Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Long non-coding RNA expression profiles predict clinical phenotypes of seminoma and yolk sac tumor†
Li Li *a,
Luo Qianga,
Liu Yufenga,
Zhang Linbcd and
Yao Wub
*a,
Luo Qianga,
Liu Yufenga,
Zhang Linbcd and
Yao Wub
aThe First Affiliated Hospital of Zhengzhou University, Zhengzhou, 450001, China. E-mail: llzdyfy@163.com; Tel: +86-731-67781922
bDepartment of Occupational and Environmental Health, College of Public Health, Zhengzhou University, Zhengzhou, China
cCenter for Reproductive Medicine, Shandong Provincial Hospital Affiliated to Shandong University, 157 Jingliu Road, Jinan 250001, China
dKey Laboratory of Reproductive Endocrinology, Shandong University, Ministry of Education, National Research Center for Assisted Reproductive Technology and Reproductive Genetics, 157 Jingliu Road, Jinan 250001, China
First published on 13th December 2017
Abstract
Malignant germ cell tumors (GCTs) such as seminoma and yolk sac tumor cause serious health problems but with favorable prognosis if they were diagnosed timely. To investigate potential biomarkers used for GCTs diagnosis and phenotype distinguishment, we first applied a lncRNA classification pipeline to identify 368 lncRNAs represented on the Affymetrix Human Genome U133A Array. We then comprehensively analyzed the lncRNA expression patterns in a set of previously published gene expression profiles of seminoma and yolk sac tumor stratified by different age groups (children and adults). The lncRNAs expression signatures between children and adults in different GCTs phenotypic groups were identified respectively, five aberrantly expressed lncRNAs were shared by children and adults, indicating a role for them in distinguishing seminoma from yolk sac tumor regardless of age. In parallel, nine distinctive lncRNAs were also determined between seminoma and yolk sac tumor, which suggested that people may face a high risk of suffering from GCTs. Our findings may contribute to the early diagnosis and prognosis of GCTs regardless of patients' age and other diseases.
Introduction
In recent years, cancer is emerging as a major public health problem worldwide. According to the statistical data released by the American Cancer Society and the National Cancer Center of China, cancer has become the second leading cause of death in both countries.1,2 Over the past decades, the number of patients dying from cancer has substantially dropped, whereas cancer death numbers related to the brain and central nervous system (CNS) are increasing, for now, they have surpassed leukemia becoming the leading cause of cancer deaths in children and adolescents (aged birth to 19 years old).1 Moreover, the 5 year survival rate (%) of brain and CNS cancers has risen from 57% to 74% among patients who were less than 20 years old, whereas the overall survival rate regardless of patient age was still under 35% and showed a decreasing trend.3Germ cell tumors (GCTs), one phenotype of the brain and CNS cancer, accounted for 11.8% of pediatric tumors in China,4 and the malignant GCTs accounted for 2.9% of all malignant tumors in children who were younger than 15 years old worldwide.5 In general, GCTs are characterized by a high heterogeneity of their histological differentiation, but they show a similar histological pattern independent of their primary site or sex.6 As indicated by Teilum in 1965, the neoplastic cell that derived from gonadal or extragonadal germ cell was able to trans-differentiate into embryonal and exo-embryonal malignant carcinoma.7 The former includes mature/immature teratoma in embryo and choriocarcinoma (CHC) and yolk sac tumor (yolk sac tumor) outside the embryo. Meanwhile, the exo-embryonal carcinoma such as seminoma (testis), dysgerminoma (ovary) and germinoma (brain) are all malignant tumors (Fig. 1).
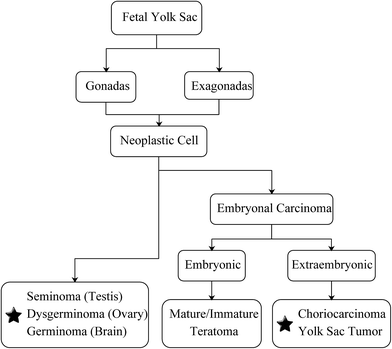 | ||
| Fig. 1 Classification and development of germ cell tumors. Malignant tumors are labeled with pentagram. | ||
In recent years, biomarkers including al-fetoprotein (AFP) and human choriogonadotropin (HCG) have been used for diagnosis of yolk sac tumor and CHC, and a moderate elevation of β-HCG was considered to occur in seminoma. Despite great progress achieved in the early diagnosis and distinguishment of different clinical phenotypes of GCTs, a great amount of misdiagnosis still occurred every year. For example, the reference value of HCG used to diagnosis seminoma/germinoma (<50 IU L−1) was similar to syncytiotrophoblast-like giant cells.8 Additionally, in neonates and young infants, the AFP was born with a physiologically elevated level, but children older than two years old with a high AFP level (≥100 μg L−1) can be considered as malignant GCTs.9 Nevertheless, in some liver diseases such as acute liver failure, hepatocellular carcinoma, and hepatoblastoma, the APF secretion is also elevated due to hepatocellular regeneration.10 Thus, it may lead to incorrect judgment to make clinical decisions only depending on images or molecular biomarkers, and it would be of great significance to find out more stable and accurate biomarkers that were used to diagnosis and distinguish different clinical phenotypes of GCTs regardless of patient age and the disturbance from other diseases.
The emerging role of long non-coding RNA (lncRNA) as promising biomarker and critical therapeutic target has drawn considerable attentions. However, the role of lncRNA in GCTs has not been investigated. Typically, lncRNAs are non-protein coding transcripts longer than 200 nucleotides which were involved in numerous critical biological processes such as X chromosome silencing, genomic imprinting, chromosome modification, transcriptional activation, transcriptional interference, and nuclear transport.11 HOTAIR, for example, a well-studied lncRNA, was found aberrantly expressed in different subtypes of breast cancer, which highlighted the role of lncRNA in distinguishing breast cancer from different subtypes for the first time.12 In glioma and colorectal cancer, lncRNAs such as HOXA-AS, MALAT1, and NEAT1 were all found to be specifically distributed.13,14 Hence it may be a new way to distinguish malignant GCTs from embryonal to exo-embryonal via lncRNA profiling. Favorably, microarray datasets shared by previous studies can be achieved from the Gene Expression Omnibus (GEO) and used to investigate our hypothesis.
Herein, we aimed at profiling lncRNA expression signatures in embryonal malignant carcinoma (yolk sac tumor) and exo-embryonal malignant carcinoma (seminoma) by analyzing a cohort of previously published microarray datasets that achieved from the GEO. The distinctive lncRNAs were identified through comparison between groups of different age and GCTs phenotypes respectively. Our findings provide novel information on lncRNA expression profiles that may help to distinguish GCTs from different phenotypes regardless of the limitation of age and disturbance from other diseases, and the results also provided potential diagnostic biomarkers and therapeutic targets for yolk sac tumor and seminoma.
Materials and methods
GEO seminoma and yolk sac tumor expression data
All experiments were performed in compliance with the guidelines approved by the ethics committee at the Memorial Sloan-Kettering Cancer Center (New York, NY) between 1987 and 1999. Informed consents were obtained from human participants of this study. The microarray datasets of seminoma and yolk sac tumor related to children and adult were obtained from the GEO. To compare the lncRNA expression signatures according to patient of different age and GCT phenotypes, two panels of adult and pediatric GCT gene expression datasets were included in this study: GSE3218 and GSE10615. The raw files of these two datasets which were based on the platform of Affymetrix Human Genome U133A Array were downloaded from the GEO, the data quality control process including quartile normalization, background adjustment, and summarization was processed using the Robust Multichip Average software (RMA, 1.2.0 In Development), which has been proved to be more efficient in estimating lncRNA expression fold changes than other software. Also, samples with a median expression value that exceeded the control limit line in plots of normalized unscaled standard error (NUSE) and relative log expression (RLE) were excluded from the downstream analysis. With this, a set of probe ID-centric gene expression values was obtained.lncRNA classification pipeline
To evaluate lncRNA expressions in the microarray datasets that were obtained from the above step, we adopted the lncRNA classification pipeline which had been previously described to identify lncRNAs represented on the Affymetrix Genome array.15 In brief, we first mapped the ID-centric gene expression matrix to the NetAffx Annotation File (HG-U133A Annotations, CSV format, Release 35, 7 MB, 10/7/2014), which was available on the Affymetrix official website (http://www.affymetrix.com). Next, we only retained probes that labeled as “NR_” in the column of RefSeq transcripts IDs. While in the Ensembl gene IDs column, we selected probes that labeled as “lincRNA,” “processed_transcript,” “macro_lncRNA” or “misc_RNA.” Lastly, we filtered the extracted annotated lncRNAs to exclude pseudogenes, rRNAs, microRNAs or other short RNAs (tRNAs, snRNAs, and snoRNAs).Differentially expressed lncRNAs screening
Gene-e software was used to determine the differentially expressed lncRNAs between seminoma and yolk sac tumor in adult and children. Similarly, the distinctive lncRNAs between adult and children stratified by GCTs phenotypes (seminoma or yolk sac tumor) were also investigated. Conditions used to screen the differentially expressed lncRNAs were set as follows: false discovery rate (FDR) < 20%, fold change ≥ 2, permutation time 1000 and p-value < 0.01. The co-existed differentially expressed lncRNAs between seminoma and yolk sac tumor or adult and children were intersected using Venn diagram. In order to investigate the effectiveness of the pack mode, the principal component analysis (PCA) was adopted using MeV (available at http://mev.tm4.org).Validation of differentially expressed lncRNAs
The Oncomine database which was hosted by Thermo Fisher Scientific Inc. provided more than 715 datasets and 86![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 733 samples with expertly curated data. Thus we took advantage of this database to validate the expression of lncRNAs shared by adults and children, the comparison mode was selected as cancer vs. normal after uploading lncRNAs, and values of fold change, t test, and p statistics were recorded and used for further analysis.
733 samples with expertly curated data. Thus we took advantage of this database to validate the expression of lncRNAs shared by adults and children, the comparison mode was selected as cancer vs. normal after uploading lncRNAs, and values of fold change, t test, and p statistics were recorded and used for further analysis.
Statistical analysis
All statistical analyses were processed using SAS version 9.2 for windows (SAS Institute Inc., Cary, North Carolina, USA). Differentially expressed lncRNAs were investigated using Gene-e software, PCA analysis was adopted using MeV online version. The age of children and adult with normal distribution were shown as mean ± standard deviation (SD). A p value less than 0.05 was considered as statistically significant unless otherwise specified.Results
Datasets characteristics
The gene expression data of pediatric and adult seminoma and yolk sac tumors were included in this study: GES10615 and GSE3218A. The gene chip GSE10615 contained 28 pediatric samples, among which 18 samples were malignant yolk sac tumors, while 10 samples were malignant seminomas, including 1 seminoma sample which was excluded after quality control (Fig. 2 and 3). In parallel, a total number of 21 adult samples including 9 yolk sac tumors and 12 seminomas were included in GES3218A. Each sample involved in this study was purely one phenotype of tumor specified, not mixed with others. The age of the children represented by GSE10615 was 8.3 ± 5.6 years old, while the related indicator was not obtained in the adult group.lncRNA expression profiles on Affymetrix Human Genome U133A Array
With the lncRNA classification pipeline, 398 probe sets corresponding to 368 lncRNA genes were identified. Of these, 49 probe sets (40 genes) were annotated as lncRNAs by both RefSeq and Ensembl database, 267 probe sets (216 genes) were annotated by RefSeq database, and 180 probe sets (192 genes) were annotated by Ensembl database. In addition, probe sets that were annotated by both databases but had controversial definitions were excluded from this study (Tables S1 and S2†).Distinctive lncRNA expressions between seminoma and yolk sac tumor
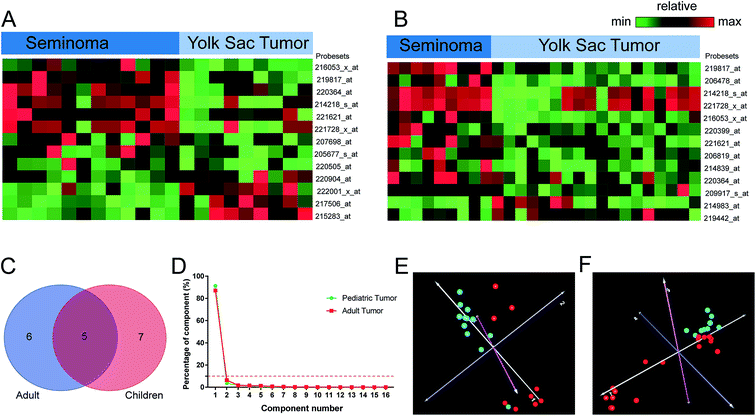
We compared the lncRNA expression patterns between seminoma and yolk sac tumor stratified by age. A total number of 13 probe sets corresponding to 11 lncRNA genes were identified in adults. Meanwhile, 13 probe sets corresponding to 12 genes were found aberrantly expressed in children, and 6 probe sets (5 genes) including XIST, C17orf86, FAM182A, FLJ11235, and C12orf47 were shared by children and adult. Moreover, 19 probe sets corresponding to 16 lncRNA genes were identified between seminoma and yolk sac tumor when the effect of age was excluded (Table 1). Interestingly, five lncRNAs shared by adult and children were all involved in this cluster, and there were 10 differentially expressed lncRNAs that were overlapped in adult and pediatric lncRNA clusters. The PCA results of seminoma and yolk sac tumor were shown in Fig. 4, the cumulative component percentage reached up to 90% when the component number was set as 3. However, it was much higher (more than 90%) when the component number was set as 2 regardless of age, and dots with different colors of the same disease tightly clustered together.| Probesets | RefSeq transcript ID | Ensembl gene ID | Gene symbol | Regulationa | Gene title |
|---|---|---|---|---|---|
| a Compared with seminoma. | |||||
| Adult | |||||
| 205677_s_at | NR_002605 | ENSG00000273541 | DLEU1 | Down | Deleted in lymphocytic leukemia 1 |
| 221728_x_at | NR_001564 | ENSG00000274655 | XIST | Down | X (inactive)-specific transcript |
| 214218_s_at | NR_001564 | ENSG00000274655 | XIST | Down | X (inactive)-specific transcript |
| 207698_at | NR_026773 | ENSG00000146521 | C6orf123 | Down | Chromosome 6 open reading frame 123 |
| 222001_x_at | NR_024510 | ENSG00000277147 | LOC728855 | Up | Hypothetical LOC728855 |
| 205677_s_at | NR_002605 | ENSG00000176124 | DLEU1 | Down | Deleted in lymphocytic leukemia 1 |
| 221621_at | NR_027058 | ENSG00000234912 | C17orf86 | Down | Chromosome 17 open reading frame 86 |
| 220904_at | NR_026780 | ENSG00000231690 | C6orf208 | Up | Chromosome 6 open reading frame 208 |
| 222001_x_at | NR_024510 | ENSG00000226067 | LOC728855 | Down | Hypothetical LOC728855 |
| 220505_at | NR_024274 | ENSG00000224854 | C9orf53 | Down | Chromosome 9 open reading frame 53 |
| 220904_at | NR_026780 | ENSG00000281305 | C6orf208 | Down | Chromosome 6 open reading frame 208 |
| 214218_s_at | NR_001564 | ENSG00000229807 | XIST | Down | X (inactive)-specific transcript |
| 221728_x_at | NR_001564 | ENSG00000229807 | XIST | Down | X (inactive)-specific transcript |
| 216053_x_at | NR_026713 | ENSG00000125804 | FAM182A | Down | Family with sequence similarity 182 |
| 216053_x_at | NR_026713 | ENSG00000175170 | FAM182A | Down | Family with sequence similarity 182 |
| 220364_at | NR_027706 | ENSG00000278921 | FLJ11235 | Down | Hypothetical FLJ11235 |
| 215283_at | NR_015389 | ENSG00000263753 | LOC339290 | Up | Hypothetical LOC339290 |
| 217506_at | NR_015389 | ENSG00000263753 | LOC339290 | Up | Hypothetical LOC339290 |
| 219817_at | NR_015404 | ENSG00000234608 | C12orf47 | Down | Chromosome 12 open reading frame 47 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||
| Child | |||||
| 221728_x_at | NR_001564 | ENSG00000274655 | XIST | Down | X (inactive)-specific transcript |
| 214218_s_at | NR_001564 | ENSG00000274655 | XIST | Down | X (inactive)-specific transcript |
| 206478_at | NR_026800 | ENSG00000226777 | KIAA0125 | Down | KIAA0125 |
| 221621_at | NR_027058 | ENSG00000234912 | C17orf86 | Down | Chromosome 17 open reading frame 86 |
| 206819_at | NR_003714 | ENSG00000197210 | POM121L9P | Down | POM121 membrane glycoprotein-like 9 |
| 209917_s_at | NR_015381 | ENSG00000182165 | TP53TG1 | Down | TP53 target 1 |
| 206819_at | NR_003714 | ENSG00000161103 | POM121L9P | Down | POM121 membrane glycoprotein-like 9 |
| 214218_s_at | NR_001564 | ENSG00000229807 | XIST | Down | X (inactive)-specific transcript |
| 221728_x_at | NR_001564 | ENSG00000229807 | XIST | Down | X (inactive)-specific transcript |
| 216053_x_at | NR_026713 | ENSG00000125804 | FAM182A | Down | Family with sequence similarity 182 |
| 220399_at | NR_024321 | ENSG00000225880 | NCRNA00115 | Down | Non-protein coding RNA 115 |
| 216053_x_at | NR_026713 | ENSG00000175170 | FAM182A | Down | Family with sequence similarity 182 |
| 214839_at | NR_024281 | ENSG00000253230 | LOC157627 | Down | Hypothetical LOC157627 |
| 206819_at | NR_003714 | ENSG00000128262 | POM121L9P | Down | POM121 membrane glycoprotein-like 9 |
| 206478_at | NR_026800 | ENSG00000277059 | KIAA0125 | Down | KIAA0125 |
| 220364_at | NR_027706 | ENSG00000278921 | FLJ11235 | Down | Hypothetical FLJ11235 |
| 219817_at | NR_015404 | ENSG00000234608 | C12orf47 | Down | Chromosome 12 open reading frame 47 |
| 220399_at | NR_024321 | ENSG00000272812 | NCRNA00115 | Down | Non-protein coding RNA 115 |
| 219442_at | NR_024034 | ENSG00000276867 | C16orf67 | Up | Chromosome 16 open reading frame 67 |
| 219442_at | NR_024034 | ENSG00000131797 | C16orf67 | Up | Chromosome 16 open reading frame 67 |
| 214983_at | NR_001545 | ENSG00000233864 | TTTY15 | Up | Testis-specific transcript, Y-linked 15 |
Distinctive lncRNA expressions between children and adult
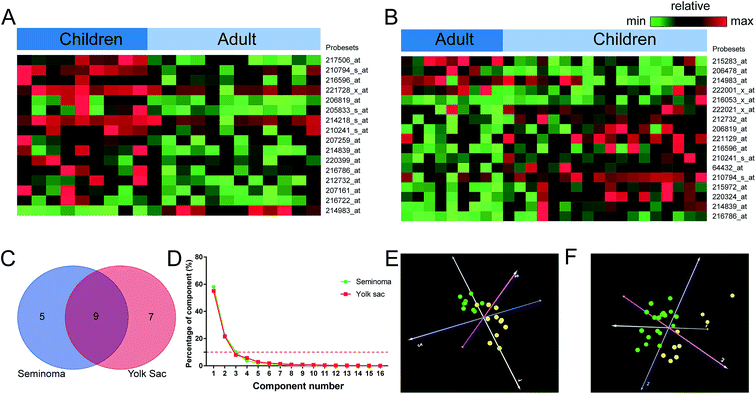
We also compared the lncRNA expression patterns between children and adult stratified by seminoma and yolk sac tumor. In seminoma, 16 probe sets corresponding to 14 lncRNAs were identified, while in yolk sac tumor, 17 probe sets corresponding to 16 lncRNAs were shown to be aberrantly expressed (Table 2). A total number of 9 lncRNAs such as PART1, NCRNA00230A, POM121L9P, MEG3, TP53TG1, LOC157627, LOC339290, DKFZP434L187, and TTTY15 were shared by seminoma and yolk sac tumor (Table 3). The PCA results of children and adult were shown in Fig. 5 with a validity ≥90%, and 2 components were clustered, dots of the same age group were clustered together.| Probesets | RefSeq transcript ID | Ensembl gene ID | Gene symbol | Regulationa | Gene title |
|---|---|---|---|---|---|
| a Compared with children. | |||||
| Seminoma | |||||
| 221728_x_at | NR_001564 | ENSG00000274655 | XIST | Down | X (inactive)-specific transcript |
| 214218_s_at | NR_001564 | ENSG00000274655 | XIST | Down | X (inactive)-specific transcript |
| 205833_s_at | NR_024617 | ENSG00000152931 | PART1 | Down | Prostate androgen-regulated transcript 1 |
| 205833_s_at | NR_024617 | ENSG00000273701 | PART1 | Down | Prostate androgen-regulated transcript 1 |
| 205833_s_at | NR_024617 | ENSG00000275634 | PART1 | Down | Prostate androgen-regulated transcript 1 |
| 216786_at | NR_002161 | ENSG00000233522 | NCRNA00230A | Down | Non-protein coding RNA 230A |
| 207161_at | NR_022006 | ENSG00000122548 | KIAA0087 | Down | KIAA0087 |
| 206819_at | NR_003714 | ENSG00000197210 | POM121L9P | Down | POM121 membrane glycoprotein-like 9 |
| 210794_s_at | NR_002766 | ENSG00000214548 | MEG3 | Down | Maternally expressed 3 |
| 212732_at | NR_002766 | ENSG00000214548 | MEG3 | Down | Maternally expressed 3 |
| 216722_at | NR_001559 | ENSG00000230265 | VENTXP1 | Down | VENT homeobox (Xenopus laevis) 1 |
| 210241_s_at | NR_015381 | ENSG00000182165 | TP53TG1 | Down | TP53 target 1 |
| 206819_at | NR_003714 | ENSG00000161103 | POM121L9P | Down | POM121 membrane glycoprotein-like 9 |
| 207259_at | NR_024626 | ENSG00000167117 | C17orf73 | Down | Chromosome 17 open reading frame 73 |
| 214218_s_at | NR_001564 | ENSG00000229807 | XIST | Down | X (inactive)-specific transcript |
| 221728_x_at | NR_001564 | ENSG00000229807 | XIST | Down | X (inactive)-specific transcript |
| 220399_at | NR_024321 | ENSG00000225880 | NCRNA00115 | Down | Non-protein coding RNA 115 |
| 214839_at | NR_024281 | ENSG00000253230 | LOC157627 | Down | Hypothetical LOC157627 |
| 206819_at | NR_003714 | ENSG00000128262 | POM121L9P | Down | POM121 membrane glycoprotein-like 9 |
| 216786_at | NR_002161 | ENSG00000230663 | NCRNA00230A | Down | Non-protein coding RNA 230A |
| 217506_at | NR_015389 | ENSG00000263753 | LOC339290 | Down | Hypothetical LOC339290 |
| 216722_at | NR_001559 | ENSG00000259849 | VENTXP1 | Down | VENT homeobox (Xenopus laevis) 1 |
| 216596_at | NR_026771 | ENSG00000225930 | DKFZP434L187 | Down | Hypothetical LOC26082 |
| 220399_at | NR_024321 | ENSG00000272812 | NCRNA00115 | Down | Non-protein coding RNA 115 |
| 214983_at | NR_001545 | ENSG00000233864 | TTTY15 | Up | Testis-specific transcript, Y-linked 15 |
| 216596_at | NR_026771 | ENSG00000282096 | DKFZP434L187 | Down | Hypothetical LOC26082 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||
| Yolk sac | |||||
| 206478_at | NR_026800 | ENSG00000226777 | KIAA0125 | Up | KIAA0125 |
| 215972_at | NR_024617 | ENSG00000152931 | PART1 | Down | Prostate androgen-regulated transcript 1 |
| 222001_x_at | NR_024510 | ENSG00000277147 | LOC728855 | Up | Hypothetical LOC728855 |
| 221129_at | NR_026770 | ENSG00000267496 | C17orf88 | Down | Chromosome 17 open reading frame 88 |
| 222021_x_at | NR_003264 | ENSG00000281237 | SDHAP1 | Down | Succinate dehydrogenase complex |
| 222021_x_at | NR_003264 | ENSG00000242086 | SDHAP1 | Down | Succinate dehydrogenase complex |
| 222021_x_at | NR_003264 | ENSG00000281794 | SDHAP1 | Down | Succinate dehydrogenase complex |
| 222021_x_at | NR_003264 | ENSG00000215837 | SDHAP1 | Down | Succinate dehydrogenase complex |
| 222021_x_at | NR_003264 | ENSG00000280912 | SDHAP1 | Down | Succinate dehydrogenase complex |
| 216786_at | NR_002161 | ENSG00000233522 | NCRNA00230A | Down | Non-protein coding RNA 230A |
| 206819_at | NR_003714 | ENSG00000197210 | POM121L9P | Down | POM121 membrane glycoprotein-like 9 |
| 210794_s_at | NR_002766 | ENSG00000214548 | MEG3 | Down | Maternally expressed 3 |
| 212732_at | NR_002766 | ENSG00000214548 | MEG3 | Down | Maternally expressed 3 |
| 222021_x_at | NR_003264 | ENSG00000280512 | SDHAP1 | Down | Succinate dehydrogenase complex |
| 222001_x_at | NR_024510 | ENSG00000226067 | LOC728855 | Up | Hypothetical LOC728855 |
| 222021_x_at | NR_003264 | ENSG00000281687 | SDHAP1 | Down | Succinate dehydrogenase complex |
| 210241_s_at | NR_015381 | ENSG00000182165 | TP53TG1 | Down | TP53 target 1 |
| 206819_at | NR_003714 | ENSG00000161103 | POM121L9P | Down | POM121 membrane glycoprotein-like 9 |
| 222021_x_at | NR_003264 | ENSG00000281334 | SDHAP1 | Down | Succinate dehydrogenase complex |
| 222021_x_at | NR_003264 | ENSG00000073578 | SDHAP1 | Down | Succinate dehydrogenase complex |
| 222021_x_at | NR_003264 | ENSG00000282953 | SDHAP1 | Down | Succinate dehydrogenase complex |
| 216053_x_at | NR_026713 | ENSG00000125804 | FAM182A | Down | Family with sequence similarity 182 |
| 222021_x_at | NR_003264 | ENSG00000281915 | SDHAP1 | Down | Succinate dehydrogenase complex |
| 216053_x_at | NR_026713 | ENSG00000175170 | FAM182A | Down | Family with sequence similarity 182 |
| 214839_at | NR_024281 | ENSG00000253230 | LOC157627 | Down | Hypothetical LOC157627 |
| 206819_at | NR_003714 | ENSG00000128262 | POM121L9P | Down | POM121 membrane glycoprotein-like 9 |
| 216786_at | NR_002161 | ENSG00000230663 | NCRNA00230A | Down | Non-protein coding RNA 230A |
| 206478_at | NR_026800 | ENSG00000277059 | KIAA0125 | Up | KIAA0125 |
| 222021_x_at | NR_003264 | ENSG00000280521 | SDHAP1 | Down | Succinate dehydrogenase complex |
| 222021_x_at | NR_003264 | ENSG00000280993 | SDHAP1 | Down | Succinate dehydrogenase complex |
| 215283_at | NR_015389 | ENSG00000263753 | LOC339290 | Up | Hypothetical LOC339290 |
| 222021_x_at | NR_003264 | ENSG00000280909 | SDHAP1 | Down | Succinate dehydrogenase complex |
| 222021_x_at | NR_003264 | ENSG00000281060 | SDHAP1 | Down | Succinate dehydrogenase complex |
| 222021_x_at | NR_003264 | ENSG00000281603 | SDHAP1 | Down | Succinate dehydrogenase complex |
| 222021_x_at | NR_003264 | ENSG00000281418 | SDHAP1 | Down | Succinate dehydrogenase complex |
| 216596_at | NR_026771 | ENSG00000225930 | DKFZP434L187 | Down | Hypothetical LOC26082 |
| 64432_at | NR_015404 | ENSG00000234608 | C12orf47 | Down | Chromosome 12 open reading frame 47 |
| 220324_at | NR_026807 | ENSG00000233237 | C6orf155 | Down | Chromosome 6 open reading frame 155 |
| 222021_x_at | NR_003264 | ENSG00000185485 | SDHAP1 | Down | Succinate dehydrogenase complex |
| 214983_at | NR_001545 | ENSG00000233864 | TTTY15 | Up | Testis-specific transcript, Y-linked 15 |
| 216596_at | NR_026771 | ENSG00000282096 | DKFZP434L187 | Down | Hypothetical LOC26082 |
| Probesets | RefSeq transcript ID | Ensembl gene ID | Gene symbol | Regulationa | Gene title |
|---|---|---|---|---|---|
| a Compared with seminoma. | |||||
| 205677_s_at | NR_002605 | ENSG00000273541 | DLEU1 | Down | Deleted in lymphocytic leukemia 1 |
| 221728_x_at | NR_001564 | ENSG00000274655 | XIST | Down | X (inactive)-specific transcript |
| 214218_s_at | NR_001564 | ENSG00000274655 | XIST | Down | X (inactive)-specific transcript |
| 206478_at | NR_026800 | ENSG00000226777 | KIAA0125 | Down | KIAA0125 |
| 205834_s_at | NR_024617 | ENSG00000152931 | PART1 | Down | Prostate androgen-regulated transcript 1 |
| 207698_at | NR_026773 | ENSG00000146521 | C6orf123 | Down | Chromosome 6 open reading frame 123 |
| 222001_x_at | NR_024510 | ENSG00000277147 | LOC728855 | Up | Hypothetical LOC728855 |
| 205677_s_at | NR_002605 | ENSG00000176124 | DLEU1 | Down | Deleted in lymphocytic leukemia 1 |
| 205834_s_at | NR_024617 | ENSG00000275634 | PART1 | Down | Prostate androgen-regulated transcript 1 |
| 221621_at | NR_027058 | ENSG00000234912 | C17orf86 | Down | Chromosome 17 open reading frame 86 |
| 206819_at | NR_003714 | ENSG00000197210 | POM121L9P | Down | POM121 membrane glycoprotein-like 9 |
| 220904_at | NR_026780 | ENSG00000231690 | C6orf208 | Up | Chromosome 6 open reading frame 208 |
| 222001_x_at | NR_024510 | ENSG00000226067 | LOC728855 | Up | Hypothetical LOC728855 |
| 210886_x_at | NR_015381 | ENSG00000182165 | TP53TG1 | Down | TP53 target 1 |
| 209917_s_at | NR_015381 | ENSG00000182165 | TP53TG1 | Down | TP53 target 1 |
| 220904_at | NR_026780 | ENSG00000281305 | C6orf208 | Up | Chromosome 6 open reading frame 208 |
| 206819_at | NR_003714 | ENSG00000161103 | POM121L9P | Down | POM121 membrane glycoprotein-like 9 |
| 214218_s_at | NR_001564 | ENSG00000229807 | XIST | Down | X (inactive)-specific transcript |
| 221728_x_at | NR_001564 | ENSG00000229807 | XIST | Down | X (inactive)-specific transcript |
| 216053_x_at | NR_026713 | ENSG00000125804 | FAM182A | Down | Family with sequence similarity 182 |
| 220399_at | NR_024321 | ENSG00000225880 | NCRNA00115 | Down | Non-protein coding RNA 115 |
| 216053_x_at | NR_026713 | ENSG00000175170 | FAM182A | Down | Family with sequence similarity 182 |
| 206819_at | NR_003714 | ENSG00000128262 | POM121L9P | Down | POM121 membrane glycoprotein-like 9 |
| 206478_at | NR_026800 | ENSG00000277059 | KIAA0125 | Down | KIAA0125 |
| 220364_at | NR_027706 | ENSG00000278921 | FLJ11235 | Down | Hypothetical FLJ11235 |
| 215283_at | NR_015389 | ENSG00000263753 | LOC339290 | Up | Hypothetical LOC339290 |
| 217506_at | NR_015389 | ENSG00000263753 | LOC339290 | Up | Hypothetical LOC339290 |
| 219817_at | NR_015404 | ENSG00000234608 | C12orf47 | Down | Chromosome 12 open reading frame 47 |
| 220399_at | NR_024321 | ENSG00000272812 | NCRNA00115 | Down | Non-protein coding RNA 115 |
| 219442_at | NR_024034 | ENSG00000276867 | C16orf67 | Up | Chromosome 16 open reading frame 67 |
| 219442_at | NR_024034 | ENSG00000131797 | C16orf67 | Up | Chromosome 16 open reading frame 67 |
Validation of differentially expressed lncRNAs
The expression levels of lncRNAs shared by yolk sac tumor and seminoma were investigated using Oncomine database and compared between yolk sac tumor and seminoma, yolk sac tumor or seminoma and normal respectively. Apart from lncRNA FAM182A and FLJ11235 which cannot be found in Oncomine database, the expressions of lncRNA XIST and C17orf86 were all of statistical significance between cancer and normal except for C17orf86 in seminoma (Table 4), which were highly consistent with our assumptions.| lncRNA | Seminoma | Yolk sac tumor | ||||
|---|---|---|---|---|---|---|
| Fold change | t value | p value | Fold change | t value | p value | |
| XIST | 12.12 | 8.34 | 9.28 × 10−7 | 2.19 | 2.03 | 0.04 |
| C17orf86 | 1.23 | 2.90 | 0.01 | 1.00 | 0.02 | 0.49 |
Discussion
To date, different phenotypes of malignant GCTs were determined mainly depending on histological changes, but it may be misdiagnosed when facing patients of different ages or suffering from other diseases. As epidemiological and clinical evidence indicated, the histological changes of malignant GCTs among different clinical phenotypes were partly overlapped,16,17 thus biomarkers that were currently used also led to misunderstandings under certain conditions. Therefore, the emerging role of lncRNAs as potential diagnostic biomarkers maybe shine insight on the antidiastole on GCTs phenotypes.In this study, two groups of patients (children and adult) which were stratified by two phenotypes of malignant GCTs (seminoma and yolk sac tumor) were involved. With the lncRNA classification pipeline, we firstly analyzed the differentially expressed lncRNAs between seminoma and yolk sac tumor. A set of 11 aberrantly expressed lncRNAs were identified in adult and 12 lncRNAs were determined in children. Five distinctive lncRNAs including XIST, C17orf86, FAM182A, FLJ11235, and C12orf47 were involved in the intersection of children and adult, indicating a potential role of these lncRNAs in distinguishing seminoma from yolk sac tumor regardless of age. As reported previously, XIST expressions were widely detected in seminomatous testicular germ cell tumors, and the presence of the unmethylated XIST were frequent in testicular germ cell tumors.18 To our best knowledge, the role of the other four lncRNAs in malignant GCTs has not been investigated. However, the lncRNA C17orf86 that was also known as SNHG20 was associated with the metastasis of hepatocellular carcinoma, and the elevated expression level of SNHG20 could promote carcinoma cellular invasion.19,20 Besides, the function of the left three lncRNAs including FAM182A, FLJ11235, and C12orf47 has not been explored even in other diseases.
We also processed comparisons between children and adult, which were stratified by two phenotypes of GCTs (seminoma and yolk sac tumor). In total, 14 expressed lncRNAs were identified in seminoma and 16 lncRNAs were determined in yolk sac tumor. Of these, 9 lncRNAs including PART1, NCRNA00230A, POM121L9P, MEG3, TP53TG1, LOC157627, LOC339290, DKFZP434L187, and TTTY15 were overlapped between seminoma and yolk sac tumor, which suggested that people with one or a certain number of the overlapped lncRNAs may be facing high risk in suffering from seminoma or yolk sac tumor. Coincident with previous studies, MEG3, which has been widely found in many cancers, also regulated the growth of testicular germ cell tumor through PTEN/PI3K/AKT pathway.21 And the lncRNA POM121L9P was pointed out to be associated with male sterility via binding to Piwi proteins in mammalian.22 Another lncRNA, PART1, is a novel human prostate-specific and androgen-regulated gene that loci in chromosome 5q12, the available studies have proved that the expression level of this lncRNA was elevated by approximate 73.1% detected using specimens of stage I–III non-small cell lung cancer.23 Moreover, TP53TG1 was an important regulator of cellular homeostasis, which could undergo cancer-specific promoter hypermethylation-associated silencing and inhibit the occurrence and development of cancer.24 Another lncRNA, TTTY15, was highly cited in prostate cancer, the fusion action mode of this gene with USP9Y was identified in a large cohort study of prostate cancer.25–27 The function of the left lncRNAs is still lack of annotations.
To further explore identical biomarkers that were unaffected by age and GCTs phenotypes, we collaborated lncRNAs shared by groups of different ages or GCTs phenotypes, but none lncRNA fed back when we loaded these two datasets into the statistical software to compare for the overlapped region. However, in the validation process conducted by investigating Oncomine database, we found lncRNA XIST was differentially expressed between cancer and normal, indicating a lncRNA screening criteria of high strict and accurate hold by this study.
In summary, five differentially expressed lncRNAs shared by adult and child were identified in comparison between seminoma and yolk sac tumor, while nine lncRNAs shared by seminoma and yolk sac tumor were determined in comparison between adult and child. The lncRNAs identified in this study may be of great potential in distinguishing GCTs of different phenotypes (seminoma and yolk sac tumor), and they can also be used as promising biomarkers in indicating risk levels from which patients of seminoma or yolk sac tumor may suffer regardless of age. Although some of the lncRNAs had been validated, the majority of them have not been investigated, further studies are still needed.
Conflicts of interest
There are no conflicts of interests.Acknowledgements
This research was supported by the National Natural Science Foundation of China (No. 81273039, 81472954 and 81773404). We thank those who helped us a lot in School of Public Health, Zhengzhou University.References
- R. L. Siegel, K. D. Miller and A. Jemal, Cancer statistics, 2016, Ca-Cancer J. Clin., 2016, 66, 7–30 CrossRef PubMed.
- W. Chen, R. Zheng and P. D. Baade, et al., Cancer statistics in China, 2015, Ca-Cancer J. Clin., 2016, 66, 115–132 CrossRef PubMed.
- K. D. Miller, R. L. Siegel and C. C. Lin, et al., Cancer treatment and survivorship statistics, 2016, Ca-Cancer J. Clin., 2016, 66, 271–289 CrossRef PubMed.
- S. Zhang, G. Liang, Y. Ju and C. You, Clinical and Radiologic Features of Pediatric Basal Ganglia Germ Cell Tumors, World Neurosurg., 2016, 95, 516–524 CrossRef PubMed.
- U. Gobel, D. T. Schneider, G. Calaminus, R. J. Haas, P. Schmidt and D. Harms, Germ-cell tumors in childhood and adolescence. GPOH MAKEI and the MAHO study groups, Ann. Oncol., 2000, 11, 263–271 CrossRef CAS PubMed.
- F. J. Rescorla, Pediatric germ cell tumors, Semin. Surg. Oncol., 1999, 16, 144 CrossRef CAS PubMed.
- G. Teilum, Classification of endodermal sinus tumour (mesoblastoma vitellinum) and so-called “embryonal carcinoma” of the ovary, Acta Pathol. Microbiol. Scand., 1965, 64, 407 CrossRef CAS PubMed.
- I. Juric and N. Basic-Jukic, Testicular Seminoma Occurring After Kidney Transplantation in a Patient Previously Treated for Teratoma: De Novo Malignancy or Recurrence in a Different Histologic Form?, Transplant. Proc., 2016, 48, 3128–3129 CrossRef CAS PubMed.
- H. Zhang, P. Zhang and J. Fan, et al., Determining an Optimal Cutoff of Serum beta-Human Chorionic Gonadotropin for Assisting the Diagnosis of Intracranial Germinomas, PLoS One, 2016, 11, e0147023 Search PubMed.
- Y. Huang, Z. Jia, J. Tu, T. Shen, F. Tian and G. Jiang, Supplemental conventional transarterial embolization/chemoembolization therapy via extrahepatic arteries for hepatocellular carcinoma, J. Cancer Res. Ther., 2017, 13, 720–724 CrossRef PubMed.
- J. J. Quinn and H. Y. Chang, Unique features of long non-coding RNA biogenesis and function, Nat. Rev. Genet., 2016, 17, 47–62 CrossRef CAS PubMed.
- X. Su, G. G. Malouf and Y. Chen, et al., Comprehensive analysis of long non-coding RNAs in human breast cancer clinical subtypes, Oncotarget, 2014, 5, 9864–9876 Search PubMed.
- F. Wu, C. Zhang and J. Cai, et al., Upregulation of long noncoding RNA HOXA-AS3 promotes tumor progression and predicts poor prognosis in glioma, Oncotarget, 2017, 8, 53110–53123 Search PubMed.
- J. Li, Z. Li and W. Zheng, et al., LncRNA-ATB: an indispensable cancer-related long noncoding RNA, Cell Proliferation, 2017, 50, e12381 CrossRef PubMed.
- X. Zhang, S. Sun and J. K. Pu, et al., Long non-coding RNA expression profiles predict clinical phenotypes in glioma, Neurobiol. Dis., 2012, 48, 1–8 CrossRef PubMed.
- J. M. Bieniek, T. Juvet, M. Margolis, E. D. Grober, K. C. Lo and K. A. Jarvi, Prevalence and management of incidental small testicular masses discovered on ultrasonographic evaluation of male infertility, J. Urol. Search PubMed , in press.
- W. Xu and Y. Li, Is Omentectomy Mandatory Among Early Stage (I, II) Malignant Ovarian Germ Cell Tumor Patients? A Retrospective Study of 223 Cases, Int. J. Gynecol. Cancer, 2017, 27, 1373–1378 CrossRef PubMed.
- G. E. Lind, R. I. Skotheim and R. A. Lothe, The epigenome of testicular germ cell tumors, APMIS, 2007, 115, 1147–1160 CrossRef CAS PubMed.
- D. Zhang, C. Cao, L. Liu and D. Wu, Up-regulation of LncRNA SNHG20 Predicts Poor Prognosis in Hepatocellular Carcinoma, J. Cancer, 2016, 7, 608–617 CrossRef PubMed.
- J. Liu, C. Lu, M. Xiao, F. Jiang, L. Qu and R. Ni, Long non-coding RNA SNHG20 predicts a poor prognosis for HCC and promotes cell invasion by regulating the epithelial-to-mesenchymal transition, Biomed. Pharmacother., 2017, 89, 857–863 CrossRef CAS PubMed.
- N. Q. Yang, X. J. Luo, J. Zhang, G. M. Wang and J. M. Guo, Crosstalk between Meg3 and miR-1297 regulates growth of testicular germ cell tumor through PTEN/PI3K/AKT pathway, Am. J. Transl. Res., 2016, 8, 1091–1099 Search PubMed.
- A. Girard, R. Sachidanandam, G. J. Hannon and M. A. Carmell, A germline-specific class of small RNAs binds mammalian Piwi proteins, Nature, 2006, 442, 199–202 Search PubMed.
- M. Li, W. Zhang, S. Zhang, C. Wang and Y. Lin, PART1 expression is associated with poor prognosis and tumor recurrence in stage I–III non-small cell lung cancer, J. Cancer, 2017, 8, 1795–1800 CrossRef PubMed.
- A. Diaz-Lagares, A. B. Crujeiras and P. Lopez-Serra, et al., Epigenetic inactivation of the p53-induced long noncoding RNA TP53 target 1 in human cancer, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, E7535–E7544 CrossRef CAS PubMed.
- Y. Zhang, X. Y. Mao and X. Liu, et al., High frequency of the SDK1:AMACR fusion transcript in Chinese prostate cancer, Int. J. Clin. Exp. Med., 2015, 8, 15127–15136 Search PubMed.
- Y. Zhu, S. Ren and T. Jing, et al., Clinical utility of a novel urine-based gene fusion TTTY15-USP9Y in predicting prostate biopsy outcome, Urol. Oncol., 2015, 33, 384 Search PubMed.
- S. Ren, Z. Peng and J. H. Mao, et al., RNA-seq analysis of prostate cancer in the Chinese population identifies recurrent gene fusions, cancer-associated long noncoding RNAs and aberrant alternative splicings, Cell Res., 2012, 22, 806–821 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra12131h |
| This journal is © The Royal Society of Chemistry 2017 |