Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Self-assembled hybrid nanomaterials with alkaline protease and a variety of metal ions†
Muzi Jingab,
Xu Fei *a,
Weifan Renb,
Jing Tian*b,
Hui Zhib,
Longquan Xua,
Xiuying Wanga and
Yi Wangb
*a,
Weifan Renb,
Jing Tian*b,
Hui Zhib,
Longquan Xua,
Xiuying Wanga and
Yi Wangb
aInstrumental Analysis Center, Dalian Polytechnic University, 1# Qinggongyuan Road, Dalian 116034, P. R. China. E-mail: feixudlpu@163.com; Fax: +86-411-86322038; Tel: +86-411-86323691 ext. 201
bSchool of Biological Engineering, Dalian Polytechnic University, 1# Qinggongyuan Road, Dalian 116034, P. R. China. E-mail: tianjing@dlpu.edu.cn; Tel: +86-411-86324485
First published on 16th October 2017
Abstract
Enzyme immobilization is a well-known essential process to improve enzyme stability and enable the industrial reuse of enzymes for more reaction cycles. In this work, we used alkaline protease and a variety of metal ions (Cu2+, Zn2+, Mn2+ and Ag+) to synthesize hybrid nanomaterials by self-assembly method respectively, but only two kinds of hierarchical flower-like hybrid nanomaterials were formed. These hybrid nanomaterials' structures were characterized by Fourier transform infrared spectroscopy, X-ray diffraction and energy-dispersive X-ray spectroscopy. Compared with free alkaline protease, two kinds of hybrid nanomaterials exhibited higher enzyme activity (∼927% for alkaline protease–Cu3(PO4)2·3H2O hybrid nanomaterials, ∼201% for alkaline protease–Zn3(PO4)2·4H2O hybrid nanomaterials). Enzyme concentration could affect the size and petal density of nanomaterials, so that hampered mass transfer. Furthermore, we concluded that the formation of flower-like hybrid nanomaterials was influenced by atomic radius and the outermost electron orbit of metal ions. These findings have great significance in the synthesis of the hybrid nanomaterials.
1. Introduction
Enzymes have been commonly studied because of their unique properties as industrial catalysts, such as high water-soluble, high reaction specificity, high catalytic activity, low toxicity and biocompatible.1–5 However, loss of activity in solution during the reaction, poor stability of free enzymes in aqueous solution, high cost and lack of reusability strictly hamper their applications in the field of agriculture, food and pharmaceutical industry.6–8 In order to overcome these difficulties, the methods of enzyme immobilization have been developed by many researchers in the past decades. Although most of the immobilized enzymes show increased stability by comparing with free enzymes, several weaknesses exist, including mass-transfer limitation, activity loss due to the harsh chemical synthesis conditions, and unfavorable conformational change in the enzyme structure.9–12Unlike conventional enzyme immobilization methods, a novel and facile method about improving the enzymatic properties of immobilized enzymes has been first developed by Zare's group.13 They have reported a flower-shaped organic–inorganic nanomaterial using copper(II) ions as the inorganic component and various enzymes as the organic component. In recent years, bovine serum albumin,14–16 horseradish peroxidase,2,10,17,18 glucose oxidase,17 papain,19–21 lipase,21–23 urease,24 soybean peroxidase,25 lactoperoxidase,4 trypsin,26 α-amylase12 and chymotrypsin27 have been chosen as organic component. And the inorganic component is focused on Cu3(PO4)2·3H2O, Zn3(PO4)2·4H2O, and CaHPO4 in previous works.14–27 All of these organic–inorganic hybrid nanomaterials exhibit much higher enzyme activity and stability than free and conventional immobilized enzymes. Moreover, this kind of hybrid nanomaterial has attracted widespread interest because of its potential applications in biocatalysis, biosensing, energy storage, gas sensing, proteomic analysis and so on.16,28–38
However, the formation mechanism of the enzyme–inorganic hybrid nanomaterials is still not clear. The effect of different metal ions on the formation of nanomaterials is very important for nano-enzyme immobilization. In the present study, we employed to prepare hybrid nanomaterials with alkaline protease, which can catalyze reactions of hydrolysis of peptide, ester and amide bonds,39–41 as organic component. Alkaline protease–Cu3(PO4)2·3H2O and alkaline protease–Zn3(PO4)2·4H2O hybrid nanomaterials were prepared respectively by a eco-friendly one step procedure and the activity of the as-synthesized hybrid nanomaterials were evaluated. Both of hybrid nanomaterials showed higher enzyme activity than that of free alkaline protease under the optimal enzyme concentration because of their high specific area, which is favorable for mass transfer. In addition, we compared copper with zinc to explore the effect of different patterns on enzyme activity.
2. Experimental section
2.1 Materials
Alkaline protease and α-amylase were obtained from Beijing Aoboxing Bio-tech Co., Ltd. Papain was purchased from Shanghai Yuanye Bio Technology Co., Ltd. Lipase was supplied from Sigma-Aldrich-Fluka Chemical Co., Ltd. N-α-Benzoyl-L-arginine ethyl ester (BAEE) was purchased from Aladdin Reagent Co., Ltd. DTT was obtained from Beijing Solarbio Science and Technology Co., Ltd. ZnSO4, NaCl, KCl, Na2HPO4, KH2PO4 was supplied by Tianjin Damao Chemical Reagent company. CuCl2, MnSO4 and AgNO3 were procured from Shenyang Reagent Industry. All the above mentioned chemical reagents were analytical grade and used as received without further purification. Deionized water was used throughout all experiments.2.2 Measurement
The UV-vis absorption spectra were taken by a Perkin-Elmer LAMBDA35 (USA). Scanning electron microscopy (SEM) was conducted on a JEOL JSM 7800F electron microscope with primary electron energy of 15 kV and energy dispersive X-ray spectroscopy (EDS) was recorded with Oxford INCA. The X-ray diffraction (XRD) was measured on a Shimadzu XRD-7000S diffractometer with Cu Kα radiation (50 kV, 200 mA, λ = 0.154 nm) and a scanning step of 0.02°. The FT-IR spectrum was measured by a Perkin-Elmer Spectrum 10 infrared spectrophotometer (USA).2.3 Preparation of enzyme–inorganic hybrid nanomaterials
Enzyme–Cu3(PO4)2·3H2O hybrid nanomaterials were synthesized by self-assembly method as follow: 20 μL of aqueous CuCl2 solution (120 mM) was mixed with 3 mL of PBS (0.01 M, pH 7.4) containing different concentrations of alkaline protease. The mixture was incubated at 25 °C for three days, then the precipitate was collected by centrifugation (3500 rpm for 5 min) and washed for three times. The final products were dried by vacuum freeze-drier.Enzyme–Zn3(PO4)2·4H2O hybrid nanomaterials were prepared as follow: 80 μL of aqueous ZnSO4 solution (0.05 g mL−1) was added to 1 mL of PBS (0.01 M, pH 7.4) contained different concentrations of alkaline protease. After being mechanical stirring for 3 h, the product was collected as same as the above mentioned procedures.
Similarly, the above mentioned two self-assembly methods were applied to prepare the Mn2+ or Ag+-containing products with papain, lipase and α-amylase respectively.
2.4 Determination of the weight percentage of enzyme in hybrid nanomaterials
For determining the enzyme content, the dried hybrid nanomaterials were calcined at 700 °C for 2 h by muffle furnace. After removing the organic enzyme of the hybrid nanomaterials, Cu3(PO4)2 or Zn3(PO4)2 as the inorganic metal salt was obtained. The weight percentage of enzyme in hybrid nanomaterials was calculated as follow:where W is the weight percentage of enzyme (%), GN is the weight of the hybrid nanomaterials (g), and Go is the weight of Cu3(PO4)2·3H2O or Zn3(PO4)2·4H2O (g).
2.5 Evaluation of catalytic activity of enzyme–inorganic hybrid nanomaterials
The catalytic activity of the alkaline protease–inorganic hybrid nanomaterials and free alkaline protease was evaluated using N-α-benzoyl-L-arginine ethyl ester (BAEE) as the substrate under the same conditions (25 °C, pH 7.4). Alkaline protease could catalyze the hydrolytic cleavage of the ester linkage in BAEE and produce N-α-benzoyl-L-arginine (BA) which can be detected at 253 nm. Briefly, 1 mL activated enzyme solution DTT (30 mM) was added to 2 mL PBS (0.01 M, pH 7.4) contained the alkaline protease–inorganic hybrid nanomaterials (the amount of alkaline protease embedded in nanomaterials was equivalent to 0.25 mg free alkaline protease). After incubating 10 min at 25 °C, 3 mL BAEE (2 mM) was added to the mixture and the reaction time was 5 min. Then the alkaline protease–inorganic hybrid nanomaterials were isolated by centrifugation (3500 rpm, 5 min) rapidly and the supernatant was detected at 253 nm by UV-vis absorption spectroscopy. One activity unit of alkaline protease was defined as the amount of enzyme that produce a ΔA253 of 0.001 per minute with BAEE as substrate under the experimental conditions. The catalytic activity of enzyme–inorganic hybrid nanomaterials and free enzyme was calculated as follow:where U is the enzymatic activity of alkaline protease (U mg−1), ΔA253 is the absorbance changes at 253 nm, GE is the amount of alkaline protease in hybrid nanomaterials (mg), T is the reaction time (min).
3. Results and discussion
3.1 Synthesis and characterization of enzyme–inorganic hybrid nanomaterials
To synthesize the enzyme–inorganic hybrid nanomaterials, aqueous CuCl2 or ZnSO4 solution was added to PBS (0.01 M, pH 7.4) that contained different concentrations of alkaline protease at 25 °C. After rinsing and freeze drying, we obtained a series of hybrid nanomaterials, and the formation mechanism is illustrated in Fig. 1. | ||
| Fig. 1 Proposed growth mechanism of enzyme–Cu3(PO4)2·3H2O hybrid nanomaterials and enzyme–Zn3(PO4)2·4H2O hybrid nanomaterials. | ||
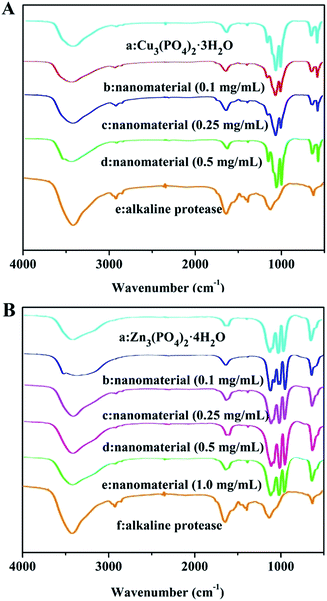
In order to define the structure and compositions of as-prepared hybrid nanomaterials contained different concentrations of alkaline protease. Clearly, FTIR, XRD, EDS were used for systematical analysis of the products. As shown in Fig. 2A(a), the strong characteristic absorptions at 1049 cm−1 (asymmetric stretching), 990 cm−1 (symmetric stretching), and 630 cm−1 (bending), all of which were attributed to P–O vibrations and indicating the existence of phosphate groups. In Fig. 2A(e) and B(f), typical alkaline protease absorption peaks occurred at 1638 cm−1 and 1543 cm−1 for –CONH, 2800 cm−1 to 3000 cm−1 for –CH2 and –CH3, and around 3300 cm−1 for –OH. Comparison of Fig. 2A(b–d) indicated that the characteristic absorption peaks of alkaline protease and Cu3(PO4)2·3H2O were maintained in IR spectra. With the increase of alkaline protease concentration, the characteristic absorption of alkaline protease in the nanomaterials increased gradually until 1.0 mg mL−1 in Fig. 2B(b–d). Over this concentration, excessive enzymes were not able to participate in the hybrid nanomaterials. Similarly, it was shown in Fig. 2B(b–e) that the spectra include main characteristic absorption peaks of alkaline protease and Zn3(PO4)2·4H2O. The strong characteristic absorption peaks at 1016 cm−1, 954 cm−1, 632 cm−1, which were attributed to P–O vibrations [Fig. 2B(a)]. Therefore, the alkaline protease–Zn3(PO4)2·4H2O hybrid nanomaterial was composited by the above two materials.
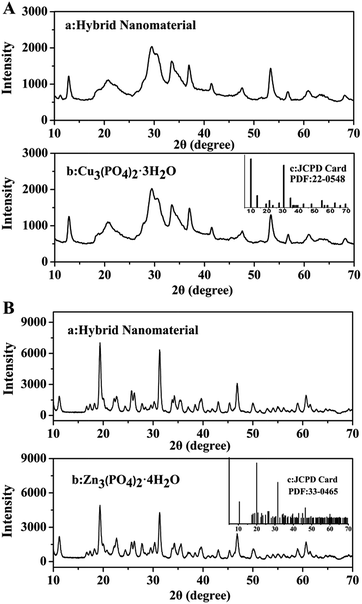
The XRD patterns in Fig. 3 illustrated that the crystallographic structures of two kinds of hybrid nanomaterials were Cu3(PO4)2·3H2O and Zn3(PO4)2·4H2O respectively. All the diffraction peaks presented in Fig. 3A and B could be indexed to Cu3(PO4)2·3H2O (JCPDS card no. 22-0548) and Zn3(PO4)2·4H2O (JCPDS card no. 33-0465) separately. Therefore, the two kinds of hybrid nanomaterials were well crystallized and featured high crystallinity after incorporating alkaline protease.
The EDS patterns of hybrid nanomaterials were shown in Fig. 4A and B respectively. The EDS results revealed the presence of zinc (Zn), copper (Cu), phosphorus (P), oxygen (O), carbon (C) in the sample.
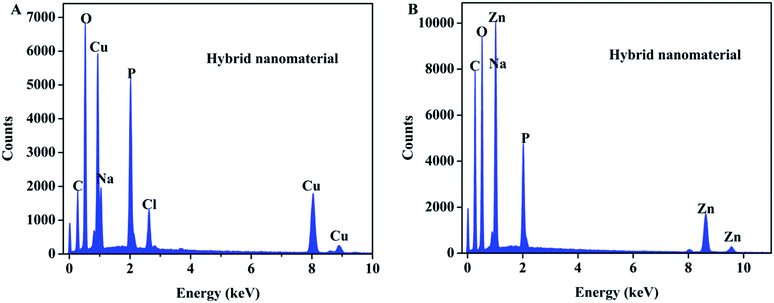 | ||
| Fig. 4 EDS patterns of (A) enzyme–Cu3(PO4)2·3H2O hybrid nanomaterials and (B) enzyme–Zn3(PO4)2·4H2O hybrid nanomaterials. | ||
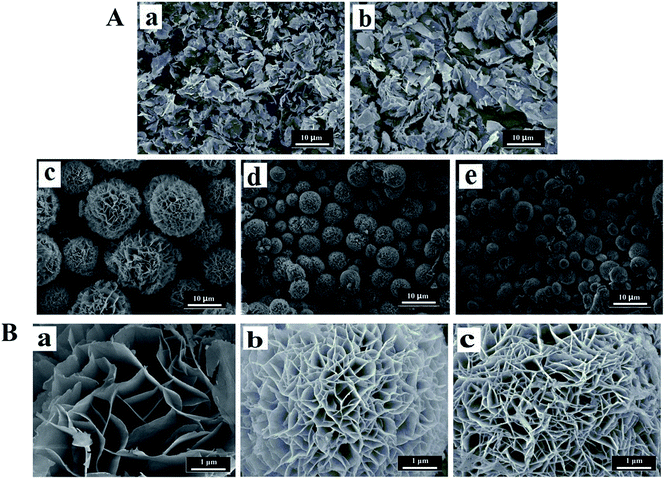
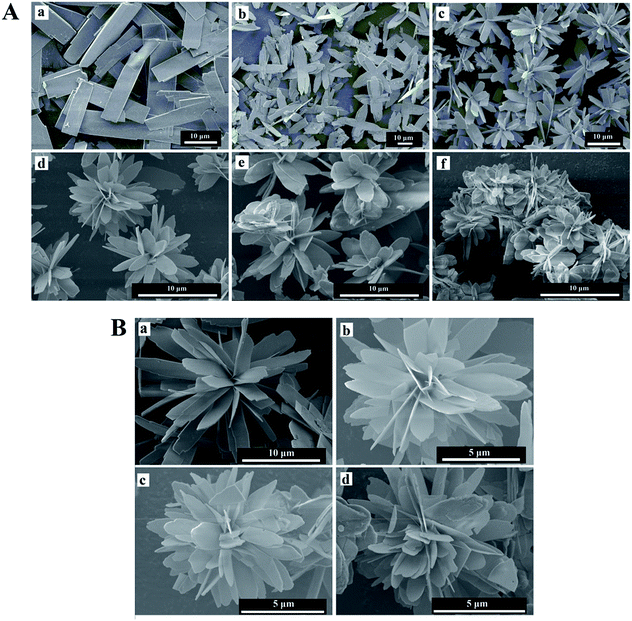
The general morphology of the enzyme–inorganic hybrid nanomaterials were determined by scanning electron microscopy (SEM). The SEM images of hybrid nanomaterials in Fig. 5 and 6, exhibited the flower-like structures and uniform morphology. The alkaline protease–Cu3(PO4)2·3H2O hybrid nanomaterials (Fig. 5A) were like peony. Compared with pure Cu3(PO4)2·3H2O nanomaterials [Fig. 5A(a)], the morphology of the products in the presence of enzyme changed distinctly. However, when the concentration of enzyme was 0.05 mg mL−1, only irregular sheet structures were formed [Fig. 5A(b)]. With increased concentration of alkaline protease (i.e., 0.1, 0.25, 0.5 mg mL−1), the average diameters of these nanomaterials decreased approximately from 12 μm to 4 μm in Fig. 5A(c–e). In addition, the high-resolution SEM images (Fig. 5B) showed that the “petals” of hybrid nanomaterials became more compact and intense.
In the Fig. 6, the morphology of the alkaline protease–Zn3(PO4)2·4H2O hybrid nanomaterials were different from the alkaline protease–Cu3(PO4)2·3H2O hybrid nanomaterials. This kind of hybrid nanomaterial was formed by innumerable nanoplates. The nanoplates were “petals” for assembling of hybrid nanomaterials like daisy. As shown in Fig. 6A(c–f) and B, with the increasing of enzyme concentration (i.e., 0.1, 0.25, 0.5, 1.0 mg mL−1) the size of hybrid nanomaterials decreased obviously (from 15 μm to 8 μm), the layer number of nanoplates increased and the edges of the “petals” were not sharp. Notably, large nanoplate crystals was formed in the absence of alkaline protease [Fig. 6A(a)]. The Above results revealed that enzyme concentration could influence the number of nucleation sites and thus affect the size and structures of hybrid nanomaterials.19,21
The different morphology of enzyme–inorganic hybrid nanomaterials may be due to the difference in arrangement of extranuclear electrons of metal ions (including the number of extranuclear electron layer and the number of electron per layer). Especially, some metal ions, such as Mn2+ and Ag+ couldn't form the hybrid nanomaterials containing regular flower-like structure with enzyme. The alkaline protease, papain, lipase, α-amylase were self-assembled with Mn2+ and Ag+ respectively and the morphology of the resulting materials were shown in Fig. S1.†
The outermost electron orbit of Cu2+ is 3d94s0, and its atomic radius is the important factor to form the nanomaterials with flower-like structure. As congener, Ag+ (the outermost electron orbit of Ag+ is 4d105s0) has too large atomic radius to form flower-like structure. However, the reason for formation of the nanomaterials may be not only the moderate atomic radius, but also the distribution of electrons in the outermost orbital. The outermost electron orbit of Zn2+ is 3d104s0, and the full 3d orbit could improve the stability of electrons in the 4s orbit during coordination. Accordingly, the outermost electron orbit of Mn2+ is 3d54s0 and its half-filled 3d orbit could prevent the ions coordination with enzyme. Therefore, the synthesis of flower-like hybrid nanomaterials was influenced by atomic radius and the outermost electron orbit of metal ions.
3.2 Enzyme activity of enzyme–inorganic hybrid nanomaterials
To determine the catalytic activity of enzyme–Cu3(PO4)2·3H2O and enzyme–Zn3(PO4)2·4H2O formed with different alkaline protease concentrations. The weight percentages of alkaline protease and enzyme activity of two kinds of hybrid nanomaterials were presented in Tables 1 and 2 respectively. The results indicated that the weight percentages of alkaline protease in the two kinds of nanomaterials increased with increasing of alkaline protease concentration (from 0.1 to 0.5 and from 0.1 to 1.0 mg mL−1 separately). The enzyme activity of alkaline protease–Cu3(PO4)2·3H2O hybrid nanomaterials formed by 0.1, 0.25 and 0.5 mg mL−1 of alkaline protease in solution were determined to be 868.19, 491.07 and 236.87 U mg−1 respectively. Meanwhile, the enzyme activity of alkaline protease–Zn3(PO4)2·4H2O hybrid nanomaterials formed using different concentrations of alkaline protease (i.e., 0.1, 0.25, 0.5, 1.0 mg mL−1) were evaluated to be 158.25, 188.76, 176.31, 88.39 U mg−1 respectively. Compared the activity of free alkaline protease (93.7 U mg−1), such increase in enzyme activity can be ascribed to flower-like structures featuring large surface areas and extensive confinement, which resulted in high substrate accessibility to the active sites of alkaline protease. However, the catalytic activity of enzyme–Zn3(PO4)2·4H2O hybrid nanomaterials synthesized with 1.0 mg mL−1 of the alkaline protease was a little lower than the free alkaline protease, the reason was assumed due to transfer limitation, which may be due to high density of “nanopetal” structure. Therefore, we assumed that the enzyme concentration influenced the catalytic activity of hybrid nanomaterials by affecting the structure of hybrid nanomaterials, including size and density.| Enzyme concentration (mg mL−1) | Nanomaterials (g) | Cu3(PO4)2 (g) | Cu3(PO4)2·3H2O (g) | Weight percentage of enzyme (%) | The average value of weight percentage (%) | Enzymatic activity (U mg−1) |
|---|---|---|---|---|---|---|
| 0.1 | 0.2291 | 0.1880 | 0.2147 | 6.29 | 6.50 ± 0.20 | 868.19 ± 14.84 |
| 0.2485 | 0.2031 | 0.2319 | 6.68 | |||
| 0.2758 | 0.2257 | 0.2578 | 6.53 | |||
| 0.25 | 0.1806 | 0.1293 | 0.1477 | 18.22 | 17.40 ± 0.82 | 491.07 ± 8.11 |
| 0.3486 | 0.2546 | 0.2908 | 16.58 | |||
| 0.3171 | 0.2293 | 0.2619 | 17.41 | |||
| 0.5 | 0.2923 | 0.1820 | 0.2078 | 28.91 | 31.05 ± 1.86 | 236.87 ± 2.33 |
| 0.2864 | 0.1705 | 0.1947 | 32.02 | |||
| 0.3004 | 0.1783 | 0.2036 | 32.22 |
| Enzyme concentration (mg mL−1) | Nanomaterials (g) | Zn3(PO4)2 (g) | Zn3(PO4)2·4H2O (g) | Weight percentage of enzyme (%) | The average value of weight percentage (%) | Enzymatic activity (U mg−1) |
|---|---|---|---|---|---|---|
| 0.1 | 0.5022 | 0.4129 | 0.4900 | 2.43 | 2.68 ± 0.42 | 158.25 ± 6.81 |
| 0.5728 | 0.4709 | 0.5588 | 2.44 | |||
| 0.5439 | 0.4439 | 0.5267 | 3.16 | |||
| 0.25 | 0.5596 | 0.4470 | 0.5304 | 5.22 | 5.17 ± 0.27 | 188.76 ± 9.81 |
| 0.5984 | 0.4797 | 0.5692 | 4.88 | |||
| 0.6385 | 0.5090 | 0.6040 | 5.40 | |||
| 0.5 | 0.6902 | 0.5342 | 0.6339 | 8.16 | 8.66 ± 0.49 | 176.31 ± 2.08 |
| 0.6094 | 0.4690 | 0.5565 | 8.68 | |||
| 0.5733 | 0.4390 | 0.5209 | 9.14 | |||
| 1.0 | 0.5464 | 0.3940 | 0.4675 | 14.44 | 13.68 ± 0.97 | 88.39 ± 0.80 |
| 0.6097 | 0.4418 | 0.5243 | 14.01 | |||
| 0.5012 | 0.3692 | 0.4381 | 12.59 |
Based above results, the enzyme–Cu3(PO4)2·3H2O hybrid nanomaterials (formed with 0.1 mg mL−1 alkaline protease) and enzyme–Zn3(PO4)2·4H2O hybrid nanomaterials (formed with 0.25 mg mL−1 alkaline protease) were chosen to determine the reusability of the hybrid nanomaterials respectively. After reacting with substrate each time, the hybrid nanomaterials were centrifuged and washed three times with deionized water. Then the resulting product was applied in a subsequent catalytic cycle. The value of enzyme activity for first reaction cycle was regarded as 100%. As shown in Fig. 7A, after the sixth cycle, although enzyme activity of enzyme–Cu3(PO4)2·3H2O hybrid nanomaterials was reduced to 21.21%, the activity still much higher than free alkaline protease. When enzyme–Zn3(PO4)2·4H2O hybrid nanomaterials reacted with substrate after three times, the relative activity was reduced to 23.04% (Fig. 7B). However, after the third cycle, the activity was lower than free alkaline protease. From the SEM images of two kinds of hybrid nanomaterials after reacting each time (Fig. S2†), we found that the petals density of nanomaterials have decreased. Therefore, we assumed that the main reason of decreasing enzyme activity may be due to the reduction of hierarchical structure.
 | ||
| Fig. 7 Effect of recycling on the activity of (A) enzyme–Cu3(PO4)2·3H2O hybrid nanomaterials and (B) enzyme–Zn3(PO4)2·4H2O hybrid nanomaterials. | ||
Significantly, in the previous similar studies, several methods have been developed to immobilize the alkaline protease on different supports, such as core–mesoporous shell silica nanospheres, TiO2 nanoparticles, gold–TiO2 core–shell nanowires, alginate calcium chloride core–shell microcapsules and so on.42–46 Compared with these strategies, in our works, alkaline protease–inorganic hybrid nanomaterials were prepared by a feasible self-assembly method. This method belongs to a physical process without any toxic organic solvents, thus the activity of alkaline protease can be maintained and exhibit higher than free enzyme. Moreover, this facile and low cost method can overcome the existing weaknesses about low stability and activity loss or inactivation of enzyme, which can make industrial production easy to realize.
4. Conclusions
We synthesized two kinds of composite materials with different flower-like morphology. After calculating the actual weight percentage of alkaline protease in hybrid nanomaterials by calcination method, the accurate enzymatic activity of them were obtained and much higher than that of the free alkaline protease in solution. And we found that enzyme concentration affected the size and petal density of nanomaterials, so that hampered mass transfer, which then directly affected the catalytic activity of hybrid nanomaterials. In addition, we concluded that the formation of flower-like hybrid nanomaterials was influenced by atomic radius and the outermost electron orbit of metal ions. These findings have great significance in the synthesis of these hybrid nanomaterials, which have potential in industrial applications.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (31771914, 21204007), Natural Science Foundation of Liaoning Province of China (20170540076, 20170540079).Notes and references
- X. Wu, M. Hou and J. Ge, Catal. Sci. Technol., 2015, 5, 5077–5085 CAS.
- I. Ocsoy, E. Dogru and S. Usta, Enzyme Microb. Technol., 2015, 75–76, 25–29 CrossRef CAS PubMed.
- B. Krajewska, Enzyme Microb. Technol., 2004, 35, 126–139 CrossRef CAS.
- C. Altinkaynak, I. Yilmaz, Z. Koksal, H. Özdemir, I. Ocsoy and N. Özdemir, Int. J. Biol. Macromol., 2016, 84, 402–409 CrossRef CAS PubMed.
- P. Wang, Curr. Opin. Biotechnol., 2006, 17, 574–579 CrossRef CAS PubMed.
- A. Schmid, J. S. Dordick, B. Hauer, A. Kiener, M. Wubbolts and B. Witholt, Nature, 2001, 409, 258–268 CrossRef CAS PubMed.
- R. N. Patel, Expert Opin. Drug Discovery, 2008, 3, 187–245 CrossRef CAS PubMed.
- N. J. Turner, Nat. Chem. Biol., 2009, 5, 567–573 CrossRef CAS PubMed.
- E. T. Hwang and M. B. Gu, Eng. Life Sci., 2013, 13, 49–61 CrossRef CAS.
- Z. Lin, Y. Xiao, Y. Yin, W. Hu, W. Liu and H. Yang, ACS Appl. Mater. Interfaces, 2014, 6, 10775–10782 CAS.
- G. He, W. Hu and C. M. Li, Colloids Surf., B, 2015, 135, 613–618 CrossRef CAS PubMed.
- L. B. Wang, Y. C. Wang, R. He, A. Zhuang, X. P. Wang, J. Zeng and J. G. Hou, J. Am. Chem. Soc., 2013, 135, 1272–1275 CrossRef CAS PubMed.
- J. Ge, J. D. Lei and R. N. Zare, Nat. Nanotechnol., 2012, 7, 428–432 CrossRef CAS PubMed.
- Z. Zhang, Y. Zhang, L. He, Y. Yang, S. Liu, M. Wang, S. Fang and G. Fu, J. Power Sources, 2015, 284, 170–177 CrossRef CAS.
- B. Zhang, P. Li, H. Zhang, X. Li, L. Tian, H. Wang, X. Chen, N. Ali, Z. Ali and Q. Zhang, Appl. Surf. Sci., 2016, 366, 328–338 CrossRef CAS.
- H. Cao, D.-P. Yang, D. Ye, X. Zhang, X. Fang, S. Zhang, B. Liu and J. Kong, Biosens. Bioelectron., 2015, 68, 329–335 CrossRef CAS PubMed.
- J. Sun, J. Ge, W. Liu, M. Lan, H. Zhang, P. Wang, Y. Wang and Z. Niu, Nanoscale, 2014, 6, 255–262 RSC.
- B. Somturk, M. Hancer, I. Ocsoy and N. Ozdemir, Dalton Trans., 2015, 44, 13845–13852 RSC.
- L. Liang, X. Fei, Y. Li, J. Tian, L. Xu, X. Wang and Y. Wang, RSC Adv., 2015, 5, 96997–97002 RSC.
- B. Zhang, P. Li, H. Zhang, L. Fan, H. Wang, X. Li, L. Tian, N. Ali, Z. Ali and Q. Zhang, RSC Adv., 2016, 6, 46702–46710 RSC.
- Y. Li, X. Fei, L. Liang, J. Tian, L. Xu, X. Wang and Y. Wang, J. Mol. Catal. B: Enzym., 2016, 133, 92–97 CrossRef CAS.
- Z. Wu, X. Li, F. Li, H. Yue, C. He, F. Xie and Z. Wang, RSC Adv., 2014, 4, 33998–34002 RSC.
- B. Zhang, P. Li, H. Zhang, H. Wang, X. Li, L. Tian, N. Ali, Z. Ali and Q. Zhang, Chem. Eng. J., 2016, 291, 287–297 CrossRef CAS.
- B. Somturk, I. Yilmaz, C. Altinkaynak, A. Karatepe, N. Özdemir and I. Ocsoy, Enzyme Microb. Technol., 2016, 86, 134–142 CrossRef CAS PubMed.
- Y. Yu, X. Fei, J. Tian, L. Xu, X. Wang and Y. Wang, Colloids Surf., B, 2015, 130, 299–304 CrossRef CAS PubMed.
- Z. Lin, Y. Xiao, L. Wang, Y. Yin, J. Zheng, H. Yang and G. Chen, RSC Adv., 2014, 4, 13888–13891 RSC.
- Y. Yin, Y. Xiao, G. Lin, Q. Xiao, Z. Lin and Z. Cai, J. Mater. Chem. B, 2015, 3, 2295–2300 RSC.
- J. Zeng and Y. Xia, Nat. Nanotechnol., 2012, 7, 415–416 CrossRef CAS PubMed.
- J. Ge, D. Lu, Z. Liu and Z. Liu, Biochem. Eng. J., 2009, 44, 53–59 CrossRef CAS.
- C.-Y. Chiu, Y. Li, L. Ruan, X. Ye, C. B. Murray and Y. Huang, Nat. Chem., 2011, 3, 393–399 CrossRef CAS PubMed.
- C. K. King'ondu, A. Iyer, E. C. Njagi, N. Opembe, H. Genuino, H. Huang, R. A. Ristau and S. L. Suib, J. Am. Chem. Soc., 2011, 133, 4186–4189 CrossRef PubMed.
- M. D. Ye, H. Y. Liu, C. J. Lin and Z. Q. Lin, Small, 2013, 9, 312–321 CrossRef CAS PubMed.
- S. K. S. Patel, S. V. Otari, Y. Chan Kang and J.-K. Lee, RSC Adv., 2017, 7, 3488–3494 RSC.
- J. Jiao, X. Xin, X. Wang, Z. Xie, C. Xia and W. Pan, RSC Adv., 2017, 7, 43474–43482 RSC.
- K. Mandel, F. Dillon, A. A. Koos, Z. Aslam, F. Cullen, H. Bishop, A. Crossley and N. Grobert, RSC Adv., 2012, 2, 3748–3752 RSC.
- C. Ke, Y. Fan, Y. Chen, L. Xu and Y. Yan, RSC Adv., 2016, 6, 19413–19416 RSC.
- N. Sharma, M. Parhizkar, W. Cong, S. Mateti, M. A. Kirkland, M. Puri and A. Sutti, RSC Adv., 2017, 7, 25437–25443 RSC.
- Z. Xu, R. Wang, C. Liu, B. Chi, J. Gao, B. Chen and H. Xu, RSC Adv., 2016, 6, 30791–30794 RSC.
- A. E. Głowacka, E. Podstawka, M. H. Szczęsna-Antczak, H. Kalinowska and T. Antczak, Comp. Biochem. Physiol., Part B: Biochem. Mol. Biol., 2005, 140, 321–331 CrossRef PubMed.
- A. Anwar and M. Saleemuddin, Bioresour. Technol., 1998, 64, 175–183 CrossRef CAS.
- U. Patil and A. Chaudhari, J. Chem. Technol. Biotechnol., 2009, 84, 1255–1262 CrossRef CAS.
- M. S. Sadjadi, N. Farhadyar and K. Zare, Superlattices Microstruct., 2009, 46, 77–83 CrossRef CAS.
- A. S. Shebl Ibrahim, A. A. Al-Salamah, A. M. El-Toni, K. S. Almaary, M. A. El-Tayeb, Y. B. Elbadawi and G. Antranikian, Int. J. Mol. Sci., 2016, 17, 184 CrossRef PubMed.
- N. Farhadyar and M. S. Sadjadi, J. Biomed. Nanotechnol., 2011, 7, 466–470 CrossRef CAS PubMed.
- S. Guleria, A. Walia, A. Chauhan and C. K. Shirkot, 3 Biotech, 2016, 6, 208 CrossRef PubMed.
- A. S. Shebl Ibrahim, A. M. El-Toni, A. A. Al-Salamah, K. S. Almaary, M. A. El-Tayeb, Y. B. Elbadawi and G. Antranikian, Bioprocess Biosyst. Eng., 2016, 39, 793–805 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra10597e |
| This journal is © The Royal Society of Chemistry 2017 |