Open Access Article
Open Access ArticleEstablishment of a rapid and sensitive method based on recombinase polymerase amplification to detect mts90, a new molecular target of Mycobacterium tuberculosis†
Yunjun Moa,
Fang Cuib,
Dairong Lic,
Yi Daid,
Xinmin Lia,
Xinyuan Zhanga,
Yulan Qiua,
Yibing Yina,
Xuemei Zhanga and
Wenchun Xu *a
*a
aKey Laboratory of Laboratory Medical Diagnostics Designated by the Ministry of Education, School of Laboratory Medicine, Chongqing Medical University, No. 1 Yixueyuan Road, Yuzhong District, Chongqing, 400016, P. R. China. E-mail: 100977@cqmu.edu.cn; Fax: +86 23 68485992; Tel: +86 23 68485239
bDepartment of Laboratory Medicine, The First Affiliated Hospital of Chongqing Medical University, Chongqing 400016, P. R. China
cDepartment of Respiratory Medicine, The First Affiliated Hospital of Chongqing Medical University, Chongqing 400016, P. R. China
dInstitute of Life Sciences, Chongqing Medical University, Chongqing 400016, P. R. China
First published on 26th October 2017
Abstract
Tuberculosis (TB) remains a significant challenge to public health, especially in developing countries. Failure in early diagnosis and lack of rapid and accurate diagnostic methods lead to ongoing prevalence and transmission of TB. Recently, the recombinase polymerase amplification (RPA) technique has made it possible to rapidly amplify and detect nucleic acids without specialized devices. We developed a RPA-based method for identifying Mycobacterium tuberculosis (MTB) by detecting mts90, a more specific target identified in our previous research. Different screening methods were employed for selecting a preferred primer pair of amplification, and probes were confirmed as very fast and reliable tools in the screening of potential primer candidates. The results showed that the mts90 RPA assay was very sensitive and capable of detecting 6 copies of recombinant plasmid containing mts90 sequence per reaction. The assay was specific for detecting MTB, as it did not identify the genomic DNA from other mycobacteria and pathogens. When applied to analyze clinical samples, including sputum, bronchoalveolar lavage fluid (BALF) and tissues, the mts90 RPA assay had a coincidence rate of 96.43% (27/28) compared to the Biochip test, which has been used in clinics for diagnosing TB. The mts90 RPA assay can be completed within 20 minutes at 39 °C without thermal cycling; its simple operation and rapid detection suggest RPA-based MTB assays could be further developed for TB diagnosis in resource-poor settings.
1. Introduction
Tuberculosis (TB) is an infectious disease primarily caused by Mycobacterium tuberculosis complex (MTC) with a high morbidity and mortality, especially in TB endemic countries and regions where healthcare resources are limited and inaccessible. The World Health Organization (WHO) “global TB report 2016” estimated that there were about 10.4 million new TB cases globally in 2015 and noted a 4.3 million gap between the estimated number of incident cases and notifications of new cases,1 indicating that there is a serious misdiagnosis in the current TB diagnosis and treatment. TB still poses a global health threat due to a lack of accurate diagnosis and timely treatment, coupled with HIV co-infection and the continued emergence of multidrug resistant or extreme drug-resistant strains.2–5 Focusing on global TB control, the United Nations put an end to the global TB epidemic as one of the 2030 sustainable development goals in the post-2015 era,6 and a strategy directed by WHO is to achieve a rapid and accurate diagnosis for more TB suspected patients.7Conventional methods of TB diagnosis are sputum smear microscopy and culture. Smear microscopy is the primary test to diagnose TB. Many countries are phasing out this method because of its poor sensitivity and specificity.1,8 Bacterial culture is the current reference standard method; however, the requirement for laboratory capacities and up to 12 weeks to finalize the results and provide reports restrict its applications.1,9 Nucleic acid amplification tests (NAAT) aiming at a rapid and accurate detection of TB can avoid the limitations of conventional methods.10,11 The first rapid TB diagnostic method, Xpert MTB/RIF (Cepheid, USA), was endorsed by the WHO in 2010 and recommended as a preliminary screening test for individuals suspected of co-infection with HIV by 2015.12,13 However, the Xpert MTB/RIF device demands specialized training of operators, stable electricity supply and a suitable working environment,14 which are difficult for under-developed communities to afford. So it is worthwhile to develop a simple, rapid, sensitive and low-cost method for diagnosing TB to facilitate the need of healthcare systems in areas with a high TB burden.2,15
Unlike traditional PCR methods, isothermal amplification can amplify nucleic acids at a low incubation temperature without a precise thermal cycling, possessing the potential to develop point-of-care testing (POCT) application tools.16,17 A newly developed technique of isothermal nucleic acid amplification, recombinase polymerase amplification (RPA), mainly relies on three enzymes, i.e., recombinase, single-stranded DNA-binding protein (SSB) and strand displacement DNA polymerase, to complete testing in 20 minutes at an optimal working temperature, approximately 37 °C to 42 °C.18 The protein–DNA complex formed by recombinase and primers can find homologous sequences in the target DNA, followed by a chain exchange reaction, and then the DNA synthesis is initiated by strand displacement polymerase. RPA can become a highly specific detection with a well-designed probe and RPA is ideally suited to field use with resource limits.18,19 Due to its diverse detection formats, high detection efficiency and adaptability, RPA has been widely applied in nucleic acid detection, such as infectious diseases diagnosis and cancer research.20–22 Particularly, RPA have been described for TB diagnosis by detecting MTC and nontuberculous mycobacteria (NTM).23–25
Mycobacterium tuberculosis (MTB) is an important member of MTC, accounting for 90% to 95% of the cases of human TB,26 and therefore it is crucial to detect MTB for the clinical diagnosis of TB. mtp40 is a frequently-used molecular target for identifying MTB; however, reports showed it is absent in some MTB strains and is also present in some other members of MTC.27,28 esat-6 and cfp-10 are also used for detecting MTB, for which a RPA-based assay has been reported.29 But the esat-6 and cfp-10 also exist in some NTM, such as Mycobacterium kansasii and Mycobacterium marinum.30 In addition, IS6110 is a highly sensitive marker for detecting MTC, but some MTB strains cannot be identified due to lack of IS6110.31,32 In order to detect MTB more accurately and effectively, mts90 was identified as a more specific molecular diagnostic target of MTB in our previous study.33 Therefore, a method capable of sensitively detecting mts90 may help in improving the detection rate of TB and subsequently preventing TB transmission in the epidemic areas.
In this study, we established a real-time RPA method targeting mts90 for the rapid, sensitive and specific detection of MTB. The principle of this method is illustrated in Scheme 1. In addition, we evaluated the clinical usability of this assay by testing a variety of clinical samples.
 | ||
| Scheme 1 Schematic illustration of real-time RPA performed on the amplification and detection of mts90. | ||
2. Materials and methods
2.1. Bacterium strains
Mycobacterium tuberculosis H37Rv (ATCC27294) and Mycobacterium gastri (CMCC95006) were obtained from the National Institutes for Food and Drug Control (NIFDC, P. R. China). Mycobacterium bovis BCG and other pathogens, such as Staphylococcus aureus, Streptococcus pneumoniae D39, Pseudomonas aeruginosa and Escherichia coli, were from the Pathogenic Microbiology Laboratory of Chongqing Medical University (Chongqing, P. R. China). The genomic DNA of these bacteria was extracted according to the previous description.342.2. Generation of reference plasmid
According to the manufacturer's guidance, the plasmid standard of mts90 (2973968 to 2974057 nt of GenBank accession numbers CP009480.1) was constructed by a pMD19-T Vector Cloning Kit (TaKaRa). A fragment containing the mts90 region was ligated to pMD19-T vector, and then was transformed into cloning bacteria (DH5α). Plasmid DNA was extracted using the Plasmid Mini Kit (Omega Bio-tek) and verified by PCR as well as sequencing (TSINGKE Biological Technology, Chengdu, P. R. China). PCR reactions were performed in a T100™ Thermal Cycler (Bio-Rad, USA) with the following protocol: 94 °C for 5 min, followed by 30 cycles of 94 °C for 1 min, 63 °C for 30 s, 72 °C for 15 s, and finally 72 °C for 10 min. The DNA concentration was quantified by a NanoDrop 1000 UV/Vis spectrophotometer (Thermo Scientific, USA).2.3. RPA primer and probe design
Primers and probes for the mts90 RPA assay were designed following the manufacturer's (TwistDx, Cambridge, UK) manual (https://www.twistdx.co.uk/en/support/rpa-assay-design-2). Amplicons for the primer design were from conserved regions containing a nucleotide sequence of mts90. The screening of the primer candidates was accomplished via electrophoresis, fluorescent dye and probes for selecting a preferred primer set. All primers and probes (Table 1) were synthesized from Sangon Biotech (Shanghai, P. R. China).| Primer name | Sequence (5′ → 3′) | Nucleotidea |
|---|---|---|
| a Nucleotide corresponding to the 5′ of primer on M. tuberculosis H37Rv sequence (GenBank CP009480.1).b An optimization of the R5 reverse primer, which gradually shrank to 30 bp from 38 bp.c 90F/90R was used to amplify mts90 by PCR, referenced to Jianing Zhao et al.d (1) dT-FAM, thymidine nucleotide carrying fluorescein; (2) THF, tetrahydrofuran spacer; (3) dT-BHQ, thymidine nucleotide carrying a black hole quencher; (4) probe had a 3′ C3-spacer for blocking extension. Both dT-FAM and dT-BHQ replaced T of the sequence, THF of P1 replaced A, but C was replaced in P2. | ||
| F2 | CAAGCGGCATCTGATGTAGTCTCTTATTGC | 2974074 |
| F3 | CCTCTGACCAGGCGACATAGACAACAGTACCC | 2974123 |
| F4 | CATCTGATGTAGTCTCTTATTGCGCGTTTATTGCG | 2974067 |
| F5 | CATCTGATGTAGTCTCTTATTGCG | 2974067 |
| F6 | CTCTTATTGCGCGTTTATTGCGCGGGACAGGG | 2974054 |
| R1 | TGTAGCTGGCTTGCCAGCGGCCGGAGTTGAACTG | 2973934 |
| R2 | CACGCGGCCGTCGGGGCCGGTGTAGCTGGCTTGCC | 2973914 |
| R5 | GTCGATCTTGGCGTTGAAGGTTTTGGGGGCG | 2973878 |
| R5-1b | TCGATCTTGGCGTTGAAGGTTTTGGGGGCGATGTACAC | 2973879 |
| R5-2b | TCGATCTTGGCGTTGAAGGTTTTGGGGGCGATGTAC | 2973879 |
| R5-3b | TCGATCTTGGCGTTGAAGGTTTTGGGGGCGATGT | 2973879 |
| R5-4b | TCGATCTTGGCGTTGAAGGTTTTGGGGGCGAT | 2973879 |
| R5-5b | TCGATCTTGGCGTTGAAGGTTTTGGGGGCG | 2973879 |
| R6 | GTCGATCTTGGCGTTGAAGGTTTTGG | 2973878 |
| R7 | ATGCGACCGAATTTGCGTCTCT | 2973972 |
| R8 | CGTTGAAGGTTTTGGGGGCGATGTACAC | 2973889 |
| R10 | GTCGGTGAGCCATGCTTCGGCGTCGATCTTG | 2973857 |
| 90Fc | AGTCTCTTATTGCGCGTTTATTGCG | 2974057 |
| 90Rc | TCGGATGCGACCGAATTTGC | 2973968 |
| P1d | CATCTGATGTAGTCTCTTATTGCGCGTT(FAM-dT)1(THF)2T(BHQ-dT)3GCGCGGGACAGGGGAC-spacer C34 | 2974067 |
| P2d | CAGAGACGCAAATTCGGTCGCATCCGACAGT(FAM-dT)1(THF)2AAC(BHQ-dT)3CCGGCCGCTGGCAAG-spacer C34 | 2973994 |
2.4. RPA assays
RPA reactions analyzed by electrophoresis and fluorescent dye were performed on the TwistAmp® fpg kit (TwistDx, Cambridge, UK). Each reaction contained a 50 μL mixture. A pre-mixture (47.5 μL) was prepared firstly, including 2.1 μL of the RPA primer pair (10 nM), 29.5 μL of rehydration buffer, and 13.8 μL of template and ddH2O. Then the pre-mixture was diverted into the reaction tube to mix with TwistAmp fpg lyophilized enzyme pellets, and 2.5 μL of magnesium acetate (280 mM) were added to trigger the reaction immediately. The mixture was quickly placed in a 39 °C water bath and incubated for at least 20 min. Following purification by a High Pure PCR Product Purification Kit (Roche), its products were checked by agarose gel (2.5%) electrophoresis. When monitored with fluorescent dye, 2.5 μL of 20× EvaGreen® Dye (Biotum) were added in the pre-mixture up to 47.5 μL. After adding 2.5 μL of magnesium acetate (280 mM), RPA reactions were kept at 39 °C with the Rotor Gene Q (Qiagen, Germany) and set to cycle readings every 20 s.RPA reactions coupled with a specific probe were carried out by the TwistAmp® exo kit (TwistDx, Cambridge, UK). The 47.5 μL pre-reaction included 2.1 μL of primer pair (10 nM), 0.6 μL of probe (10 nM), 29.5 μL of rehydration buffer, 13.2 μL of template and ddH2O. After adding 2.5 μL of magnesium acetate (280 mM), the mixture was rapidly transferred to the CFX Connect™ Real-time System (Bio-Rad, USA) and the reaction procedure was to incubate at 39 °C and read fluorescence values every 20 s for 20 min.
2.5. Analytical sensitivity and specificity of mts90 RPA assay
The reference plasmid was serially diluted to 6 × 105, 6 × 104, 6 × 103, 6 × 102, 6 × 101, and 6 copies per microliter, and these dilutions were used to determine the sensitivity of the mts90 RPA assay. To determine specificity, we detected DNA templates extracted from Mycobacterium tuberculosis H37Rv (M. tuberculosis H37Rv), Mycobacterium bovis BCG (M. bovis BCG), Mycobacterium gastri (M. gastri), Escherichia coli (E. coli), Staphylococcus aureus (S. aureus), Streptococcus pneumoniae D39 (S. pneumoniae D39) and Pseudomonas aeruginosa (P. aeruginosa). 3 μL of genomic DNA of each strain was added into the RPA reactions.2.6. Clinical samples for the assessment of the mts90 RPA assay application
A total of 28 clinical samples from the First Affiliated Hospital of Chongqing Medical University (Chongqing, P. R. China) were used to evaluate the mts90 RPA assay. This protocol was approved by the Clinical Research Ethics Committee of Chongqing Medical University. These clinical samples were identified as MTC positive by a Biochip test (CapitalBio Corporation, Beijing, P. R. China). Pre-treatment for the digestion and decontamination of the clinical samples was performed by using a reference NaOH–Na citrate standard method,35 and then the supernatant was discarded after centrifugation, and the pellet was washed and resuspended repeatedly with sterile PBS.36 The DNA of the clinical samples was obtained by using the nucleic acid extraction reagents in the Mycobacteria Identification Array Kit (CapitalBio Corporation, Beijing, P. R. China). 2.5 μL of the DNA of each sample were used as the template in the RPA reactions.3. Results
3.1. Design of mts90 RPA assay
To select a preferred primer pair, we used different methods to screen for the optimal primer pair in corresponding candidate primers (Table 1) and conducted RPA reactions for the reference plasmid. The results of electrophoresis showed that the primer pair of F3/R2 had the best amplification effect among 5 pairs of primers (Fig. S1A†). Monitoring the performance of different primers in real time is a method that can minimize the risk of cross-contamination caused by aerosols when opening lids. By adding fluorescent dye (EvaGreen® Dye) to perform real-time detection, only one pair (F3/R5) of 4 primer sets was selected for acquiring the most obvious difference between a positive amplification and negative control (Fig. S1B†). In addition, two probes were designed to test different primer pairs. The results demonstrated that the combination of probe P2 and primer pair F3/R10 produced the highest fluorescence signal among 7 pairs of primers (Fig. 1A). In order to determine the suitable one for further electrophoresis analysis, three pairs of primers selected above were further verified with genomic DNA from M. tuberculosis H37Rv. The products were viewed on a 2.5% agarose gel after purification. The band of the F3/R10 positive lane showed the highest amplification performance (Fig. 1B). Therefore, probe P2 and primer pair F3/R10 were used for subsequent experiments.3.2. Sensitivity analysis of mts90 RPA assay
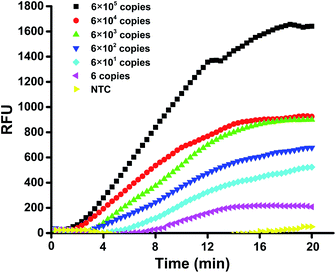
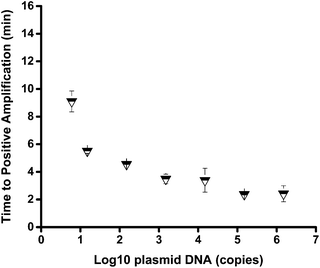
The sensitivity of the mts90 RPA assay was investigated by detecting a serial dilution of the reference plasmid. The results showed that the 10-fold dilutions of plasmid from 6 × 105 to 6 × 100 copies per reaction could produce an amplification signal within 20 minutes (Fig. 2), and the fluorescence intensity of 6 copies per reaction was approximately 200 RFU, which was significantly higher than that of the background. It was proved by repeated tests (n ≥ 3) on different concentrations that the sensitivity and repeatability of the mts90 RPA assay were valid (Table 2). Six (6)-copy preparation was amplified in 5 of 5 (100%), and the time needed for a positive amplification signal was after about 8 minutes (Fig. 3). The results above demonstrated that the sensitivity of this assay was as low as 6 copies per reaction.| mts90 RPA (copies per reaction) | Positive reactions/replicates |
|---|---|
| a The data used for the sensitivity analysis of mts90 RPA assay were obtained from testing on the plasmid. The sensitivity was as low as 6 copies per reaction for a 100% positive rate of detection (5/5). | |
| 1.5 × 106 | 3/3 |
| 1.5 × 105 | 3/3 |
| 1.5 × 104 | 3/3 |
| 1.5 × 103 | 3/3 |
| 1.5 × 102 | 3/3 |
| 1.5 × 101 | 4/4 |
| 6 | 5/5 |
| 0 | 0/3 |
3.3. Specificity of mts90 RPA assay
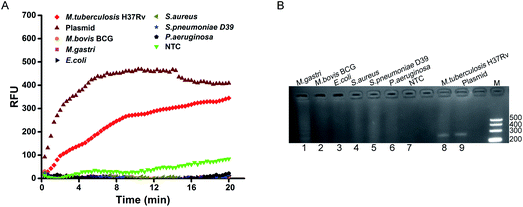
To analyze the specificity of the mts90 RPA assay for MTB, we detected the genomic DNA extracted from seven strains. As expected, only M. tuberculosis H37Rv was found to be positive with the positive control of the mts90 plasmid in real-time detection (Fig. 4A). Although M. bovis BCG and M. gastri are members of MTC, the strains tested were negative. Similarly, the remaining other pathogenic bacteria did not produce any positive amplification signal. Furthermore, the results analyzed by electrophoresis were consistent with the above. The bands of M. tuberculosis H37Rv and plasmid are shown at the same size (Fig. 4B). Thus, the mts90 RPA assay established for detecting MTB had the desired specificity.3.4. Performance evaluation of mts90 RPA assay with clinical samples
The performance of the mts90 RPA assay was evaluated by testing clinical samples. A total of 28 clinical samples were collected that were identified as positive by a Biochip test, including sputum (15), bronchoalveolar lavage fluid (BALF – 9) and lung tissue (4). The mts90 RPA assay could successfully detect MTB from the clinical samples within 20 minutes (Table 3). There was a 100% detection rate when testing samples of sputum and tissue (15/15 and 4/4, respectively). Although for the samples of BALF, 8 in 9 cases were detected positive by the mts90 RPA assay and one of them was negative, the total coincidence rate was high, up to 96.43% (27/28) (Table 4).| Sample # | Sample type | RPA(TT)a | Sample # | Sample type | RPA(TT)a |
|---|---|---|---|---|---|
| a TT, threshold time in minutes. TT (min) = (CT values × 20 s per cycle)/60, CT was calculated by using Bio-Rad CFX Manager software 3.1.b Sample # 24, the sample of BALF, was detected in the negative by the mts90 RPA assay, NG (negative). | |||||
| 1 | Sputum | 3.81 | 15 | Sputum | 12.64 |
| 2 | Sputum | 7.60 | 16 | BALF | 9.82 |
| 3 | Sputum | 19.64 | 17 | BALF | 7.90 |
| 4 | Sputum | 4.36 | 18 | BALF | 8.73 |
| 5 | Sputum | 9.17 | 19 | BALF | 10.20 |
| 6 | Sputum | 9.78 | 20 | BALF | 16.86 |
| 7 | Sputum | 11.67 | 21 | BALF | 17.97 |
| 8 | Sputum | 11.48 | 22 | BALF | 11.66 |
| 9 | Sputum | 8.34 | 23 | BALF | 11.79 |
| 10 | Sputum | 15.28 | 24b | BALF | NG |
| 11 | Sputum | 15.00 | 25 | Tissue | 16.61 |
| 12 | Sputum | 7.62 | 26 | Tissue | 15.22 |
| 13 | Sputum | 12.49 | 27 | Tissue | 19.28 |
| 14 | Sputum | 10.06 | 28 | Tissue | 13.23 |
| Sample type | Case | Coincidence rate (n/N) |
|---|---|---|
| a All the clinical samples were identified as MTC positive by Biochip test (CapitalBio Corporation). The results showed that these samples were detected totally in 27 of 28 (96.43%) positive cases by mts90 RPA assay, n (number of positive amplifications by real-time RPA detection), N (total number of samples). | ||
| Sputum | 15 | 100% (15/15) |
| BALF (bronchoalveolar lavage fluid) | 9 | 88.89% (8/9) |
| Tissue (lung) | 4 | 100% (4/4) |
| Total | 28 | 96.43% (27/28) |
4. Discussion
A rapid and sensitive mts90 RPA assay for detecting MTB was established and it was successfully applied to detect clinical samples. According to the manufacturer's guidelines, primer selection is critical to the development of RPA methods, and it is necessary to use a long primer (30–35 bases) to obtain the desired amplification effect. However, based on a recent review paper, some published RPA studies used primers below 30 bases or over 35 bases,37 showing some differences from the guidelines. Therefore, different primer lengths (22–38 bases) were designed and tested in our experiments.The primer set of F3/R2 (32/35 bases) was selected based on electrophoresis analysis. F3/R5 (R5, 31 bases) was selected by fluorescent dye (EvaGreen® Dye); while F3/R10 (R10, 31 bases) was screened using well-designed probes. The lengths of the primers obtained from the above screening methods were within the recommended range, which explains that the appropriately long primers might be preferred for RPA amplification. Certainly, a RPA reaction executed at a single temperature (37 °C) within 20 minutes also relies on the initial template concentration and amplicon size.38 In this study, we spent much of the time for the purification of RPA products before electrophoresis analysis, and it remains a challenge for analytical specificity using intercalating dye to monitor real-time fluorescence; however, probes could rapidly generate comparative data after 16 minutes and improve the specificity among the screening. Moreover, the primer set of F3/R10 selected by the probes showed the best amplification effect when tested with genomic DNA of M. tuberculosis H37Rv. Thus, we confirmed that it is more efficient and reliable to use probes for selecting a preferred primer pair in the development of RPA methods.
The sensitivity of the mts90 RPA assay was high with only 6 copies per reaction for detecting the reference plasmid. The sensitivity was slightly higher than that of the TaqMan qPCR method developed previously; its sensitivity was 10 copies of plasmid.33 Fig. 3 shows that the time needed to reach a positive amplification depends on the input concentrations of the plasmid, but this proportional relation was not strict. As for the analytical specificity of the mts90 RPA assay, it could successfully detect M. tuberculosis H37Rv but not M. bovis BCG, M. gastri and other pathogens. The probe (P2) used for the mts90 RPA assay was 52 oligonucleotides in length with homologous sequences of mts90, which ensured that the assay was capable of specifically detecting MTB in the RPA reactions. The structure of probe P2 was also analyzed online (http://www.nupack.org/), seen in Fig. S2B.†
The clinical performance of the mts90 RPA assay was evaluated by detecting clinical samples. The results showed that the accuracy of this assay was 96.43% (27/28), with only a unique case of BALF that tested inconsistently with clinical results. These specimens were identified as MTC positive using a Biochip system that is currently the only approved one certified by the China Food and Drug Administration (CFDA) for the rapid identification of mycobacteria. The Biochip test can identify MTC and 16 common NTMs;39 but it cannot identify specific types of MTC. Therefore, the sample detected as negative by the mts90 RPA assay might be a strain other than MTB within MTC; thus, further sequencing is required. However, the clinical utility of this assay requires more clinical samples to be further evaluated.
Compared to the Biochip test, our real-time RPA assay only requires basic operation technical skills and typically 20 minutes of reaction time, which is much simpler than the complicated testing procedures of about 6 hours for the Biochip system.39,40 Although other well-established techniques for the diagnosis of TB by detecting nucleic acids exist, RPA has its own special advantages for POCT diagnosis in particular. Unlike many PCR assays (such as Xpert MTB/RIF), RPA has the potential for miniaturization and is cost effective, as it could complete its amplification and detection with existing facilities. Furthermore, fluorescent detection is really not reliant on costly special equipment. A simple battery powered device is also feasible.41,42 The loop-mediated isothermal amplification test for TB (TB-LAMP) reviewed and recommended by WHO in 2016 is a manual test that takes less than 1 hour and its results can be read with the naked eye under ultraviolet light.43 However, RPA further reduces the testing time and only two primers are necessary, which is simpler than the four to six primers required for the LAMP reactions.44 Besides, RPA requires a lower energy and may proceed at temperatures between 25 °C and 42 °C compared with around 65 °C for LAMP.18,45 More significantly, most required reagents form as a freeze-dried pellet to simplify the testing and all the reagents involved in the RPA reactions are cold-chain independent.46 Overall, RPA is more suitable for healthcare in remote parts of the globe.
5. Conclusion
In summary, the mts90 RPA assay showed a high sensitivity and specificity for detecting MTB, and the assay was simple to operate, as well as quick with a reaction that could be completed within 20 minutes, greatly improving the detection efficiency. Therefore, the mts90 RPA assay developed in this study can be a complement to the existing nucleic acid diagnostic methods of TB. Further in-depth studies may be necessary to design a simpler readout method for the mts90 RPA assay and apply the assay to diagnose TB in low-income and TB high-burden areas.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This study was supported by Society and Livelihood Foundation of Chongqing (grant No. cstc2015shmszx120098).References
- W. H. Organization, Global tuberculosis report 2016, World Health Organization, 2016 Search PubMed.
- M. Pai and M. Schito, J. Infect. Dis., 2015, 211(suppl. 2), S21–S28 CrossRef PubMed.
- N. R. Gandhi, P. Nunn, K. Dheda, H. S. Schaaf, M. Zignol, D. van Soolingen, P. Jensen and J. Bayona, Lancet, 2010, 375, 1830–1843 CrossRef.
- G. B. Migliori, A. Matteelli, D. Cirillo and M. Pai, Canadian Journal of Infectious Diseases and Medical Microbiology, 2016, 19, 169 CrossRef.
- J. Bruchfeld, M. Correia-Neves and G. Kallenius, Cold Spring Harbor Perspect. Med., 2015, 5, a017871 CrossRef PubMed.
- G. A. UN, Transforming our world: The 2030 agenda for sustainable development, General Assembly of the United Nations, 2015 Search PubMed.
- W. H. Organization, Draft Report of the Sixty-Seventh World Health Assembly on Global Strategy and Targets for Tuberculosis Prevention, Care and Control after 2015, World Health Organization, 2015 Search PubMed.
- S. D. Lawn and A. I. Zumla, Lancet, 2011, 378, 57–72 CrossRef.
- S. Asmar and M. Drancourt, Front. Microbiol., 2015, 6, 1184 Search PubMed.
- Y. Derese, E. Hailu, T. Assefa, Y. Bekele, A. Mihret, A. Aseffa, J. Hussien, I. Ali and M. Abebe, J. Infect. Dev. Countries, 2012, 6, 53–57 CAS.
- C. C. Boehme, M. P. Nicol, P. Nabeta, J. S. Michael, E. Gotuzzo, R. Tahirli, T. G. Ma, R. Blakemore, W. Worodria and C. Gray, Lancet, 2011, 377, 1495–1505 CrossRef.
- C. C. Boehme, P. Nabeta, D. Hillemann, M. P. Nicol, S. Shenai, F. Krapp, J. Allen, R. Tahirli, R. Blakemore, R. Rustomjee, A. Milovic, M. Jones, S. M. O'Brien, D. H. Persing, S. Ruesch-Gerdes, E. Gotuzzo, C. Rodrigues, D. Alland and M. D. Perkins, N. Engl. J. Med., 2010, 363, 1005–1015 CrossRef CAS PubMed.
- W. H. Organization, Global tuberculosis report 2015, World Health Organization, 2015 Search PubMed.
- C. M. Denkinger, I. Nicolau, A. Ramsay, P. Chedore and M. Pai, Eur. Respir. J., 2013, 42, 544–547 CrossRef PubMed.
- S. A. Cheon, H. H. Cho, J. Kim, J. Lee, H. J. Kim and T. J. Park, J. Microbiol. Methods, 2016, 123, 51–61 CrossRef PubMed.
- L. Yan, J. Zhou, Y. Zheng, A. S. Gamson, B. T. Roembke, S. Nakayama and H. O. Sintim, Mol. BioSyst., 2014, 10, 970–1003 RSC.
- J. Li and J. Macdonald, Biosens. Bioelectron., 2015, 64, 196–211 CrossRef CAS PubMed.
- O. Piepenburg, C. H. Williams, D. L. Stemple and N. A. Armes, PLoS Biol., 2006, 4, e204 Search PubMed.
- Z. A. Crannell, B. Rohrman and R. Richardskortum, PLoS One, 2014, 9, e112146 Search PubMed.
- D. S. Boyle, D. A. Lehman, L. Lillis, D. Peterson, M. Singhal, N. Armes, M. Parker, O. Piepenburg and J. Overbaugh, mBio, 2013, 4, 49–52 CrossRef PubMed.
- M. D. Moore and L. A. Jaykus, Sci. Rep., 2017, 7, 40244 CrossRef CAS PubMed.
- Y. Liu, T. Lei, Z. Liu, Y. Kuang, J. Lyu and Q. Wang, Int. J. Mol. Sci., 2016, 17, 792 CrossRef PubMed.
- D. S. Boyle, R. McNerney, H. Teng Low, B. T. Leader, A. C. Perez-Osorio, J. C. Meyer, D. M. O'Sullivan, D. G. Brooks, O. Piepenburg and M. S. Forrest, PLoS One, 2014, 9, e103091 Search PubMed.
- Q. Ma, H. Liu, F. Ye, G. Xiang, W. Shan and W. Xing, Mol. Cell. Probes, 2017 DOI:10.1016/j.mcp.2017.08.004.
- S. Hansen, J. Schafer, K. Fechner, C. P. Czerny and A. Abd El Wahed, PLoS One, 2016, 11, e0168733 Search PubMed.
- L. M. Parsons, R. Brosch, S. T. Cole, A. Somoskovi, A. Loder, G. Bretzel, D. Van Soolingen, Y. M. Hale and M. Salfinger, J. Clin. Microbiol., 2002, 40, 2339–2345 CrossRef CAS PubMed.
- A. Weil, B. B. Plikaytis, W. R. Butler, C. L. Woodley and T. M. Shinnick, J. Clin. Microbiol., 1996, 34, 2309–2311 CAS.
- E. Liébana, A. Aranaz, B. Francis and D. Cousins, J. Clin. Microbiol., 1996, 34, 933–938 Search PubMed.
- B. Y. C. Ng, E. J. H. Wee, N. P. West and M. Trau, Sci. Rep., 2015, 5, 15027 CrossRef CAS.
- S. M. Arend, K. E. van Meijgaarden, K. de Boer, E. C. de Palou, D. van Soolingen, T. H. Ottenhoff and J. T. van Dissel, J. Infect. Dis., 2002, 186, 1797–1807 CrossRef CAS PubMed.
- S. D. Van, P. E. de Haas, P. W. Hermans, P. M. Groenen and J. D. van Embden, J. Clin. Microbiol., 1993, 31, 1987 Search PubMed.
- M. N. Huyen, E. W. Tiemersma, K. Kremer, P. de Haas, N. T. Lan, T. N. Buu, C. Sola, F. G. Cobelens and D. van Soolingen, International Journal of Tuberculosis and Lung Disease, 2013, 17, 1479–1485 CrossRef CAS PubMed.
- J. Zhao, Y. Wang, D. Li, J. Liu, X. Zhang, Y. He, H. Wang, J. Cao, Y. Yin and W. Xu, World J. Microbiol. Biotechnol., 2014, 30, 2189–2197 CrossRef CAS PubMed.
- C. Bouakaze, C. Keyser, S. J. D. Martino, W. Sougakoff, N. Veziris, H. Dabernat and B. Ludes, J. Clin. Microbiol., 2010, 48, 1758 CrossRef CAS PubMed.
- P. T. Kent and G. P. Kubica, Public health mycobacteriology. A guide to the level III laboratory, US Department of Health and Human Services, 1985 Search PubMed.
- D. I. Gomez, C. S. Mullin, F. Mora-Guzmán, J. G. Crespo-Solis, S. P. Fisher-Hoch, J. B. Mccormick and B. I. Restrepo, Tuberculosis, 2011, 91, S43–S48 CrossRef CAS PubMed.
- R. K. Daher, G. Stewart, M. Boissinot and M. G. Bergeron, Clin. Chem., 2016, 62, 947–958 CAS.
- G. A. Hill-Cawthorne, L. O. Hudson, M. F. El Ghany, O. Piepenburg, M. Nair, A. Dodgson, M. S. Forrest, T. G. Clark and A. Pain, PLoS One, 2014, 9, e101419 Search PubMed.
- L. Zhu, G. Jiang, S. Wang, C. Wang, Q. Li, H. Yu, Y. Zhou, B. Zhao, H. Huang, W. Xing, K. Mitchelson, J. Cheng, Y. Zhao and Y. Guo, J. Clin. Microbiol., 2010, 48, 3654–3660 CrossRef CAS PubMed.
- Y. Pang, Y. Zhou, S. Wang, Y. Tan, J. Yue, B. Zhao, L. Wang, Y. Zhao and K. M. Kam, International Journal of Tuberculosis and Lung Disease, 2011, 15, 1680–1685 CrossRef CAS PubMed.
- A. E. W. Ahmed, W. Manfred and F. T. Hufert, J. Clin. Virol., 2015, 69, 16–21 CrossRef PubMed.
- D. Mondal, P. Ghosh, M. A. A. Khan, F. Hossain, S. Böhlken-Fascher, G. Matlashewski, A. Kroeger, P. Olliaro and A. A. E. Wahed, Parasites Vectors, 2016, 9, 281 CrossRef PubMed.
- W. H. Organization, The Use of Loop-Mediated Isothermal Amplification (TB-LAMP) for the Diagnosis of Pulmonary Tuberculosis: Policy Guidance, World Health Organization, 2016 Search PubMed.
- K. Nagamine, T. Hase and T. Notomi, Mol. Cell. Probes, 2002, 16, 223 CrossRef CAS PubMed.
- T. Notomi, H. Okayama, H. Masubuchi, T. Yonekawa, K. Watanabe, N. Amino and T. Hase, Nucleic Acids Res., 2000, 28, E63 CrossRef CAS PubMed.
- O. Faye, O. Faye, B. Soropogui, P. Patel, A. A. E. Wahed, C. Loucoubar, G. Fall, D. Kiory, N. F. Magassouba, S. Keita, M. K. Kondé, A. A. Diallo, L. Koivogui, H. Karlberg, A. Mirazimi, O. Nentwich, O. Piepenburg, M. Niedrig, M. Weidmann and A. A. Sall, Eurosurveillance, 2015, 20, 30053 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra09999a |
| This journal is © The Royal Society of Chemistry 2017 |