Investigating extracellular electron transfer of Rikenella microfusus: a recurring bacterium in mixed-species biofilms†
M.
Grattieri
 a,
K.
Hasan
a,
K.
Hasan
 a,
R. D.
Milton
a,
R. D.
Milton
 a,
S.
Abdellaoui
a,
M.
Suvira
b,
B.
Alkotaini
a,
S.
Abdellaoui
a,
M.
Suvira
b,
B.
Alkotaini
 a and
S. D.
Minteer
a and
S. D.
Minteer
 *a
*a
aDepartment of Chemistry, University of Utah, 315 S 1400 E Room 2020, Salt Lake City, Utah 84112, USA. E-mail: minteer@chem.utah.edu
bDepartment of Chemistry and Biochemistry, Loyola University Chicago, 1032 W Sheridan Rd, Chicago, IL 60660, USA
First published on 29th June 2017
Abstract
The optimization of bioelectrochemical systems operating with microorganisms requires a deep understanding of the extracellular electron transfer (EET) processes, however, EET studies have been reported for few bacterial species. Herein, the bioelectrocatalytic properties of Rikenella microfusus, an anaerobic bacterium commonly found in electrode-colonizing biofilms, have been investigated for the first time.
The capability of microorganisms to transfer electrons to/from an electrode surface is critical for the development of bioelectrochemical systems.1,2 This process is known as extracellular electron transfer (EET), since electrons must be transferred outside of the cellular membrane of the microorganisms to/from an insoluble electron acceptor (i.e. an electrode or minerals).3,4 Numerous technologies rely on this EET, such as microbial fuel cells (MFCs), microbial electrolysis cells, microbial desalination cells and microbial solar cells.5–8 However, established EET mechanisms are available only for a few microorganisms, for example, Geobacter sulfurreducens,9,10Shewanella oneidensis,11–13Rhodopseudomonas palustris.14 Reporting and clarifying EET mechanisms for different microorganisms would be beneficial not only for the understanding of bacterial metabolism and their interactions, but would also impact the development and optimization of bioelectrochemical systems.14 A MFC is a bioelectrochemical system able to convert chemical energy from civil and industrial wastewater into electrical energy.15 The possible applications of this technology span from wastewater treatment, energy production and biosensors.16,17 Pure cultures of bacteria are often applied during the development of MFCs at the lab scale,18–20 in order to control biofilm formation and EET properties.21 However, for practical applications, a mixed bacterial community forms a biofilm on the electrodes, since wastewater solutions and other types of biomass are commonly utilized as inoculum.22–26 It has been recently shown that mixed species biofilms can achieve conductivity similar to pure culture biofilms of Geobacter sulfurreducens, although microorganisms other than Geobacteraceae constituting nearly half of the microbial community have not been accounted to contribute to the EET.19,27 Recently, different reports have shown a relevant presence of Bacteroidetes in the microbial community of bioelectrochemical systems inoculated with wastewater or digestate.19,28,29 Croese et al. analyzed the bacterial population of a biofilm developed in a microbial electrolysis cell, where Bacteroidetes comprised 17% of the total population and showed similarity (below 92%) to cultured Rikenella microfusus (R. microfusus).28R. microfusus belongs to the phylum Bacteroidetes, genus of Bacteroides, order of Bacteroidales. The latter was isolated from fecal materials, it is an obligated anaerobic, nonmotile and Gram-negative bacterium,30 which is capable of oxidizing a number of substrates such as glucose, mannose and lactose.31 The oxidizing agent that is ultimately reduced by the bacterium remains unclear. Despite the potential role of this bacterium in the electrochemical processes characterizing the MFCs, its contribution has not been studied and there is a lack of information regarding its metabolism.30,32 Herein, we report the bioelectrocatalytic properties of pure R. microfusus biofilms and their role in EET processes within mixed-culture MFCs. Pure biofilms of R. microfusus were necessary in this study in order to avoid contribution of other bacteria to the electrochemical response under investigation. However, the obtained results are discussed taking into consideration the interaction between different bacterial species in mixed consortium.
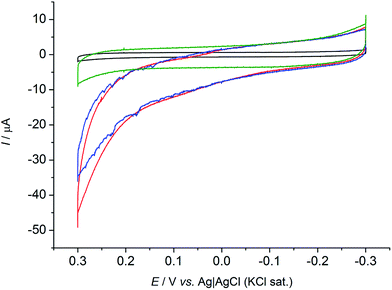
Cyclic voltammetric tests (CVs) were performed under anaerobic conditions to explore the capability of R. microfusus to communicate with the electrode surface. The responses obtained from carbon cloth electrodes exposed for two days to R. microfusus before the electrochemical experiments to allow bacterial colonization of the electrode are reported in Fig. 1. A catalytic current due to oxidation of the substrate (50 mM glucose) can be clearly identified when R. microfusus was present (red and blue lines). The result was confirmed by replicate experiments (Fig. S2, ESI†). The absence of bacterial cells (Fig. 1 black line) revealed no catalytic current, confirming the ability of R. microfusus to oxidize glucose. Riboflavin (RB) has been reported as a self-secreted redox mediator used by bacterial cells during EET processes,33 specifically with Shewanella oneidensis MR-1 or Shewanella sp. MR-4.34 Accordingly, possible effects of exogenous RB (6.25 μM, Fig. 1 red line) were explored. The onset potential for glucose oxidation (approximately −0.12 V vs. Ag|AgCl KCl sat.) was not significantly modified, however, the maximum current achieved for glucose oxidation increased from 35 ± 2 to 44 ± 4 μA (+25%). To investigate if the obtained current response was due only to the cells in direct contact with the electrode surface, with no electrode mediator involved in the process, an electrode was transferred to a fresh sterile electrolyte (50 mM glucose) and immediately tested by CV (Fig. 1 green line). The change of electrolyte caused the loss of the catalytic current previously detected. This result confirms that redox substances secreted by the bacterial cells are necessary for EET.
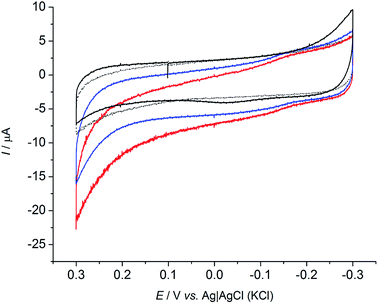
To further confirm the presence of redox substances in solution, a set of experiments was conducted using a carbon cloth electrode that was not previously exposed to R. microfusus. The electrode was directly placed in the electrochemical cell containing bacterial cells with the same setup as the previous experiments and immediately tested (Fig. 2). A clear redox couple with a formal reduction potential (E0′) of −0.039 V vs. Ag|AgCl (KCl sat.) was obtained when bacterial cells are present in solution (blue line). Evaluation of a clean electrode in sterile electrolyte did not yield a redox couple, indicating that the origin of the peak is related with bacterial metabolism. The inset of Fig. 2 shows a comparison of the CVs obtained with the fresh electrodes (blue and black lines) and the CV obtained for an electrode exposed to bacterial cells (red line). Interestingly, the onset potential for the oxidation of glucose corresponds to the onset potential of the anodic peak (Epa) indicating that the EET might be mediated by the redox species detected. Further studies will be dedicated to determine the redox species involved in the EET mechanism of R. microfusus.
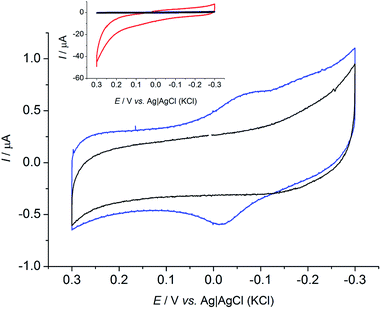 | ||
| Fig. 2 Cyclic voltammograms with a fresh carbon cloth electrode in presence of bacterial cells (blue) and in sterile electrolyte (black). Inset shows the CVs with the fresh electrodes compared to CV obtained with pre-acclimated electrode (red) as in Fig. 1. Scan rate: 1 mV s−1. | ||
To confirm the biotic origin of the obtained current, in a different experiment CVs were recorded (Fig. 3) after chronoamperometric tests (70 hours with an applied potential of +0.2 V vs. Ag|AgCl (KCl sat.)), in condition of glucose depletion (blue) and after addition of glucose (red), to a final concentration of 50 mM. The current response clearly increased after addition of substrate to the electrolyte when R. microfusus was present in solution. Additionally, it is interesting to note that R. microfusus immediately responded to the presence of substrate, since the CV was performed immediately after the addition. Further glucose additions did not lead to increased current response, indicating that bacterial cells are saturated by 50 mM glucose concentration. Conversely, in the absence of bacterial cells, the current response was substantially unaltered by the addition of substrate (black continuous and dashed lines for absence and presence of substrate respectively). This result further confirms that the electrochemical signal was due to glucose oxidation catalyzed by metabolically active bacterial cells and it was not due bacterial cells that had burst. It is also interesting to note that the maximum current achieved for glucose oxidation was decreased to 22 ± 3 μA (−50%), which might be related with oxidative stress due to the applied potential during chronoamperometric analysis.
The effects of addition of glucose over time were investigated by chronoamperometric analysis were a potential of +0.2 V (vs. Ag|AgCl (KCl sat.)) was applied on the electrode after the cyclic voltammetric analysis reported in Fig. 1 (Fig. S3, ESI†). When glucose was added (to a final concentration of 50 mM) a significant increase of the catalytic current output was obtained only for the experiments that contained additional exogenous RB. The effects of the addition of substrate lasted for approximately 7 hours, after which the current decreased to a lower value. Interestingly, the addition of glucose to R. microfusus that did not contain additional RB only yield an small increase in the oxidative current up to 3 hours, when the baseline current was restored, implicating the importance of RB in electrocatalytic activity of R. microfusus.
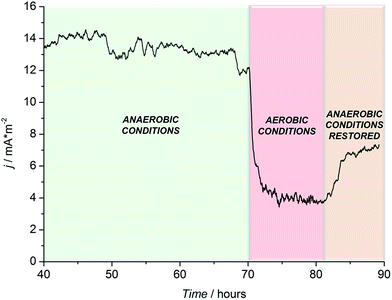
To further confirm the microbial origin of the detected current, N2 purging was suspended, allowing oxygen to diffuse in solution (where R. microfusus is an obligate anaerobe). As shown in Fig. 4, the establishment of aerobic conditions inside the electrochemical cells caused a sharp decrease of the current, to values comparable with the control experiment (control experiment in Fig. S3, ESI†), confirming that the current obtained was generated by the bioelectrocatalytic activity of R. microfusus. After 12 hours of continuous operation under aerobic conditions, N2 purging was re-established to flush O2 from the solution. Only a small increase of current generation was obtained, suggesting that some bacterial cells might have been irreversibly inhibited in the presence of oxygen.
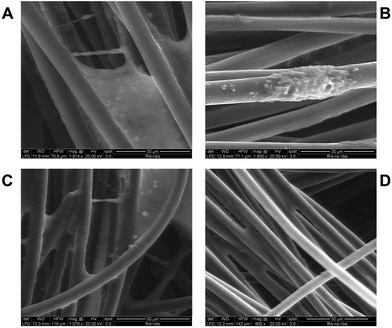
Scanning electron microscopy (SEM) analysis was performed for the electrodes after 7 days with an applied potential of +0.2 V (vs. Ag|AgCl (KCl sat.)) with (Fig. 5A and C) and without the addition of exogenous RB (Fig. 5B and D). Interestingly, when exogenous RB was present in the electrolyte, a more uniform coverage of the carbon fibers by bacterial cells was obtained. Moreover, an increased amount of extracellular polymeric substances can be noted (Fig. 5C and S6†), which could facilitate EET.35 These results well correlate with the 25% increase in the current response obtained with exogenous RB in solution. SEM analysis for control (negative) experiments, where no R. microfusus was inoculated (Fig. S5, ESI†), revealed electrode surfaces completely free from bacteria cells, confirming the origin of the cells colonizing the electrode in Fig. 5.
To investigate the capability of R. microfusus to secrete redox enzymes in solution, protein electrophoresis was conducted on the growth supernatant. The gel electrophoresis experiments revealed the presence of two bands, corresponding to proteins with masses higher than 30 kDa. Gel Proteomic analysis was performed to determine the peptide sequences associated with the obtained bands and the results are reported in Table 1. Mascot™ analysis of the sequences allowed to associate the results to two enzymes: protease IV and phosphopyruvate hydratase respectively. The latter is an enzyme involved in the glycolysis process that catalyse the reaction (eqn (1)):
| 2-Phospho-D-glycerate ↔ phosphoenolpyruvate + H2O | (1) |
| NCBI accession no. | Mass | Score | Protein | Detected peptides sequences |
|---|---|---|---|---|
| gi|488469612 | 45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 773 773 |
71 | Phosphopyruvate hydratase | K. IQIVGDDLFVTNPK. R |
| R. AAVPSGASTGQFEAVELR. D | ||||
| R. IEEELGDSAEYAGASAFPR. F | ||||
| K. VNQIGSLSETIDAVELAHR. N | ||||
| R. AAVPSGASTGQFEAVELR. D | ||||
| gi|85681833 | 48![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 565 565 |
161 | Protease IV | R. GGLYGGPSYCGAPTSQR. N |
| R. AAPLAPKPGTPLQVGVGLK. T | ||||
| K. YSQGNVSAVGVTYDGHTALTR. V | ||||
| K. KYSQGNVSAVGVTYDGHTALTR. V |
The secretion of this enzyme by bacterial cells has been previously reported36 and its detection in the supernatant is important due to its involvement in the glucose oxidation in the glycolytic process. Although not being a redox enzyme, its presence, together with the redox species detected in Fig. 2, suggests that R. microfusus is capable of performing partial glycolysis in solution.
Conclusions
This electrochemical evaluation clearly demonstrates the bioelectrocatalytic properties of R. microfusus for the first time, where R. microfusus is capable of undergoing EET with a carbon cloth electrode. A clear redox couple was detected, with E0′ = −0.039 V vs. Ag|AgCl (KCl sat.), suggesting that R. microfusus can secrete redox mediators to facilitate EET. Additionally, an improvement of glucose oxidation currents was obtained in the presence of exogenous RB (+25% current response) suggesting that RB enhances bacterial activity. Relevant concentrations of R. microfusus have been reported in mixed-species biofilms, together with other bacteria able to secrete flavins in solution, as Shewanella oneidensis MR-1 or Shewanella sp. MR-4.34 Accordingly, the interaction between different bacterial cells in a mixed-species biofilm is suggested. The results obtained by SEM analysis are consistent with the electrochemical results. The increased coverage of carbon fibers by bacterial cells and biofilm in the presence of exogenous RB further indicates that its presence might be involved in the biofilm formation. These findings are of great interest for the study of mixed-species microbial consortium, which develop when MFCs are operated in real conditions using wastewater and biomasses as inoculum. For the development of MFCs technology, and for their optimization and commercialization, further research is required to clarify the contribution of different bacterial species to the EET process.Notes and references
- A. P. Borole, G. Reguera, B. Ringeisen, Z.-W. Wang, Y. Feng and B. H. Kim, Energy Environ. Sci., 2011, 4, 4813 CAS.
- R. Nakamura, F. Kai, A. Okamoto, G. J. Newton and K. Hashimoto, Angew. Chem., Int. Ed., 2009, 48, 508–511 CrossRef CAS PubMed.
- D. K. Newman and R. Kolter, Nature, 2000, 405, 94–97 CrossRef CAS PubMed.
- L. Shi, H. Dong, G. Reguera, H. Beyenal, A. Lu, J. Liu, H. Q. Yu and J. K. Fredrickson, Nat. Rev. Microbiol., 2016, 14, 651–662 CrossRef CAS PubMed.
- U. Schröder, F. Harnisch and L. T. Angenent, Energy Environ. Sci., 2015, 8, 513–519 Search PubMed.
- K. Rabaey and R. A. Rozendal, Nat. Rev. Microbiol., 2010, 8, 706–716 CrossRef CAS PubMed.
- A. ElMekawy, H. M. Hegab and D. Pant, Energy Environ. Sci., 2014, 7, 3921–3933 CAS.
- A. J. McCormick, P. Bombelli, R. W. Bradley, R. Thorne, T. Wenzel and C. J. Howe, Energy Environ. Sci., 2015, 8, 1092–1109 CAS.
- N. S. Malvankar and D. R. Lovley, Curr. Opin. Biotechnol., 2014, 27, 88–95 CrossRef CAS PubMed.
- C. E. Levar, C. H. Chan, M. G. Mehta-Kolte and D. R. Bond, mBio, 2014, 5, e02034 CrossRef CAS PubMed.
- C. M. Ding, M. L. Lv, Y. Zhu, L. Jiang and H. Liu, Angew. Chem., Int. Ed., 2015, 54, 1446–1451 CrossRef CAS PubMed.
- D. Millo, F. Harnisch, S. A. Patil, H. K. Ly, U. Schroder and P. Hildebrandt, Angew. Chem., Int. Ed., 2011, 50, 2625–2627 CrossRef CAS PubMed.
- A. Okamoto, K. Hashimoto and K. H. Nealson, Angew. Chem., Int. Ed., 2014, 53, 10988–10991 CrossRef CAS PubMed.
- A. Kumar, L. Huan-Hsuan Hsu, P. Kavanagh, F. Barriere, P. N. L. Lens, L. Lapinsonniere, J. H. V. Lienhard, U. Schroder, X. Jiang and D. Leech, Nat. Rev. Chem., 2017, 1, 0024 CrossRef.
- D. R. Lovley, Curr. Opin. Biotechnol., 2008, 19, 564–571 CrossRef CAS PubMed.
- M. Grattieri, K. Hasan and S. D. Minteer, ChemElectroChem, 2017, 4, 834–842 CrossRef CAS.
- C. Santoro, C. Arbizzani, B. Erable and I. Ieropoulos, J. Power Sources, 2017, 356, 225–244 CrossRef CAS.
- H. R. Luckarift, S. R. Sizemore, J. Roy, C. Lau, G. Gupta, P. Atanassov and G. R. Johnson, Chem. Commun., 2010, 6048–6050 RSC.
- N. S. Malvankar, J. Lau, K. P. Nevin, A. E. Franks, M. T. Tuominen and D. R. Lovley, Appl. Environ. Microbiol., 2012, 78, 5967–5971 CrossRef CAS PubMed.
- R. B. Song, C. E. Zhao, L. P. Jiang, E. S. Abdel-Halim, J. R. Zhang and J. J. Zhu, ACS Appl. Mater. Interfaces, 2016, 8, 16170–16177 CAS.
- Y. C. Yong, Y. Y. Yu, X. Zhang and H. Song, Angew. Chem., Int. Ed., 2014, 53, 4480–4483 CrossRef CAS PubMed.
- T. Ewing, J. T. Babauta, E. Atci, N. Tang, J. Orellana, D. Heo and H. Beyenal, J. Power Sources, 2014, 269, 284–292 CrossRef CAS.
- E. Martinucci, F. Pizza, D. Perrino, A. Colombo, S. P. M. Trasatti, A. Lazzarini Barnabei, A. Liberale and P. Cristiani, Int. J. Hydrogen Energy, 2015, 40, 14683–14689 CrossRef CAS.
- A. Schievano, A. Colombo, M. Grattieri, S. P. Trasatti, A. Liberale, P. Tremolada, C. Pino and P. Cristiani, J. Power Sources, 2017, 340, 80–88 CrossRef CAS.
- C. Santoro, Y. Lei, B. Li and P. Cristiani, Biochem. Eng. J., 2012, 62, 8–16 CrossRef CAS.
- Y. Feng, X. Wang, B. E. Logan and H. Lee, Appl. Microbiol. Biotechnol., 2008, 78, 873–880 CrossRef CAS PubMed.
- K. P. Nevin, H. Richter, S. F. Covalla, J. P. Johnson, T. L. Woodard, A. L. Orloff, H. Jia, M. Zhang and D. R. Lovley, Environ. Microbiol., 2008, 10, 2505–2514 CrossRef CAS PubMed.
- E. Croese, M. A. Pereira, G. J. Euverink, A. J. Stams and J. S. Geelhoed, Appl. Microbiol. Biotechnol., 2011, 92, 1083–1093 CrossRef CAS PubMed.
- M. Daghio, I. Gandolfi, G. Bestetti, A. Franzetti, E. Guerrini and P. Cristiani, New Biotechnol., 2015, 32, 79–84 CrossRef CAS PubMed.
- C. Kaneuchi and T. Mitsuoka, Int. J. Syst. Bacteriol., 1978, 478–481 CrossRef.
- X. L. Su, Q. Tian, J. Zhang, X. Z. Yuan, X. S. Shi, R. B. Guo and Y. L. Qiu, Int. J. Syst. Evol. Microbiol., 2014, 64, 2986–2991 CrossRef CAS PubMed.
- N. R. Krieg, L. Wolfgang, J. Euzéby and W. B. Whitman, in Bergey's Manual of Systematic Bacteriology, eds. N. R. Krieg, W. Ludwig, W. Whitman, B. P. Hedlund, B. J. Paster, J. T. Staley, D. Brown and A. Parte, Springer, New York, 2010, pp. 25–469 Search PubMed.
- H. Jensen, M. TerAvest, M. Kokish and C. M. Ajo-Franklin, ACS Synth. Biol., 2016, 5, 679–688 CrossRef CAS PubMed.
- E. Marsili, D. B. Baron, I. D. Shikhare, D. Coursolle, J. A. Gralnick and D. R. Bond, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 3968–3973 CrossRef CAS PubMed.
- D. R. Bond, S. M. Strycharz-Glaven, L. M. Tender and C. I. Torres, ChemSusChem, 2012, 5, 1099–1105 CrossRef CAS PubMed.
- C. K. Yang, H. E. Ewis, X. Zhang, C. D. Lu, H. J. Hu, Y. Pan, A. T. Abdelal and P. C. Tai, J. Bacteriol., 2011, 193, 5607–5615 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Materials and methods, SEM analysis, bacterium growth curve. See DOI: 10.1039/c7se00270j |
| This journal is © The Royal Society of Chemistry 2017 |