal Society of Chemistry 2018 |