Open Access Article
Open Access ArticleGut microbiota as important modulator of metabolism in health and disease
Xiang-qian Wangab,
Ai-hua Zhangb,
Jian-hua Miaoa,
Hui Sunb,
Guang-li Yanb,
Fang-fang Wua and
Xi-jun Wang *ab
*ab
aNational Engineering Laboratory for the Development of Southwestern Endangered Medicinal Materials, Guangxi Botanical Garden of Medicinal Plant, Nanning Guangxi, China. E-mail: xijunwangls@126.com; Fax: +86-451-82110818; Tel: +86-451-82110818
bNational Chinmedomics Research Center, Sino-America Chinmedomics Technology Collaboration Center, National TCM Key Laboratory of Serum Pharmacochemistry, Chinmedomics Research Center of State Administration of TCM, Laboratory of Metabolomics, Department of Pharmaceutical Analysis, Heilongjiang University of Chinese Medicine, Heping Road 24, Harbin, China
First published on 19th December 2018
Abstract
The human gastrointestinal tract colonizes a large number of microbial microflora, forms a host-microbiota co-metabolism structure with the host to participate in various metabolic processes in the human body, and plays a major role in the host immune response. In addition, the dysbiosis of intestinal microbial homeostasis is closely related to many diseases. Thus, an in-depth understanding of the relationship between them is of importance for disease pathogenesis, prevention and treatment. The combined use of metagenomics, transcriptomics, proteomics and metabolomics techniques for the analysis of gut microbiota can reveal the relationship between microbiota and the host in many ways, which has become a hot topic of analysis in recent years. This review describes the mechanism of co-metabolites in host health, including short-chain fatty acids (SCFA) and bile acid metabolism. The metabolic role of gut microbiota in obesity, liver diseases, gastrointestinal diseases and other diseases is also summarized, and the research methods for multi-omics combined application on gut microbiota are summarized. According to the studies of the interaction mechanism between gut microbiota and the host, we have a better understanding of the use of intestinal microflora in the treatment of related diseases. It is hoped that the gut microbiota can be utilized to maintain human health, providing a reference for future research.
1 Introduction
The human gastrointestinal tract is the most important digestive organ and immune organ of the human body. There are more than 1000 types of microbiota living in the intestinal tract, and their number exceeds 1000 trillion, which is 10 times the total number of human cells.1 Gut microbiota and the host have formed a close symbiotic relationship with the long history of evolution.2 This cannot be only to realize the exchange of its own material and information, but also to participate in the host's nutrition, metabolism, excretion process and conversion of metabolites. Host metabolism is actually the combination of the body's own genome and the gut microbial genome. Factors such as host genotypes, physiological status, diet, drugs, and living conditions can affect the composition of the gut microbiota.3 The nutritional metabolites of normal gut microbiota enter various tissues and organs with the circulation of blood, which are used for the physiological activities of the host, thus maintaining the integrity of the intestinal mucosal barrier and playing a role in the host immune regulation.4 Therefore, the intestinal microflora is considered to be another necessary “organ” of the host.5 The community formed by the host and microorganism is also known as a “superorganism”.6A growing number of studies have confirmed that gut microbiota is associated with multiple host diseases.7–9 Gut microbiota can be divided into three types: beneficial bacteria, conditional pathogenic bacteria and pathogenic bacteria. The beneficial bacteria are the dominant bacteria in the intestinal tract, which play a role in maintaining the intestinal homeostasis and immune regulation. When the number of harmful bacteria increases, it leads to an imbalance in the beneficial bacteria and causes disease.10 Gut microbiota is metabolized under homeostasis to produce numerous beneficial substances for the host. Thus, under unbalanced conditions, the growth and health of the host are affected.11 With the increasingly clear mechanism of gut microbiota to host diseases, probiotics and prebiotics have become hot topics in the treatment of diseases.12 Therefore, it is of significance to fully understand the relationship between gut microbiota metabolites and host diseases.
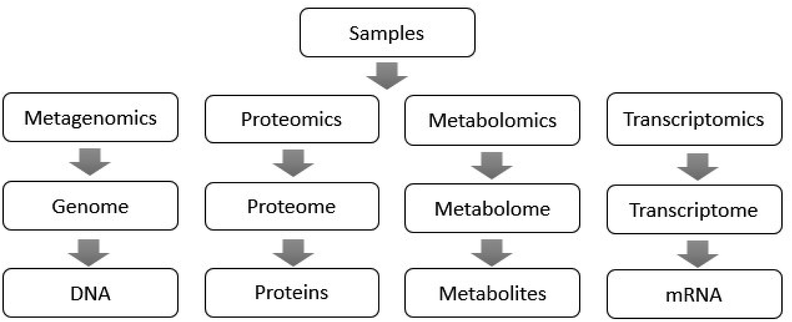
In recent years, metagenomics using high-throughput sequencing technology as an analytical tool has been continuously applied for the study of gut microbiota.13 Microgenomics, which is based on the microbial genome, analyzes the microbial diversity, population structure and functional activity, which provides a new research basis for microbial phenotype and disease.14 For the superorganism of human beings, it is possible to use transcriptomics and proteomics to analyze the entire human genome sequence, and the corresponding encoded protein functions can also provide a comprehensive understanding of the genes covered by the gut microbiota.15,16 Metabolomics can characterize all the metabolites of the host genome and microbial genome, discovering biomarkers and their changes under disease or normal physiological conditions, revealing disease-related changes and mechanisms.17 Metabolomics provides key metabolic information for the study of gut microbiota when metagenomics, transcriptomics and proteomics fail to measure intestinal microbial metabolites accurately. The functions of these technologies in the study of gut microbiota are shown in Fig. 1,18 which overcome the shortcomings of traditional biological research methods, and analyze the microflora from the aspects of intestinal microbial function, structure, genes and metabolites.19
2 Effects of microbial metabolites on host health
Host metabolism is comprised of the host's own gene metabolism and intestinal microbial gene metabolism.20 The gut microbiota connects the intestine, liver, brain and other organs through the host-microbiota co-metabolism structure to form a metabolic axis, regulating the systemic metabolism of the host.21 After being influenced by factors such as the host's genetics, diet, and living conditions, the gut microbiota can regulate the metabolism of the host.22 There are interaction mechanisms between the gut microbiota and its metabolites and host epithelial cells. Changes in the microbial microflora structure are accompanied by changes in the physiological functions of the intestine, which in turn play a part in regulating the overall metabolic disorder of the body.23 Therefore, it is extremely important to fully analyze the metabolites produced by host-microbiota co-metabolism for the assessment of disease mechanisms, including SCFA, tryptophan, and tyrosine.242.1 Gut microbiota and SCFA
Gut microbiota provides nutrients for the host by increasing the expression of intestinal nutrient transporters. Dietary fiber or complex carbohydrates is fermented into SCFA via the action of gut microbiota, further promoting the process of intestinal gluconeogenesis, and supporting the formation of lipids.25 This reveals how dietary fiber helps ensure the health of intestine. SCFA plays a significant physiological role in the host organism, which can improve intestinal function, resist pathogenic microorganisms, fight tumors, maintain the electrolyte balance of the host, and provide energy for the host epithelial cells.26 For example, gut microbiota ferments dietary fiber into SCFA, which is involved in the immune response through the G protein-coupled receptor in the intestinal tract and participates in the regulation of acute inflammation in the body.27 A study has shown that the peroxisome proliferator receptor-γ (PPAR-γ) signal induced by gut microbiota is responsible for maintaining homeostasis.28 Butyrate is mainly produced by the metabolism of Clostridia. Antibiotic treatment reduces the content of butyrate, which subsequently reduces the transduction of the PPAR-γ signal. In the absence of the PPAR-γ signal, the expression of the Nos2 inducible nitric oxide synthase gene increases, which increases the level of nitrate in the colonic lumen. Therefore, gut microbiota is of great importance in maintaining the health of the host. Oligofructose weakens weight gain, reduces fat accumulation, and improves glucose tolerance in mice, and ameliorates metabolic disorders induced by a high-fat diet (HFD) in mice by restoring the number of Bifidobacteriaceae in the intestine.29 Probiotic VSL#3 is administered to HFD mice to prevent weight gain and insulin resistance. In addition, VSL#3 can reverse obesity and diabetes in mice with leptin deficiency. These beneficial effects are linked to the increase of butyrate levels, which confirms that gut microbiota can promote metabolic efficiency and prevent the harmful effects of disease.30 Akkermansia muciniphila is a strictly anaerobic gut bacteria isolated from feces and its abundance in the intestine usually ranges from 1% to 3%.31 It can use intestinal mucin as the only carbon and nitrogen source for growth and its main metabolite is propionate. The colonization of the bacteria in the intestinal tract is closely related to the health of the host. They can participate in the inflammatory response of obese and diabetic patients, improve adverse symptoms such as insulin resistance and glucose tolerance, and also regulate the immune response of the body to maintain the metabolic balance.32 Studies in vitro have shown that butyrate can reduce the fat content in adipocytes by increasing hormone sensitivity, lipase and triglyceride lipase activity.33 In addition, butyrate decreases the production of TGF-β1 and IL-6, up-regulates the activity of anti-inflammatory cytokines, produces anti-inflammatory effects by inducing the activity of regulatory T cells and enhances body immunity.34 SCFA enters various organs of the body through blood circulation and participates in lipid synthesis and energy metabolism. This proves that by regulating the production of SCFA in the intestine, immune function and barrier protection are improved. In conclusion, SCFA is one of the most important metabolites of gut microbiota and has a series of effects on the host.2.2 Gut microbiota and bile acid metabolism
In the liver, hepatocytes convert cholesterol into primary bile acids such as cholate (CA) and chenodeoxycholate (CDCA). Under the effect of 7α-dehydroxylation of the gut microbiota, CA and CDCA are further converted to secondary bile acids, such as deoxycholate (DCA) and lithocholate, which are mainly involved in the metabolism of fat in the diet, the absorption of cholesterol and the regulation of inflammation and other processes.35 Bile acids are transferred to the liver through the portal vein after being reabsorbed in the intestine, which are continuously recycled between the liver and the intestine to form an enterohepatic circulation. The gut microbiota plays a key role in this process.36 Secondary bile acids regulate bile acid metabolism by activating farnesoid X receptor (FXR), further regulating the composition of the gut microbiota to participate in multiple host metabolic pathways.37 The results confirmed that the intestine-restricted FXR agonist can up-regulate the level of Acetatifactor and Bacteroides in gut microbiota.38 In turn, bile salt hydrolase and 7α- and 7β-dehydroxylase increase the content of lithocholic acid and further activate the G protein-coupled bile acid receptor-1 (TGR-5) signaling pathway, thus improving glucose and insulin tolerance and promoting effective browning of adipose tissue. These benefits disappeared completely after antibiotic treatment, which revealed that FXR plays a major role in bile acid metabolism by regulating gut microbiota. Bile acid was given to cirrhotic rats to increase the concentration of bile acids in their intestine, which appeared to inhibit intestinal bacterial overgrowth and reduce the occurrence of bacterial translocation.39 In summary, there is a bidirectional regulation between gut microbiota and bile acid metabolism, which can serve as the basis for more disease treatment by regulating the homeostasis between the two.2.3 Gut microbiota and other metabolic pathways
Choline is a necessary nutrient for humans and an essential substance for the synthesis of acetylcholine, which is an important neurotransmitter synthesized by choline and acetyl-CoA in nerve cells.40 Humans obtain choline through the diet or it is synthesized by the liver, which participates in normal metabolism. Gut microbiota can hydrolyze choline to trimethylamine (TMA) and further metabolize it to dimethylamine (DMA) or methylamine (MA). Abnormal choline metabolism leads to impaired glucose homeostasis.41 Gut microbiota converts choline to MA, which reduces the bioavailability of choline and plays a positive role in insulin resistance. A study has shown that gut microbiota metabolizes choline to TMA N-oxide, further reducing the risk of chronic kidney disease and cardiovascular disease.42 The metabolism of gut microbiota also includes tryptophan metabolism. Tryptophan is an essential amino acid that participates in many physiological functions in the human body. Nikolaus S. et al. researched that the composition of fecal microorganisms is related to tryptophan levels.43 Increased tryptophan can reduce the severity of colitis in mice. Indole is a tryptophan metabolite produced by intestinal bio-derivatization, acting as a channel between bacteria and microorganisms.44 Intestinal microbiota can reduce the production of indole derivatives, thereby improving the production of intestinal inflammation.453 Effects of microbial metabolites on disease
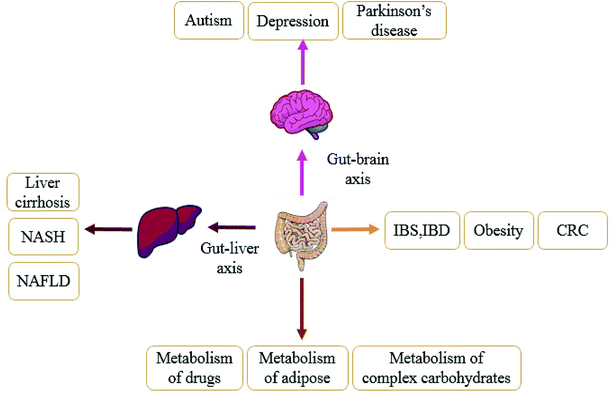
There is homeostasis between the gut microbiota and the host environment.46 Normally, the intestinal microbiota attaches to the mucosal layer on the inner wall of the intestinal tract to form a barrier, which affects the body's immunity through its own barrier function to inhibit the growth of pathogenic bacteria and prevents pathogens from entering the body. Once under the influence of antibiotics,47 diet48 and other factors, the breakdown of intestinal homeostasis will lead to disease. In the past decade, more and more studies have shown that the gut microbiota is closely related to obesity, type 2 diabetes, liver diseases, hypertension, nervous system diseases and other related metabolic diseases,49,50 as shown in Fig. 2. In following section we use obesity, liver disease, and gastrointestinal diseases as examples to fully explain the relationship between the disorder of gut microbiota and the development of diseases.3.1 Gut microbiota and obesity
Firmicutes and Bacteroidetes are two predominant bacteria in the gut microbiota, whose quantity is impacted by diet and body fat content. Louis et al. implemented a weight loss program for 16 obese patients.51 The results showed that the Firmicutes/Bacteroidetes ratio in the obese patients was higher than that in the healthy group. In addition, the abundance of Akkermansia, an intestinal microflora, was increased in the successful weight loss patients. This bacteria can improve the metabolism of the host, reduce inflammation in obese patients, and have a protective effect on insulin resistance. Ferrer et al. observed that the abundance of Firmicutes is greater than Bacteroidetes in obese patients; whereas in thin people, the results are conflicting.52 Although the gut microbial genes of the two populations are roughly similar, they are associated with the synthesis of different metabolites. Clostridium ramosum is a type of Firmicutes that is closely related to metabolic syndrome of the human body. In germ-free mice with HFD, the mice colonized with that bacteria developed symptoms of obesity.53 The mechanism is that the bacteria accelerate the transport of glucose and fat in the small intestine and expedite the deposition of body fat. In addition, the intestinal microbial diversity was significantly altered in the obese patients. The microbial diversity of obese patients is lower than that in healthy individuals, and probiotics such as Bifidobacterium and Lactobacillus spp. are reduced, whereas Clostridia and Bacilli rapidly increased, accompanied by metabolic disorders in the body.54 Al-Ghalith et al. found that gut microbiota changes the function of the intestinal barrier. A thinner inner layer of the intestine leads to nutrients entering the body and causes obesity.55 Kasselman et al. showed that gut microbiota can cause systemic low-grade inflammation in obese patients by regulating innate immune responses and inflammatory signals.56 The proliferation of Bacteroidetes in the gut microbiota of obese patients increases the production of lipopolysaccharide (LPS) and activates Toll-like receptors (TLR), which combine with LPS to induce low-grade inflammation of obesity.57 The concentration of SCFA in the gut microbiota is associated with obesity and metabolic diseases. Obese mice have higher concentrations of SCFA than normal mice,58 and their gut microbiota are rich in enzymes-encoding genes that use non-digestible dietary carbohydrates to produce SCFA. Therefore, the use of targeted therapeutic agents to modulate microbial composition can reduce obesity and even restore it to a healthy state. A study showed that Lactobacillus is involved in the host metabolism, which has a significant effect on weight changes in obese patients.59 It is related to the digestion of complex carbohydrates in the host colon and the degradation of lipids and monosaccharides in the duodenum and jejunum.60 HFD-induced obese mice administered with Lactobacillus additives maintained glucose-insulin homeostasis and reduced body weight considerably. Cox et al. administered low-dose penicillin to C57BL/6J pups to study the effects of gut microbiota on their growth and development.61 The increase in body weight and fat mass and the decrease in bone minerals in the adult mice indicate that earlier penicillin exposure leads to increasing metabolic phenotypes in mice. This shows that the host is more sensitive to microbial breakdown in infancy.3.2 Gut microbiota and liver disease
Both the intestine and the liver are involved in anatomical and physiological functions. Gut microbiota plays an important role in the development of liver inflammation and fibrosis through the enterohepatic circulation and the gut–liver axis.62 A study showed that with the progress of liver cirrhosis, the dysbiosis and inflammatory reaction of gut microbiota increased, and the intestinal barrier and the liver were damaged, indicating that the degree of intestinal dysbiosis is positively correlated to the severity of liver cirrhosis.63 Compared with healthy people, there was a significant increase in the abundance of Enterobacteriaceae and Enterococcus spp. as well as Proteus spp. in patients with liver cirrhosis, and higher endotoxins in the gut eventually become intestinal endotoxemia.64 Nonalcoholic fatty liver disease (NAFLD) is the most common liver disease in the world.65 Intestinal dysbiosis and changes in intestinal permeability promote the entry of bacteria into the portal circulation, activated inflammation through TLR signaling in hepatocytes, and conversion of simple fatty degeneration to NAFLD.66 In addition, the abundance of bacteria capable of producing ethanol in the intestinal microflora of patients with NAFLD increased, and the level of ethanol in the patients was significantly higher than that in the control group, suggesting that intestinal bacteria play a role in the pathogenesis of NAFLD.67 The progress of modern sequencing technology makes it possible for in-depth analysis of the microbial composition. Del et al. found that NAFLD patients had higher Actinobacteria and lower Bacteroidetes than healthy people.68 The abundance of Bacteroides increased in nonalcoholic steatohepatitis (NASH) patients; whereas, Prevotella decreased.69 Pyrin domain-containing 3 inflammasome participates in the pathogenesis of NASH, while gut microbiota promotes the development of NASH accompanied by the occurrence of liver fibrosis.70 Chemokine receptors are also crucial molecules in the pathogenesis of NASH.71,72 The increased expression of chemokine receptors in NASH is related to the induction of pro-inflammatory factors and activation of NF-κB, which is an independent risk factor for NASH patients. Blocking these receptors can prevent hepatic steatosis and damage, and subsequently substances secreted by them may serve as potential biomarkers for NASH patients. Research has shown that the amount of cholesterol and bile acids in the liver and feces of NASH mice fed on a special HFD increased, and secondary bile acid activates the m-TOR pathway of liver cells to induce liver cancer.73 The secondary bile acid accumulation was significantly reduced after the use of antibiotics, indicating that gut microbiota play a crucial role in bile acid conversion. Thus, gut microbiota may provide new strategies and new directions in the research and treatment of liver diseases.3.3 Gut microbiota and gastrointestinal diseases
Gut microbiota is closely related to gastrointestinal diseases, including diverticular disease (DD), inflammatory bowel disease (IBD), irritable bowel syndrome (IBS) and gastrointestinal tumors.74 Crohn's disease (CD) and ulcerative colitis (UC) are two main clinical forms of IBD. Besides its pathogenesis, the inflammatory response caused by the abnormal intestinal mucosal immune system is the key factor, which is mainly related to environmental factors, genetic factors, immune factors, and gut microbiota.75 Studies have shown that changes in the abundance in gut microbiota occur in patients suffering from different gastrointestinal diseases,76,77 including the down-regulation of probiotics and up-regulation of certain pro-inflammatory bacteria, as shown in Table 1. It is suggested that every disease has its own specific microbial markers, which provide the basis for the targeted treatment of diverse diseases. Enterotoxigenic B. fragilis induced an inflammatory response to colorectal cancer (CRC) by stimulating an immune response in a colitis-associated CRC mouse model.78 Collectively, the gut microbiota can improve the practicability of clinical diagnosis, making accurate and non-invasive CRC diagnosis possible. Schaubeck et al. confirmed that there was no inflammatory reaction in TNF knockout mice under a GF environment, while the mice fed in the SPF environment exhibited opposite results, suggesting that the extent and location of the inflammation were associated with the gut microbiota.79 Feng et al. performed a metagenomics analysis of patients with advanced colorectal adenoma and CRC.80 Bacteroides and Escherichia coli were considered to be related to the stages of inflammation and tumor development. Moreover, Bifidobacterium was absent, which can accelerate colon epithelial regeneration and inhibit potential pathogens in patients. This indicates that the preventive and therapeutic effects of diet or lifestyle on CRC deserve further investigation. At present, probiotics, prebiotics and synbiotics work well in various gastrointestinal diseases such as IBD,81 IBS,82 and CRC.83 Yang et al. found that TLR3 or TLR7 significantly improves the symptoms of intestinal inflammation in mice by producing interferon-β.84 Zhao et al. discovered that there are 15 strains in the diet that can increase the level of butyrate in the gut and inhibit the production of harmful substances, such as cesium and hydrogen sulfide, which improve type 2 diabetes effectively.85 Similarly, research has found that Fusobacterium nucleatum is abundant in the CRC tissues of patients who relapse after chemotherapy.86 It can promote the resistance of chemotherapy in CRC patients through complex network mechanisms such as the TLR signaling pathway, autophagy, and microRNA, which provides valuable guidance for the prognosis of patients with CRC.| Disease | Increased microbiota | Reduced microbiota |
|---|---|---|
| DD | Ruminococcaceae | Bacteroides fragilis |
| IBS | Dialister spp. | Clostridium difficile |
| Faecalibacterium prausnitzii | Streptococcus spp. | |
| Parabacteroides distasonis | ||
| Bacteroides fragilis | ||
| UC | Bacteroides fragilis | Clostridium difficile |
| Dialister spp. | ||
| Roseburia spp. | ||
| CD | Parabacteroides distasonis | Bacteroides fragilis |
| Dialister spp. | Faecalibacterium prausnitzii | |
| Ruminococcus gnavus | ||
| Clostridium difficile | ||
| Streptococcus spp. | ||
| IBD | Enterobacteriaceae | Rikenellaceae |
| Collinsella aerofaciens | ||
| Streptococcus spp. | ||
| Faecalibacterium prausnitzii | ||
| Akkermansia muciniphila | ||
| CRC | Fusobacterium | Clostridia |
| Porphyromonas |
4 Research methods of gut microbiota
As researchers intensify their knowledge on the importance of the gut microbiota, explorations on the structure and function of the intestinal microflora, metabolic mechanisms, and the relationship with the disease have increased gradually.87,88 These studies not only provide a basis for the human prevention and treatment of diseases, but also for the development of valuable gene resources and new microbes. The initial methods of gut microbiota research are to detect and identify microbiota after isolation and culture.89 For abundant and numerous gut microbiota systems, the traditional biological method is not comprehensive enough for bacterial analysis and difficult to reflect the relationship between gut microbiota and disease.90 Consequently, this method has serious limitations for the study of gut microbiota. In recent years, metagenomics has been successfully applied for the study of gut microbiota, sequencing the intestinal microflora, and exploring the structure and function of bacteria comprehensively.13 High-throughput sequencing technology, also called second-generation sequencing technology, is not only used for the detection of gut microbiota diversity, microecological disorders and their relationship with disease, but also to identify new functional genes and discover microbial metabolic pathways and their relationship with the host.91 In a study on Chinese intestinal dysbiosis diseases, Wu et al. used metagenomic sequencing technology to screen 300 disease-related biomarkers in the microbiota and predict their effects on the disease successfully.92 To identify which type of microbiota in the human intestine can carry out a specific metabolic transformation, it is especially important that the combined analysis of the host metabolic activity and the intestinal bacterial diversity be conducted at the overall level.93 Wang et al. relied on the Illumina MiSeq sequencing platform to analyze the gut microbiota in the feces of patients with chronic hepatitis B using metabolomics to analyze the serum by gas chromatography.94 Upon enrichment of the gut microbiota information, it was found that the four key operational taxonomic units are highly correlated with the level of serum metabolites L-phenylalanine and L-tyrosine, showing that the gut microbiota is involved in the accumulation of serum metabolites. Chi et al. used high-throughput sequencing and GC-MS metabolomics to analyze the composition of microbiota and metabolites of feces collected from manganese (Mn2+)-induced intoxicated mice.95 The mechanism of the toxicity of Mn2+ is that it can disturb the intestinal microecology and promote the gut microbiota to produce pro-inflammatory mediators and neurotransmitters, including γ-aminobutyric acid and tryptophan, to affect the gut–brain axis, further influencing the central nervous system. At present, there are a series of omics methods linking the composition and distribution of gut microbiota with changes in the host metabolism.96 Considerable progress has been made in the research on gut microbiota and diseases, including metagenomics, transcriptomics, proteomics and metabolomics. Jiang et al. analyzed the blood of patients with kidney-yin deficiency diabetes syndrome based on an NMR metabolomics platform and MALDI-TOF-MS proteomics platform.97 Changes in the amino acid metabolic pathways and energy metabolism pathways have been considered to be related to the gut microbiota. Some studies combined with transcriptomics and metabolomics explored the role of gut microbiota in the metabolic disorder of human beings and the effect of bacteria on the intestinal microenvironment after intestinal colonization.98,99 Kieffer et al. utilized a multi-omics approach to study dietary mice.100 Metagenomics and metabolomics results suggest that changes in the gut microbiota affect the homeostasis of the host metabolism. Transcriptomics analysis showed that diet is related to cell proliferation and other pathways. El et al. studied germ-free mice colonized with intestinal bacteria using transcriptomics and plasma metabolomics.101 The increase in the abundance of Helicobacter, Sphingomonas and Mucispirillum in the intestine of mice is positively correlated with the increase of pro-inflammatory cytokines in plasma, which enhances the understanding of the relationship between certain specific gut microbes and pro-inflammatory responses. Thus, it can be concluded that the technique of multi-omics has become an important tool in the research of gut microbiota. Metabolomics analyzed all the final products of the host metabolism and microbial metabolism, which revealed the regulation and mechanism of metabolism in the host and gut microbiota.102–113 In the future, there will be more research devoted to the exploration of the complex relationship between intestinal microflora and host metabolism and disease pathogenesis to maintain human health through intestinal microbiota.1145 Current challenges
At present, the research on gut microbiota is still in the exploratory stage. The emergence of germ-free animals has resulted in the discovery of the significant role of gut microbiota in host metabolism. However, germ-free animals have defects in the development of their immune system and intestinal morphology, which may affect the experimental results.115 In addition, successfully transferring the results obtained in animal experiments to clinical applications has yet to be studied. Although treatment of fecal microbiota transplantation has already been applied in clinical practice, there are still exist many adverse reactions such as nausea and dizziness,116 and many people still have a hard time accepting such treatment. Thus, researchers have considered using probiotic agents instead of fecal microbiota transplantation. Therefore, the standards and techniques for probiotic agents should be continuously improved, and clinical trials should be accelerated to make it a standardized treatment method.117 By identifying the relationship between specific bacterial microflora and disease, it is possible to diagnose the disease by only detecting the composition of the gut microbiota.118 As for the function of gut microbiota developed in the intestinal tract, the relationship between microbiota and its synergistic effects on the host still require further investigation.119–125 Thus, studies on the gut microbial functional genes and the dynamic processes in different aspects, such as individual organisms and life stages, need to be further conducted.126 A large amount of data has been collected from multi-omics studies on intestinal microbiota; however, there is a lack of strict standards for data analysis.127–132 Carrying out correlative analysis of multidimensional data effectively is particularly difficult, and due to the enormous amount of data, it will inevitably lead to a waste of data. In summary, complete gut microbiota experiments combined with the multi-omics analysis method can find bacteria related to disease at the strain level. With the advancement of clinical treatment, this can pave the way for the prevention and treatment of diseases and provide an effective means for maintaining the health of the human body.6 Conclusion and future perspectives
Gut microbiota, as the largest organ and the most complex microecosystem, forms a co-metabolism system with the host, metabolizes the diet and other substances, and participates in life activities of the host by connecting organs to maintain health. In a healthy state, the host and intestinal microflora maintain homeostasis. Under the influence of internal and external factors, the stability of the intestinal microflora is destroyed, which causes liver disease, digestive system diseases, obesity and other. The combined use of multi-omics provides a completely new approach for the study of gut microbiota. Combining the multidimensional data of intestinal microbial metabolites, gene functional structure and microbial diversity, the mechanism of gut microbiota in diseased and healthy hosts can be described. Also, biomarkers and specific bacteria groups are found in the disease state, which provide the basis for the diagnosis and pathogenesis of the disease. With the deepening of research on gut microbiota, researchers have realized the vital role of the diet in human health. Gut microbiota is able to metabolize dietary fiber into nutrients to benefit the health of the host. Hence, in the future, there will be more researchers studying the relationship between diet and host health and disease.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by grants from the Key Program of Natural Science Foundation of State (Grant No. 81830110, 81430093, 81373930, 81673586, 81703685, 81302905, 81473584, 81503386), National Key Subject of Drug Innovation (Grant No. 2015ZX09101043-005, 2015ZX09101043-011), TCM State Administration Subject of Public Welfare of (Grant No. 2015468004), University Nursing Program for Young Scholars with Creative Talents in Heilongjiang Province (UNPYSCT-2015118, UNPYSCT-2016213), Major Projects of Application Technology Research and Development Plan in Heilongjiang Province (GX16C003), TCM State Administration Subject of Public Welfare (Grant No. 2015468004), Specialized Research Fund for the Doctoral Program of Higher Education (20132327130001, 20122327120006), Application Technology and Development of Youth Talents Project in Harbin (2014RFQXJ116), University Nursing Program for Young Scholars with Creative Talents in Heilongjiang Province (UNPYSCT-2015118, UNPYSCT-2016213), Young Talent Lift Engineering Project of China Association of Traditional Chinese Medicine (QNRC2-B06), Outstanding Talents Foundation of Heilongjiang University of Chinese Medicine (2018jc01), Doctoral Innovation Fund of Heilongjiang University of Chinese Medicine (2018bs02), Chinese Postdoctoral Science Foundation (2017M621319b).References
- P. Mandal, Anaerobe, 2017, 49, 63–70 CrossRef PubMed.
- R. Okumura and K. Takeda, Inflammation Regener., 2018, 38, 5 CrossRef PubMed.
- S. Tuddenham and C. L. Sears, Curr. Opin. Infect. Dis., 2015, 28(5), 464–470 CrossRef CAS PubMed.
- H. Han, Y. Li Y, J. Fang, G. Liu, J. Yin, T. Li and Y. Yin, Int. J. Mol. Sci., 2018, 19(4), 995 CrossRef PubMed.
- C. Chassard and C. Lacroix, Curr. Opin. Clin. Nutr. Metab. Care, 2013, 16(4), 453–460 CrossRef CAS PubMed.
- J. Lederberg, Science, 2000, 288(5464), 287–293 CrossRef CAS.
- K. A. Sharkey, P. L. Beck and D. M. McKay, Nat. Rev. Gastroenterol. Hepatol., 2018, 15(12), 765–784 CrossRef CAS PubMed.
- A. Zhang, H. Sun, S. Qiu and X. Wang, J. Evidence-Based Complementary Altern. Med., 2013,(3), 78–85 Search PubMed.
- S. R. Sharpton, V. Ajmera and R. Loomba, Clin. Gastroenterol. Hepatol., 2018, S1542-3565(18)30944-3 Search PubMed.
- P. D. Cani and A. Everard, Mol. Nutr. Food Res., 2016, 60(1), 58–66 CrossRef CAS PubMed.
- A. Zhang, H. Sun, G. Yan, P. Wang and X. Wang, Biomed. Chromatogr., 2016, 30(1), 7–12 CrossRef PubMed.
- L. R. McCabe and N. Parameswaran, Calcif. Tissue Int., 2018, 102(4), 480–488 CrossRef CAS PubMed.
- M. K. Verma, V. Ahmed, S. Gupta, J. Kumar, R. Pandey, V. Mandhan and N. S. Chauhan, Sci. Rep., 2018, 8(1), 1397 CrossRef PubMed.
- T. Korem, D. Zeevi, J. Suez, A. Weinberger, T. Avnit-Sagi, M. Pompan-Lotan, E. Matot, G. Jona, A. Harmelin, N. Cohen, A. Sirota-Madi, C. A. Thaiss, M. Pevsner-Fischer, R. Sorek, R. Xavier, E. Elinav and E. Segal, Science, 2015, 349(6252), 1101–1106 CrossRef CAS PubMed.
- W. H. Pan, F. Sommer, M. Falk-Paulsen, T. Ulas, P. Best, A. Fazio, P. Kachroo, A. Luzius, M. Jentzsch, A. Rehman, F. Müller, T. Lengauerm, J. Walter, S. Künzel, J. F. Baines, S. Schreiber, A. Franke, J. L. Schultze, F. Bäckhed and P. Rosenstiel, Genome Med., 2018, 10, 27 CrossRef PubMed.
- L. Ruiz, C. Hidalgo, A. Blanco-Míguez, A. Lourenço, B. Sánchez and A. Margolles, J. Proteomics, 2016, 147, 28–39 CrossRef CAS PubMed.
- K. S. Smirnov, T. V. Maier, A. Walker, S. S. Heinzmann, S. Forcisi, I. Martinez, J. Walter and P. Schmitt-Kopplin, Int. J. Med. Microbiol., 2016, 306(5), 266–279 CrossRef CAS PubMed.
- H. Huang, P. Vangay, C. E. McKinlay and D. Knights, Immunol. Lett., 2014, 162(2 Pt A), 62–68 CrossRef CAS PubMed.
- F. Turroni, C. Milani, S. Duranti, L. Mancabelli, M. Mangifesta, A. Viappiani, G. A. Lugli, C. Ferrario, L. Gioiosa, A. Ferrarini, J. Li, P. Palanza, M. Delledonne, D. van Sinderen and M. Ventura, ISME J., 2016, 10, 1656–1668 CrossRef CAS PubMed.
- A. Baars, A. Oosting, M. Lohuis, M. Koehorst, S. El Aidy, F. Hugenholtz, H. Smidt, M. Mischke, M. V. Boekschoten, H. J. Verkade, J. Garssen, E. M. van der Beek, J. Knol, P. de Vos, J. van Bergenhenegouwen and F. Fransen, Sci. Rep., 2018, 8, 13426 CrossRef PubMed.
- J. K. Nicholson, E. Holmes, J. Kinross, R. Burcelin, G. Gibson, W. Jia and S. Pettersson, Science, 2012, 336(6086), 1262–1267 CrossRef CAS PubMed.
- G. Xie, S. Zhang, X. Zheng and W. Jia, Electrophoresis, 2013, 34(19), 2787–2798 CAS.
- M. A. Fischbach and J. A. Segre, Cell, 2016, 164(6), 1288–1300 CrossRef CAS.
- M. Yu, H. Jia, C. Zhou, Y. Yang, Y. Zhao, M. Yang and Z. Zou, J. Pharm. Biomed. Anal., 2017, 138, 231–239 CrossRef CAS PubMed.
- A. Woting and M. Blaut, Nutrients, 2016, 8(4), 202 CrossRef PubMed.
- Q. Feng, W. D. Chen and Y. D. Wang, Front. Microbiol., 2018, 9, 151 CrossRef PubMed.
- E. Alvarez-Curto and G. Milligan, Biochem. Pharmacol., 2016, 114, 3–13 CrossRef CAS PubMed.
- M. X. Byndloss, E. E. Olsan, F. Rivera-Chávez, C. R. Tiffany, S. A. Cevallos, K. L. Lokken, T. P. Torres, A. J. Byndloss, F. Faber, Y. Gao, Y. Litvak, C. A. Lopez, G. Xu, E. Napoli, C. Giulivi, R. M. Tsolis, A. Revzin, C. B. Lebrilla and A. J. Bäumler, Science, 2017, 357(6351), 570–575 CrossRef CAS PubMed.
- A. Woting, N. Pfeiffe, L. Hanske, G. Loh, S. Klaus and M. Blaut, Mol. Nutr. Food Res., 2015, 59(11), 2267–2278 CrossRef CAS PubMed.
- H. Yadav, J. H. Lee, J. Lloyd, P. Walter and S. G. Rane, J. Biol. Chem., 2013, 288(35), 25088–25097 CrossRef CAS.
- J. P. Ouwerkerk, S. Aalvink, C. Belzer and W. M. de Vos, Int. J. Syst. Evol. Microbiol., 2016, 66, 4614–4620 CrossRef CAS PubMed.
- R. El Hage, E. Hernandez-Sanabria and T. Van de Wiele, Front. Microbiol., 2017, 8, 1889 CrossRef.
- E. Heimann, M. Nyman, A. K. Pålbrink, K. Lindkvist-Petersson and E. Degerman, Adipocyte, 2016, 5(4), 359–368 CrossRef CAS PubMed.
- M. Asarat, V. Apostolopoulos, T. Vasiljevic and O. Donkor, Immunol. Invest., 2016, 45(3), 205–222 CrossRef CAS.
- S. Krishnan, N. Alden and K. Lee, Curr. Opin. Biotechnol., 2015, 36, 137–145 CrossRef CAS PubMed.
- A. Di Ciaula, G. Garruti, R. Lunardi Baccetto, E. Molina-Molina, L. Bonfrate, D. Q. Wang and P. Portincasa, Ann. Hepatol., 2017, 16(S1), 4–15 CrossRef PubMed.
- A. Wahlström, S. I. Sayin, H. U. Marschall and F. Bäckhed, Cell Metab., 2016, 24(1), 41–50 CrossRef PubMed.
- P. Pathak, X. Cen, R. G. Nichols, J. M. Ferrell, S. Boehme, K. W. Krausz, A. D. Patterson, F. J. Gonzalez and J. Y. L. Chiang, Hepatology, 2018, 68(4), 1574–1588 CrossRef CAS PubMed.
- B. Stanimirov, K. Stankov and M. Mikov, Hepatobiliary Pancreatic Dis. Int., 2015, 14(1), 18–33 CrossRef.
- M. D. Spencer, T. J. Hamp, R. W. Reid, L. M. Fischer, S. H. Zeisel and A. A. Fodor, Gastroenterology, 2011, 140(3), 976–986 CrossRef CAS PubMed.
- M. E. Dumas, R. H. Barton, A. Toye, O. Cloarec, C. Blancher, A. Rothwell, J. Fearnside, R. Tatoud, V. Blanc, J. C. Lindon, S. C. Mitchell, E. Holmes, M. I. McCarthy, J. Scott, D. Gauguier and J. K. Nicholson, Proc. Natl. Acad. Sci. U. S. A., 2006, 103(33), 12511–12516 CrossRef CAS.
- D. Y. Li and W. H. W. Tang, Semin. Nephrol., 2018, 38(2), 193–205 CrossRef CAS.
- S. Nikolaus, B. Schulte, N. Al-Massad, F. Thieme, D. M. Schulte, J. Bethge, A. Rehman, F. Tran, K. Aden, R. Häsler, N. Moll, G. Schütze, M. J. Schwarz, G. H. Waetzig, P. Rosenstiel, M. Krawczak, S. Szymczak and S. Schreiber, Gastroenterology, 2017, 153(6), 1504–1516 CrossRef CAS PubMed.
- C. M. Whitfield-Cargile, N. D. Cohen, R. S. Chapkin, B. R. Weeks, L. A. Davidson, J. S. Goldsby, C. L. Hunt, S. H. Steinmeyer, R. Menon, J. S. Suchodolski, A. Jayaraman and R. C. Alaniz, Gut Microbes, 2016, 7(3), 246–2261 CrossRef CAS.
- B. Lamas, M. L. Richard, V. Leducq, H. P. Pham, M. L. Michel, G. Da Costa, C. Bridonneau, S. Jegou, T. W. Hoffmann, J. M. Natividad, L. Brot, S. Taleb, A. Couturier-Maillard, I. Nion-Larmurier, F. Merabtene, P. Seksik, A. Bourrier, J. Cosnes, B. Ryffel, L. Beaugerie, J. M. Launay, P. Langella, R. J. Xavier and H. Sokol, Nat. Med., 2016, 22, 598–605 CrossRef CAS.
- V. Lazar, L. M. Ditu, G. G. Pircalabioru, I. Gheorghe, C. Curutiu, A. M. Holban, A. Picu, L. Petcu and M. C. Chifiriuc, Front. Immunol., 2018, 9, 1830 CrossRef.
- R. C. Vemuri, R. Gundamaraju, T. Shinde and R. Eri, Benefic. Microbes, 2017, 8(2), 179–192 CrossRef.
- S. Sirisinha, Asian Pac. J. Allergy Immunol., 2016, 34, 249–264 CAS.
- H. Fukui, T. Oshima, Y. Tanaka, Y. Oikawa, Y. Makizaki, H. Ohno, T. Tomita, J. Watari and H. Miwa, Sci. Rep., 2018, 8, 12384 CrossRef PubMed.
- C. L. Boulangé, A. L. Neves, J. Chilloux, J. K. Nicholson and M. E. Dumas, Genome Med., 2016, 8, 42 CrossRef.
- S. Louis, R. M. Tappu, A. Damms-Machado, D. H. Huson and S. C. Bischoff, PLoS One, 2016, 11(2), e0149564 CrossRef PubMed.
- M. Ferrer, A. Ruiz, F. Lanza, S. B. Haange, A. Oberbach, H. Till, R. Bargiela, C. Campoy, M. T. Segura, M. Richter, M. von Bergen, J. Seifert and A. Suarez, Environ. Microbiol., 2013, 15(1), 211–226 CrossRef CAS PubMed.
- A. Woting, N. Pfeiffer, G. Loh, S. Klaus and M. Blaut, mBio, 2014, 5(5), e01530-14 CrossRef PubMed.
- T. Sen, C. R. Cawthon, B. T. Ihde, A. Hajnal, P. M. DiLorenzo, C. B. de La Serre and K. Czaja, Physiol. Behav., 2017, 173, 305–317 CrossRef CAS.
- G. A. Al-Ghalith, P. Vangay and D. Knights, Discov. Med., 2015, 19(103), 81–88 Search PubMed.
- L. J. Kasselman, N. A. Vernice, J. DeLeon and A. B. Reiss, Atherosclerosis, 2018, 271, 203–213 CrossRef CAS.
- C. Graham, A. Mullen and K. Whelan, Nutr. Rev., 2015, 73(6), 376–385 CrossRef PubMed.
- Y. Qin, J. D. Roberts, S. A. Grimm, F. B. Lih, L. J. Deterding, R. Li, K. Chrysovergis and P. A. Wade, Genome Biol., 2018, 19, 7 CrossRef PubMed.
- J. Wang, H. Tang, C. Zhang, Y. Zhao, M. Derrien, E. Rocher, J. E. van-Hylckama Vlieg, K. Strissel, L. Zhao, M. Obin and J. Shen, ISME J., 2015, 9, 1–15 CrossRef CAS PubMed.
- F. Drissi, D. Raoult and V. Merhej, Microb. Pathog., 2017, 106, 182–194 CrossRef PubMed.
- L. M. Cox, S. Yamanishi, J. Sohn, A. V. Alekseyenko, J. M. Leung, I. Cho, S. G. Kim, H. Li, Z. Gao, D. Mahana, J. G. Zárate Rodriguez, A. B. Rogers, N. Robine, P. Loke and M. J. Blaser, Cell, 2014, 158(4), 705–721 CrossRef CAS PubMed.
- S. Bibbò, G. Ianiro, M. P. Dore, C. Simonelli, E. E. Newton and G. Cammarota, Mediators Inflammation, 2018, 2018, 9321643 Search PubMed.
- Y. X. Chen, L. N. Lai, H. Y. Zhang, Y. H. Bi, L. Meng, X. J. Li, X. X. Tian, L. M. Wang, Y. M. Fan, Z. F. Zhao, D. W. Han and C. Ji, World J. Gastroenterol., 2016, 22(10), 2949–2959 CrossRef CAS PubMed.
- V. Giannelli, V. Di Gregorio, V. Iebba, M. Giusto, S. Schippa, M. Merli and U. Thalheimer, World J. Gastroenterol., 2014, 20(45), 16795–16810 CrossRef PubMed.
- A. Zhang, H. Sun and X. Wang, Eur. J. Med. Chem., 2013, 63, 570–577 CrossRef CAS PubMed.
- M. Poeta, L. Pierri and P. Vajro, Children, 2017, 4(8), 66 CrossRef.
- V. Volynets, M. A. Küper, S. Strahl, I. B. Maier, A. Spruss, S. Wagnerberger, A. Königsrainer, S. C. Bischoff and I. Bergheim, Dig. Dis. Sci., 2012, 57(7), 1932–1941 CrossRef CAS.
- F. Del Chierico, V. Nobili, P. Vernocchi, A. Russo, C. Stefanis, D. Gnani, C. Furlanello, A. Zandonà, P. Paci, G. Capuani, B. Dallapiccola, A. Miccheli, A. Alisi and L. Putignani, Hepatology, 2017, 65(2), 451–464 CrossRef CAS PubMed.
- J. Boursier, O. Mueller, M. Barret, M. Machado, L. Fizanne, F. Araujo-Perez, C. D. Guy, P. C. Seed, J. F. Rawls, L. David, G. Hunault, F. Oberti, P. Calès and A. M. Diehl, Hepatology, 2016, 63(3), 764–775 CrossRef CAS PubMed.
- J. C. Ralston, C. L. Lyons, E. B. Kennedy, A. M. Kirwan and H. M. Roche, Annu. Rev. Nutr., 2017, 37, 77–102 CrossRef CAS.
- H. Cao, A. Zhang, H. Zhang, H. Sun and X. Wang, Phytother. Res., 2015, 29(2), 159–166 CrossRef.
- X. N. Li, A. Zhang, H. Sun, Y. Song, D. Zou and X. Wang, J. Sep. Sci., 2016, 39(24), 4700–4711 CrossRef CAS.
- S. Yamada, Y. Takashina, M. Watanabe, R. Nagamine, Y. Saito, N. Kamada and H. Saito, Oncotarget, 2018, 9, 9925–9939 Search PubMed.
- H. Chu, A. Zhang, Y. Han, S. Lu, L. Kong, J. Han, Z. Liu, H. Sun and X. Wang, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2016, 1015–1016, 50–61 CrossRef CAS PubMed.
- A. Zhang, D. Zou, G. Yan, Y. Tan, H. Sun and X. Wang, J. Sep. Sci., 2014, 37(14), 1742–1747 CrossRef CAS PubMed.
- L. R. Lopetuso, V. Petito, C. Graziani, E. Schiavoni, F. Paroni Sterbini, A. Poscia, E. Gaetani, F. Franceschi, G. Cammarota, M. Sanguinetti, L. Masucci, F. Scaldaferri and A. Gasbarrini, J. Dig. Dis., 2018, 36(1), 56–65 CrossRef.
- J. Ahn, R. Sinha, Z. Pei, C. Dominianni, J. Wu, J. Shi, J. J. Goedert, R. B. Hayes and L. Yang, J. Natl. Cancer Inst., 2013, 105(24), 1907–1911 CrossRef CAS.
- S. Wu, K. J. Rhee, E. Albesiano, S. Rabizadeh, X. Wu, H. R. Yen, D. L. Huso, F. L. Brancati, E. Wick, F. McAllister, F. Housseau, D. M. Pardoll and C. L. Sears, Nat. Med., 2009, 15, 1016–1022 CrossRef CAS.
- M. Schaubeck, T. Clavel, J. Calasan, I. Lagkouvardos, S. B. Haange, N. Jehmlich, M. Basic, A. Dupont, M. Hornef, M. von Bergen, A. Bleich and D. Haller, Gut, 2016, 65(2), 225–237 CrossRef CAS.
- Q. Feng, S. Liang S, H. Jia, A. Stadlmayr, L. Tang, Z. Lan, D. Zhang, H. Xia, X. Xu, Z. Jie, L. Su, X. Li, X. Li, J. Li, L. Xiao, U. Huber-Schönauer, D. Niederseer, X. Xu, J. Y. Al-Aama, H. Yang, J. Wang, K. Kristiansen, M. Arumugam, H. Tilg, C. Datz and J. Wang, Nat. Commun., 2015, 6, 6528 CrossRef CAS.
- P. Markowiak and K. Śliżewska, Nutrients, 2017, 9(9), 1021 CrossRef.
- A. Harper, M. M. Naghibi and D. Garcha, Foods, 2018, 7(2), 13 CrossRef PubMed.
- P. Ambalam, M. Raman, R. K. Purama and M. Doble, Best Pract. Res., Clin. Gastroenterol., 2016, 30(1), 119–131 CrossRef PubMed.
- J. Y. Yang, M. S. Kim, E. Kim, J. H. Cheon, Y. S. Lee, Y. Kim, S. H. Lee, S. U. Seo, S. H. Shin, S. S. Choi, B. Kim, S. Y. Chang, H. J. Ko, J. W. Bae and M. Kweon, Immunity, 2016, 44(4), 889–900 CrossRef CAS PubMed.
- L. Zhao, F. Zhang, X. Ding, G. Wu, Y. Y. Lam, X. Wang, H. Fu, X. Xue, C. Lu, J. Ma, L. Yu, C. Xu, Z. Ren, Y. Xu, S. Xu, H. Shen, X. Zhu, Y. Shi, Q. Shen, W. Dong, R. Liu, Y. Ling, Y. Zeng, X. Wang, Q. Zhang, J. Wang, L. Wang, Y. Wu, B. Zeng, H. Wei, M. Zhang, Y. Peng and C. Zhang, Science, 2018, 359(6380), 1151–1156 CrossRef CAS.
- T. Yu, F. Guo, Y. Yu, T. Sun, D. Ma, J. Han, Y. Qian, I. Kryczek, D. Sun, N. Nagarsheth, Y. Chen, H. Chen, J. Hong, W. Zou and J. Y. Fang, Cell, 2017, 170(3), 548–563 CrossRef CAS PubMed.
- X. Wang, A. Zhang, X. Zhou, Q. Liu, Y. Nan, Y. Guan, L. Kong, Y. Han, H. Sun and G. Yan, Sci. Rep., 2016, 6, 18997 CrossRef CAS.
- Q. Liu, A. Zhang, L. Wang, G. Yan, H. Zhao, H. Sun, S. Zou, J. Han, C. W. Ma, L. Kong, X. Zhou, Y. Nan and X. Wang, Sci. Rep., 2016, 6, 38437 CrossRef CAS.
- G. L. Yan, A. H. Zhang, H. Sun, Y. Han, H. Shi, Y. Zhou and X. J. Wang, J. Sep. Sci., 2013, 36(19), 3191–3199 CAS.
- H. Cao, A. Zhang, F. M. Zhang, Q. Q. Wang, H. Zhang, Y. H. Song, Y. Zhou, H. Sun, G. L. Yan, Y. Han and X. Wang, Biomed. Chromatogr., 2014, 28(12), 1774–1781 CrossRef CAS PubMed.
- H. Sun, W. Dong, A. Zhang, W. Wang and X. Wang, J. Sep. Sci., 2012, 35(24), 3477–3485 CrossRef CAS.
- H. Wu, L. Cai, D. Li, X. Wang, S. Zhao, F. Zou and K. Zhou, BioMed Res. Int., 2018, 2018, 2936257 Search PubMed.
- A. Zhang, H. Sun, G. Yan, P. Wang and X. Wang, BioMed Res. Int., 2015, 2015, 354671 Search PubMed.
- J. Wang, Y. Wang, X. Zhang, J. Liu, Q. Zhang, Y. Zhao, J. Peng, Q. Feng, J. Dai, S. Sun, Y. Zhao, L. Zhao, Y. Zhang, Y. Hu and M. Zhang, Front. Microbiol., 2017, 8, 2222 CrossRef PubMed.
- L. Chi, B. Gao, X. Bian, P. Tu, H. Ru and K. Lu, Toxicol. Appl. Pharmacol., 2017, 331, 142–153 CrossRef CAS PubMed.
- B. A. Petriz and O. L. Franco, Front. Chem., 2017, 5, 4 Search PubMed.
- N. Jiang, H. F. Liu, S. D. Li, W. X. Zhou, Y. X. Zhang, Q. Zhang and X. Z. Yan, Acta Pharmacol. Sin., 2015, 36, 689–698 CrossRef CAS.
- V. Marzano, L. Mancinelli, G. Bracaglia, F. Del Chierico, P. Vernocchi, F. Di Girolamo, S. Garrone, H. Tchidjou Kuekou, P. D'Argenio, B. Dallapiccola, A. Urbani and L. Putignani, PLoS Neglected Trop. Dis., 2017, 11(11), e0005916 CrossRef.
- J. R. Fletcher, S. Erwin, C. Lanzas and C. M. Theriot, mSphere, 2018, 3, e00089-18 CrossRef PubMed.
- D. A. Kieffer, B. D. Piccolo, M. L. Marco, E. B. Kim, M. L. Goodson, M. J. Keenan, T. N. Dunn, K. E. Knudsen, R. J. Martin and S. H. Adams, J. Nutr., 2016, 146(12), 2476–2490 CrossRef CAS PubMed.
- S. El Aidy, M. Derrien, R. Aardema, R. Hooiveld, S. E. Richards, A. Dane, J. Dekker, R. Vreeken, F. Levenez, J. Doré, E. G. Zoetendal, P. van Baarlen and M. Kleerebezem, Benefic. Microbes, 2014, 5(1), 67–77 CrossRef CAS PubMed.
- A. Zhang, H. Sun, Z. Wang, W. Sun, P. Wang and X. Wang, Planta Med., 2010, 76(17), 2026–2035 CrossRef CAS.
- J. Yu, L. Kong, A. Zhang, Y. Han, Z. Liu, H. Sun, L. Liu and X. Wang, J. Proteome Res., 2017, 16(9), 3219–3228 CrossRef CAS PubMed.
- H. Sun, A. Zhang and X. Wang, Phytother. Res., 2012, 26(10), 1466–1471 CAS.
- X. Wang, A. Zhang, H. Sun and G. Yan, Chin. Herb. Med., 2016, 8(4), 299–307 CrossRef.
- H. Chu, A. Zhang, Y. Han and X. Wang, World J. Tradit. Chin. Med., 2015, 1, 26–32 Search PubMed.
- X. Wang, A. Zhang, H. Sun, Y. Han and Y. Yan, TrAC, Trends Anal. Chem., 2016, 76, 86–94 CrossRef CAS.
- Q. Zhao, A. Zhang, W. Zong, N. An, H. Zhang, Y. Luan, H. Cao, H. Sun and X. Wang, RSC Adv., 2016, 6, 112534 RSC.
- X. Wang, A. Zhang, G. Yan, Y. Han and H. Sun, TrAC, Trends Anal. Chem., 2014, 63, 180–187 CrossRef CAS.
- T. Zhang, A. Zhang, S. Qiu, S. Yang and X. Wang, Crit. Rev. Anal. Chem., 2015, 46(4), 342–351 CrossRef PubMed.
- J. Ren, A. Zhang, L. Kong and X. Wang, RSC Adv., 2018, 8, 22335–22350 RSC.
- Y. Han, A. H. Zhang, Y. Z. Zhang, H. Sun, X. C. Meng and X. J. Wang, RSC Adv., 2018, 8(22), 11983–11990 RSC.
- J. Xie, A. Zhang, H. Sun, G. Yan and X. Wang, RSC Adv., 2018, 8, 812–824 RSC.
- W. Aw and S. Fukuda, J. Diabetes Invest., 2018, 9(1), 5–12 CrossRef.
- A. W. Janssen and S. Kersten, J. Physiol., 2017, 595(2), 477–487 CrossRef CAS PubMed.
- R. D. Heath, C. Cockerell, R. Mankoo, J. A. Ibdah and V. Tahan, North Clin. Istanb., 2018, 5(1), 79–88 Search PubMed.
- C. Mantegazza, P. Molinari, E. D'Auria, M. Sonnino, L. Morelli and G. V. Zuccotti, Pharmacol. Res., 2018, 128, 63–72 CrossRef PubMed.
- E. R. Davenport, J. G. Sanders, S. J. Song, K. R. Amato, A. G. Clark and R. Knight, BMC Biol., 2017, 15, 127 CrossRef PubMed.
- Q. Song, A. H. Zhang, G. L. Yan, L. Liu and X. J. Wang, RSC Adv., 2017, 7(84), 53516–53524 RSC.
- C. Liu, A. Zhang, G. L. Yan, H. Shi, H. Sun, Y. Han, Y. Zhou and X. Wang, J. Sep. Sci., 2017, 40(3), 663–670 CrossRef CAS PubMed.
- A. Zhang, H. Sun, G. Yan, P. Wang and X. Wang, Biomed. Chromatogr., 2016, 30(1), 7–12 CrossRef.
- A. Zhang, H. Sun, G. Yan, P. Wang and X. Wang, BioMed Res. Int., 2015, 2015, 354671 Search PubMed.
- X. Li, H. Sun, A. Zhang, Z. Liu, D. Zou, Y. Song, L. Liu and X. Wang, J. Sep. Sci., 2017, 40(21), 4102–4112 CrossRef CAS PubMed.
- A. Zhang, Q. Liu, H. Zhao, X. Zhou, H. Sun, Y. Nan, S. Zou, C. W. Ma and X. Wang, Sci. Rep., 2016, 6, 19333 CrossRef CAS PubMed.
- A. Zhang, H. Fang, Y. Wang, G. Yan, H. Sun, X. Zhou, Y. Wang, L. Liu and X. Wang, RSC Adv., 2017, 7, 51069–51078 RSC.
- M. Rajpoot, A. K. Sharma, A. Sharma and G. K. Gupta, Semin. Cancer Biol., 2018, 52(1), 1–8 CrossRef CAS PubMed.
- A. H. Zhang, H. Sun, S. Qiu and X. J. Wang, J. Evidence-Based Complementary Altern. Med., 2013, 2013, 402159 Search PubMed.
- X. N. Li, A. Zhang, M. Wang, H. Sun, Z. Liu, S. Qiu, T. Zhang and X. Wang, Sci. Rep., 2017, 7, 46234 CrossRef CAS.
- X. H. Wu, C. Zhao, A. H. Zhang, J. Q. Zhang, X. Wang, X. L. Sun and X. J. Wang, RSC Adv., 2018, 8(34), 19238–19250 RSC.
- H. L. Gao, A. H. Zhang, J. B. Yu, H. Sun, L. Kong, X. Q. Wang, G. L. Yan, L. Liu and X. J. Wang, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2018, 1092, 286–295 CrossRef CAS PubMed.
- A. H. Zhang, J. B. Yu, H. Sun, L. Kong, X. Q. Wang, Q. Y. Zhang and X. J. Wang, Phytomedicine, 2018, 45, 84–92 CrossRef CAS PubMed.
- A. Zhang, H. Sun and X. Wang, Mass Spectrom. Rev., 2018, 37(3), 307–320 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2018 |