Open Access Article
Open Access ArticleProtein-based fluorescent nanoparticles for super-resolution STED imaging of live cells†
Li
Shang
 *abc,
Peng
Gao
ab,
Haixia
Wang
a,
Radian
Popescu
d,
Dagmar
Gerthsen
d and
Gerd Ulrich
Nienhaus
*abc,
Peng
Gao
ab,
Haixia
Wang
a,
Radian
Popescu
d,
Dagmar
Gerthsen
d and
Gerd Ulrich
Nienhaus
 *abef
*abef
aInstitute of Applied Physics, Karlsruhe Institute of Technology (KIT), 76131 Karlsruhe, Germany
bInstitute of Nanotechnology, Karlsruhe Institute of Technology (KIT), 76344 Eggenstein-Leopoldshafen, Germany
cCenter for Nano Energy Materials, School of Materials Science and Engineering, Northwestern Polytechnical University, 710072, Xi'an, China. E-mail: li.shang@nwpu.edu.cn
dLaboratory of Electron Microscopy, Karlsruhe Institute of Technology (KIT), 76131 Karlsruhe, Germany
eInstitute of Toxicology and Genetics, Karlsruhe Institute of Technology (KIT), 76344 Eggenstein-Leopoldshafen, Germany
fDepartment of Physics, University of Illinois at Urbana-Champaign, Urbana, Illinois 61801, USA. E-mail: uli@uiuc.edu
First published on 19th December 2016
Abstract
Development of nanoparticles for super-resolution imaging (sriNPs) can greatly enrich the toolbox of robust optical probes for biological studies. Moreover, sriNPs enable us to monitor the behavior of engineered nanomaterials in complex biological environments with high spatial resolution, which is important for advancing our understanding of nano–bio interactions. Up to now, reports on sriNPs have been scarce. In this work, we report a facile strategy to prepare protein-based fluorescent NPs that can be utilized as probes in super-resolution microscopy. The method is simple and straightforward, and easily extendible to other types of fluorophores. By using Atto647N–transferrin NPs as an example, we have achieved a roughly four-fold resolution improvement by using STED nanoscopy. These protein-based sriNPs possess excellent biocompatibility, good colloidal stability and photostability, making them attractive candidates for biological studies. Moreover, STED nanoscopy enables the precise imaging of NP structures in living cells, and revealed the co-existence of multiple NPs within one endosomal vesicle.
Introduction
For successful application of nanomaterials in biology and medicine and for the assessment of their toxicity, a mechanistic understanding of their interactions with biological systems is of critical importance.1–3 Fluorescence microscopy is the method of choice to explore nano–bio interactions on the cellular level mainly because of its high sensitivity and molecular specificity. Moreover, it allows monitoring of nano–bio interactions in real time without disturbing the biological environment. Indeed, over the past decades, fluorescence microscopy-based techniques have been widely employed to investigate many important aspects in nanobiotechnology, i.e., formation of the biomolecular corona,4,5 endocytosis mechanisms and intracellular trafficking of nanoparticles (NPs),6–9 and in vivo behavior of administered NPs.10–12Conventional optical microscopy is limited by its moderate spatial resolution (ca. 250 nm), which is greater than the dimensions of many subcellular structures as well as most nanomaterials. Consequently, processes at the nano–bio interface occurring on a spatial scale smaller than the resolution cannot be reliably studied. With the rapid advancement of super-resolution fluorescence microscopy (fluorescence nanoscopy) in recent years, the spatial resolution of optical imaging has been extended far beyond the diffraction limit, approaching virtually molecular resolution.13–15 Undoubtedly, the use of super-resolution microscopy in nanobiotechnology offers vast opportunities for further revealing the intricate interactions between nanomaterials and biological entities, but is still in its infancy.16–18 Nanoscopic imaging, especially in living cells and organisms, relies heavily on suitable photophysical properties of the fluorophore, including high photostability, brightness and facile photocontrollability, i.e., the capability of light-induced switching between ‘on’ and ‘off’ states.19 Many conventional fluorophores including organic dyes and fluorescent proteins have been utilized for live-cell nanoscopy,15,20,21 whereas there are only very few reports employing nanoparticles for this purpose.22–24 Development of NPs for super-resolution imaging (sriNPs) can greatly enrich the toolbox of robust optical nanoprobes for biological studies.25–27 Moreover, they enable us to monitor the behavior of engineered NPs in complex biological environments with higher spatial resolution, which is important for advancing our understanding of nano–bio interactions.28
Here we report a facile and universal strategy of preparing such sriNPs by incorporating commercially available dyes in a controlled fashion into NPs during their synthesis. By using stimulated emission depletion (STED) nanoscopy,29 we demonstrate that these sriNPs retain the photocontrollability of the loaded dyes. In fact, the photostability of the NPs is superior to the one of the incorporated dyes, even under intense STED illumination. We chose transferrin-based protein NPs (Tf NPs) as model NPs in view of their widespread use as drug carriers in clinical medicine30 and the capability of Tf to target a variety of cancer cells that have Tf receptors overexpressed on their cell surfaces.31 By using Atto647N, a red-emitting dye often used for STED nanoscopy for labeling,32,33 we can image the internalization of our sriNPs by living cells with super-resolution.
Results and discussion
Synthesis and characterization of sriNPs
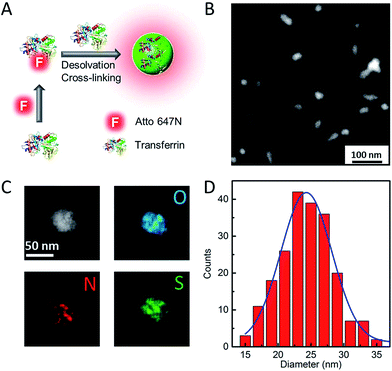
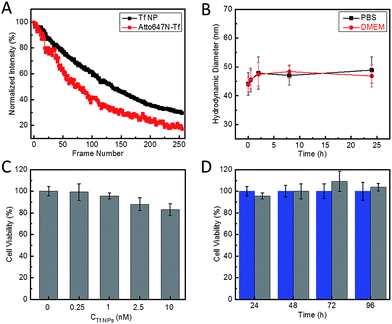
Our strategy to synthesize Atto647N–Tf NPs is fairly straightforward (Fig. 1A). We first labelled the protein by reacting Tf with Atto647N–NHS-ester. Afterwards, Atto647N–Tf was mixed with unlabeled Tf at a defined molar ratio in H2O to adjust the labeling density (brightness), and then desolvated by slowly adding acetone (for details, see the Experimental section, ESI†).34 Upon crosslinking with glutaraldehyde and further purification, Atto647N-loaded Tf NPs were obtained. We characterized the size and morphology of the NPs by using high-angle annular dark-field detector (HAADF) scanning transmission electron microscope (STEM). The NPs appear predominantly as well-isolated spheres. However, some NP clusters are visible that presumably formed during preparation of the STEM samples (Fig. 1B). Statistical analysis of >200 well-dispersed NPs from STEM images yields a diameter of 25 ± 4 nm (Fig. 1D), which is considerably smaller than the size measured by dynamic light scattering (DLS), 44 ± 6 nm (Fig. S1, ESI†). This difference can be attributed to the shrinking of biomaterials during STEM sample preparation, and to the additional hydration layer of NPs in the aqueous medium that DLS is sensitive to. The composition of Atto647N–Tf NPs was evaluated by energy dispersive X-ray spectroscopy (EDXS) elementary mapping on a single-particle level. As shown in Fig. 1C, signals from nitrogen (N Kα1), sulfur (S Kα1) and oxygen (O Kα1), the key elemental composition of protein NPs, were distributed over the whole particle.The fluorescence spectra suggest that the Atto647N–Tf NPs essentially retain the fluorescence properties of the incorporated fluorophores. Compared with the free Atto647N–Tf conjugates in phosphate buffer saline (PBS), the emission maximum of the NPs is shifted slightly from 669 to 667 nm (Fig. S2, ESI†), suggesting that the incorporated Atto647N are mostly embedded in the less polar protein nanostructure. In addition, the altered local microenvironment also modifies the fluorescence lifetime of Atto647N; it increased from 1.52 ± 0.01 ns for free Atto647N–Tf to 1.80 ± 0.03 ns for Atto647N–Tf NPs (Fig. S3, ESI†). Both lifetimes are smaller than that of Atto647N, 3.56 ± 0.01 ns. Fluorophore encapsulation often results in enhanced photostability.35,36 Indeed, upon illumination of Atto647N–Tf NPs with a STED depletion laser (780 nm, 80 MHz pulses of 300 ps width, 28 mW), after acquisition of ca. 130 image frames the fluorescence intensity dropped to half of the original intensity (Fig. 2A), cf. 80 frames for Atto647N–Tf conjugates. Improved photostability of sriNPs is advantageous for long-term imaging as well as fast particle tracking. As for free Atto647N in solution, the fluorescence of Atto647N–Tf NPs is insensitive to solvent pH in the range of 4–10 (Fig. S4, ESI†).
Laser Doppler anemometry of Atto647N–Tf NPs in PBS shows a negative ζ-potential of −38.2 ± 2.7 mV, implying a good colloidal stability in aqueous medium. The hydrodynamic diameter of these sriNPs, as measured by DLS, remains essentially constant in either PBS or cell culture medium (Dulbecco's Modified Eagle's Medium, DMEM) over 24 h (Fig. 2B). In fact, we did not observe any precipitation or agglomeration of these NPs in PBS when stored at 4 °C for more than one year. The robustness of these NPs can be attributed to the efficient chemical crosslinking of the ε-amino groups of lysine residues of Tf molecules by glutaraldehyde. Accordingly, due to removal of basic groups, glutaraldehyde crosslinking also results in a decrease of the isoelectric point (pI) of Tf from 6.0 in the free form to 5.0 in the NPs (Fig. S5, ESI†).37 Moreover, owing to the covalent linkage of Atto647N to proteins, there is no leakage of dyes from the NPs. Additionally, we have synthesized similar sriNPs by replacing Atto647N with other fluorophores, e.g., AlexaFluor 647 (data not shown).
STED imaging using sriNPs
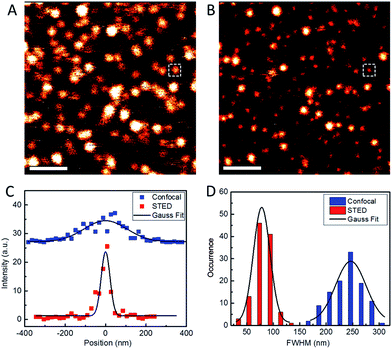
To assess the usability of our Atto647N–Tf NPs for super-resolution imaging, we dispersed the NPs on a glass coverslip and imaged them with our home-built STED confocal microscopy setup.33 Comparison of a typical confocal image with a STED image of the same region shows much sharper NP structures in the latter, implying a marked resolution improvement by using STED (Fig. 3). By analyzing >100 individual NPs and fitting their profiles by Gaussians, we obtained an average diameter of 78 ± 1 nm (FWHM, full width at half maximum) from the STED images (Fig. 3D). In contrast, the average diameter obtained from the confocal reference image was 246 ± 4 nm, as expected for the diffraction-limited confocal microscopy. Considering that the actual diameter of Tf NPs is about 44 nm, as measured by DLS, the resolution of STED and confocal microscopy using our sriNPs were calculated to be 64 and 242 nm, respectively (for details, see the Experimental section, ESI†). Thus a roughly four-fold resolution improvement was achieved by using our sriNPs for STED imaging. Apparently, our Tf NPs retain the super-resolution imaging capability of Atto647N and, therefore, can be used as nanoprobes in biology studies.We further assessed the biocompatibility of Atto647N–Tf NPs. Since they are predominantly composed of proteins, we expected a good biocompatibility. Indeed, we observed that cell viability of HeLa cells was greater than 80% after incubating with Atto647N–Tf NPs in the concentration range of 0–10 nM for 24 h (Fig. 2C), as judged from a MTT assay, which assesses the cells' metabolic activity. Moreover, we did not observe any change in cell viability or proliferation upon Atto647N–Tf NP exposure over a period of 96 h (Fig. 2D and S6, ESI†), which attests to the excellent biocompatibility of these protein-based nanomaterials.
Cellular uptake of sriNPs
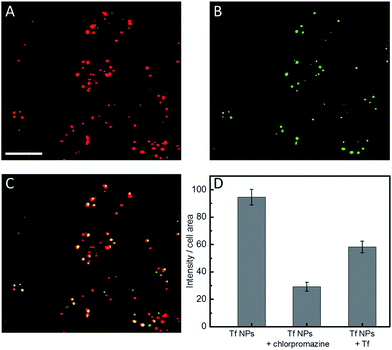
We note that a concentration of Atto647N–Tf NPs as low as 1 nM can already sufficiently stain HeLa cells, as revealed by confocal microscopy images showing bright fluorescence inside live cells after incubation with 1 nM NPs for 2 h (Fig. S7, ESI†). Tf is a well-studied glycoprotein involved in iron transport from blood to cells through internalization via receptor-mediated endocytosis. Thus, tumor cells over-expressing Tf receptors such as HeLa cells are ideal targets for Tf conjugates.38 By using specific cellular organelle markers, we found that these Atto647N–Tf NPs were mainly enriched in endosomal vesicles upon internalization, as evidenced by significant overlap of the fluorescence intensities of Tf NP with early endosome markers in confocal images (Fig. 4). Quantitative analysis revealed that 86% of Tf NPs were colocalized with early endosome structures. Apparently, the NPs have a similar fate as native Tf, which is delivered to endosomes, suggesting that Tf proteins on the NP surface still retain their recognition function.Tf is known to be predominantly internalized via clathrin-coated vesicles.39,40 By using chlorpromazine, an inhibitor known to suppress clathrin-mediated endocytosis,17 we observed 69.3 ± 3.5% decreased fluorescence intensity (normalized by the cell area) from internalized Tf NPs by HeLa cells (Fig. 4D). Moreover, by co-incubating Tf NPs with 20 μM Tf solution, the fluorescence intensity of internalized Tf NPs decreased by 38.5 ± 4.4%. Apparently, free Tf molecules in the culture medium competitively bind to Tf receptors on the cell surfaces, thus suppressing the uptake of Tf NPs. The circular dichroism spectra of Tf within the NP complex were altered from those of native Tf molecules (Fig. S8, ESI†), revealing changes of the protein secondary structure. We expect that proteins in the interior of the NPs will suffer extensive conformational changes due to desolvation and chemical crosslinking, whereas those proteins on the NP surface likely retain more native structure, certainly to an extent that they are still capable of triggering Tf-specific endocytosis processes.
STED imaging using sriNPs in live cells
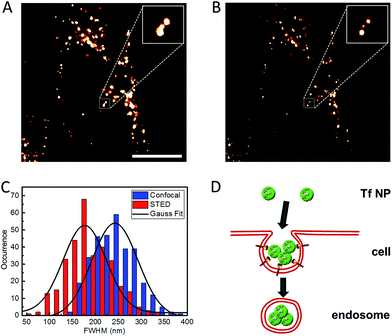
We further explored the application of Atto647N–Tf NPs for super-resolution imaging in live cells by using STED nanoscopy. HeLa cells were imaged after Atto647N–Tf NPs internalization from DMEM for 2 h. In comparison with confocal images, STED images reveal sharper structures (Fig. 5). In many cases, closely adjacent clusters, which appear as larger structures in confocal images, can be well-distinguished in the STED mode (an example is shown in the insets in Fig. 5A and B). From systematic inspection of >300 punctate fluorescent features in 18 different cells, we found that those imaged by STED had an average diameter of 176 ± 5 nm, markedly smaller than the value we obtained from confocal images of 242 ± 3 nm. However, those features are more than twice as large as the images of individual Tf NPs in STED mode (Fig. 3), suggesting that the internalized Tf NPs mostly formed clusters composed of multiple Tf NPs (Fig. 5D).Clathrin assembly typically results in formation of coated pits of about 150 nm, which will be enlarged upon cargo loading, i.e., NP incorporation.17,41,42 Indeed, by using the resolution of STED nanoscopy that we obtained for sriNPs (64 nm), the actual size of fluorescent structures in the cells was calculated to be 164 nm, which is in good agreement with the expected dimension of clathrin-coated vesicles internalizing Tf NPs via receptor-mediated endocytosis. The number of Tf NPs in each vesicle is estimated to be ca. 50, assuming that all volume of the (spherical) vesicle is densely packed by NPs, which, however, may be difficult to achieve, for example, due to solvent molecules surrounding the NPs. Although the close proximity of several NPs inside an endosome makes it challenging to distinguish individual particles, the distinctly larger size of clusters we measured by STED still reliably supports the co-existence of multiple NP inside each endosome.
Besides enhancing the spatial resolution, the temporal resolution of STED imaging can also be improved using these sriNPs. Under identical imaging conditions, the average photon number registered from individual sriNPs was more than four-fold of that registered from single dye molecules (Atto647N, Fig. S9, ESI†). Accordingly, we can reduce the pixel dwell time and therefore, speed up imaging. The resultant temporal resolution will be beneficial in sriNPs applications aimed at monitoring fast dynamics inside living cells.
Conclusions
In summary, we have developed a facile strategy to prepare protein-based fluorescent NPs that can be utilized as probes in super-resolution microscopy. The method is simple and straightforward, and easily extendible to other types of fluorophores as needed for specific applications. By using Atto647N–Tf NPs as an example, we have achieved a roughly four-fold resolution improvement by using STED nanoscopy. Moreover, these protein-based NPs possess excellent biocompatibility, good colloidal stability and photostability, making them attractive candidates for biological studies. As our cellular uptake experiments showed, STED nanoscopy enabled the precise imaging of NP structures in living cells, and revealed the co-existence of multiple NPs within one endosomal vesicle. With abundant functional groups on their surfaces, multifunctional probes based on these sriNPs should be readily achievable, e.g., by endowing them with targeting and/or therapeutic capabilities through facile conjugation. We expect that the combination of STED nanoscopy with protein-based NPs opens up exciting new opportunities for further advances in nanobiotechnology.Acknowledgements
This work was supported by the KIT within the context of the Helmholtz Program STN, and by Deutsche Forschungsgemeinschaft (DFG) grants GRK 2039 and Ni291/12-1. L. S. acknowledges support from the National 1000 Youth Talents Programme. H. W. acknowledges support by China Scholarship Council (CSC) for a PhD scholarship.References
- A. E. Nel, L. Madler, D. Velegol, T. Xia, E. M. V. Hoek, P. Somasundaran, F. Klaessig, V. Castranova and M. Thompson, Nat. Mater., 2009, 8, 543–557 CrossRef CAS PubMed.
- Y.-L. Wu, N. Putcha, K. W. Ng, D. T. Leong, C. T. Lim, S. C. J. Loo and X. Chen, Acc. Chem. Res., 2013, 46, 782–791 CrossRef CAS PubMed.
- Q. Mu, G. Jiang, L. Chen, H. Zhou, D. Fourches, A. Tropsha and B. Yan, Chem. Rev., 2014, 114, 7740–7781 CrossRef CAS PubMed.
- M. Lundqvist, J. Stigler, G. Elia, I. Lynch, T. Cedervall and K. A. Dawson, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 14265–14270 CrossRef CAS PubMed.
- C. Röcker, M. Pötzl, F. Zhang, W. J. Parak and G. U. Nienhaus, Nat. Nanotechnol., 2009, 4, 577–580 CrossRef PubMed.
- X. Jiang, C. Röcker, M. Hafner, S. Brandholt, R. M. Dörlich and G. U. Nienhaus, ACS Nano, 2010, 4, 6787–6797 CrossRef CAS PubMed.
- W. Li, R. Liu, Y. Wang, Y. Zhao and X. Gao, Small, 2013, 9, 1585–1594 CrossRef CAS PubMed.
- L. Yang, L. Shang and G. U. Nienhaus, Nanoscale, 2013, 5, 1537–1543 RSC.
- K. Welsher and H. Yang, Nat. Nanotechnol., 2014, 9, 198–203 CrossRef CAS PubMed.
- X. Gao, Y. Cui, R. M. Levenson, L. W. K. Chung and S. Nie, Nat. Biotechnol., 2004, 22, 969–976 CrossRef CAS PubMed.
- G. Chen, F. Tian, Y. Zhang, Y. Zhang, C. Li and Q. Wang, Adv. Funct. Mater., 2014, 24, 2481–2488 CrossRef CAS.
- W. G. Kreyling, A. M. Abdelmonem, Z. Ali, F. Alves, M. Geiser, N. Haberl, R. Hartmann, S. Hirn, D. J. de Aberasturi, K. Kantner, G. Khadem-Saba, J.-M. Montenegro, J. Rejman, T. Rojo, I. R. de Larramendi, R. Ufartes, A. Wenk and W. J. Parak, Nat. Nanotechnol., 2015, 10, 619–623 CrossRef CAS PubMed.
- S. W. Hell, Science, 2007, 316, 1153–1158 CrossRef CAS PubMed.
- B. Huang, M. Bates and X. Zhuang, Annu. Rev. Biochem., 2009, 78, 993–1016 CrossRef CAS PubMed.
- K. Nienhaus and G. U. Nienhaus, Chem. Soc. Rev., 2014, 43, 1088–1106 RSC.
- S. A. Johnson, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2015, 7, 266–281 CrossRef CAS PubMed.
- Y. Li, L. Shang and G. U. Nienhaus, Nanoscale, 2016, 8, 7423–7429 RSC.
- J. Chen, J. Gao, M. Zhang, M. Cai, H. Xu, J. Jiang, Z. Tian and H. Wang, Sci. Rep., 2016, 6, 30247 CrossRef CAS PubMed.
- M. Fernandez-Suarez and A. Y. Ting, Nat. Rev. Mol. Cell Biol., 2008, 9, 929–943 CrossRef CAS PubMed.
- G. T. Dempsey, J. C. Vaughan, K. H. Chen, M. Bates and X. Zhuang, Nat. Methods, 2011, 8, 1027–1036 CrossRef CAS PubMed.
- A. Byrne, C. S. Burke and T. E. Keyes, Chem. Sci., 2016, 7, 6551–6562 RSC.
- Y. K. Tzeng, O. Faklaris, B. M. Chang, Y. Kuo, J. H. Hsu and H. C. Chang, Angew. Chem., Int. Ed., 2011, 50, 2262–2265 CrossRef CAS PubMed.
- G. Lemenager, E. De Luca, Y. P. Sun and P. P. Pompa, Nanoscale, 2014, 6, 8617–8623 RSC.
- J. Hanne, H. J. Falk, F. Görlitz, P. Hoyer, J. Engelhardt, S. J. Sahl and S. W. Hell, Nat. Commun., 2015, 6, 7127 CrossRef CAS PubMed.
- M. Chen and M. Yin, Prog. Polym. Sci., 2014, 39, 365–395 CrossRef CAS.
- L. Shang, F. Stockmar, N. Azadfar and G. U. Nienhaus, Angew. Chem., Int. Ed., 2013, 52, 11154–11157 CrossRef CAS PubMed.
- X. Ji, F. Peng, Y. Zhong, Y. Su, X. Jiang, C. Song, L. Yang, B. Chu, S.-T. Lee and Y. He, Adv. Mater., 2015, 27, 1029–1034 CrossRef CAS PubMed.
- L. Shang and G. U. Nienhaus, Mater. Today, 2013, 16, 58–66 CrossRef CAS.
- S. W. Hell and J. Wichmann, Opt. Lett., 1994, 19, 780–782 CrossRef CAS PubMed.
- M. J. Hawkins, P. Soon-Shiong and N. Desai, Adv. Drug Delivery Rev., 2008, 60, 876–885 CrossRef CAS PubMed.
- J. Y. Yhee, S. J. Lee, S. Lee, S. Song, H. S. Min, S.-W. Kang, S. Son, S. Y. Jeong, I. C. Kwon, S. H. Kim and K. Kim, Bioconjugate Chem., 2013, 24, 1850–1860 CrossRef CAS PubMed.
- C. Eggeling, C. Ringemann, R. Medda, G. Schwarzmann, K. Sandhoff, S. Polyakova, V. N. Belov, B. Hein, C. von Middendorff, A. Schonle and S. W. Hell, Nature, 2009, 457, 1159–1162 CrossRef CAS PubMed.
- P. N. Hedde, R. M. Dörlich, R. Blomley, D. Gradl, E. Oppong, A. C. B. Cato and G. U. Nienhaus, Nat. Commun., 2013, 4, 2093 Search PubMed.
- W. Lin, A. Coombes, M. Davies, S. Davis and L. Illum, J. Drug Targeting, 1993, 1, 237–243 CrossRef CAS PubMed.
- X. Zhao, R. P. Bagwe and W. Tan, Adv. Mater., 2004, 16, 173–176 CrossRef CAS.
- H. S. Muddana, T. T. Morgan, J. H. Adair and P. J. Butler, Nano Lett., 2009, 9, 1559–1566 CrossRef CAS PubMed.
- K. Langer, S. Balthasar, V. Vogel, N. Dinauer, H. von Briesen and D. Schubert, Int. J. Pharm., 2003, 257, 169–180 CrossRef CAS PubMed.
- L. A. Citores, J. Miguel Ferreras, R. Muñoz, J. Benítez, P. Jiménez and T. Girbés, Cancer Lett., 2002, 184, 29–35 CrossRef CAS PubMed.
- F. Huang, A. Khvorova, W. Marshall and A. Sorkin, J. Biol. Chem., 2004, 279, 16657–16661 CrossRef CAS PubMed.
- J. A. Hanover, M. C. Willingham and I. Pastan, Cell, 1984, 39, 283–293 CrossRef CAS PubMed.
- M. Ehrlich, W. Boll, A. van Oijen, R. Hariharan, K. Chandran, M. L. Nibert and T. Kirchhausen, Cell, 2004, 118, 591–605 CrossRef CAS PubMed.
- O. Lunov, V. Zablotskii, T. Syrovets, C. Röcker, K. Tron, G. U. Nienhaus and T. Simmet, Biomaterials, 2011, 32, 547–555 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6sc04664a |
| This journal is © The Royal Society of Chemistry 2017 |