Open Access Article
Open Access ArticleSynthesis, characterization and biological evaluation of formononetin derivatives as novel EGFR inhibitors via inhibiting growth, migration and inducing apoptosis in breast cancer cell line†
Hong-Yan
Lin‡
ab,
Wen-Xue
Sun‡
ab,
Chao-Sai
Zheng
ab,
Hong-Wei
Han
ab,
Xue
Wang
ab,
Ya-Han
Zhang
ab,
Han-Yue
Qiu
ab,
Cheng-Yi
Tang
ab,
Jin-Liang
Qi
ab,
Gui-Hua
Lu
ab,
Rong-Wu
Yang
*ab,
Xiao-Ming
Wang
 *ab and
Yong-Hua
Yang
*ab and
Yong-Hua
Yang
 *ab
*ab
aState Key Laboratory of Pharmaceutical Biotechnology, NJU-NJFU Joint Institute of Plant Molecular Biology, Nanjing University, Nanjing, 210023, China. E-mail: Yangyh@nju.edu.cn; Wangxm07@nju.edu.cn; Robertyang@nju.edu.cn; Fax: +86-25-89681381; Tel: +86-25-89681381
bCo-Innovation Center for Sustainable Forestry in Southern China, Nanjing Forestry University, Nanjing, 210037, China
First published on 16th October 2017
Abstract
Over the past few decades, the human epidermal growth factor receptor (EGFR) has been established as an attractive target for non-small cell lung cancer (NSCLC) therapy. Nevertheless, the approved EGFR inhibitors, gefitinib or erlotinib have shown minimum clinical activity to breast cancer patients, who also highly expressed EGFR. In this study, we designed and synthesized a series of novel formononetin derivatives by reference to the binding mode of lapatinib to EGFR. In vitro EGFR and cell growth inhibition assay demonstrated that compound 4v exhibited the most potent anti-EGFR (IC50 = 14.5 nM) and anti-proliferation activity (IC50 = 5.44 ± 1.28 μM) against MDA-MB-231 cell line, which was comparable to that of lapatinib (EGFR, IC50 = 5.6 nM; MDA-MB-231, IC50 = 2.48 ± 1.04 μM). Further biological experiment results demonstrated that 4v could effectively induce apoptosis, inhibit proliferation and migration in MDA-MB-231 cells through targeting EGFR and then blocking the downstream signaling pathways, EGFR/PI3K/Akt/Bad, EGFR/ERK and EGFR/PI3K/Akt/β-catenin, respectively. However, it had no significant influence on cell cycle distribution and the related proteins (Cyclin A, Cyclin D1, CDK4) expression. In vivo anti-tumor results also preliminarily confirmed the effectiveness of 4v in tumor chemotherapy in mice and indicated its potential as a new EGFR inhibitor in the treatment of MDA-MB-231 malignant tumor.
Introduction
The epidermal growth factor receptor (EGFR) is a trans-membrane protein, belonging to the family of tyrosine kinases and involved in cell proliferation and signal transduction.1 It has been confirmed that EGFR is highly or abnormally expressed in many solid tumors and is correlated to tumor cell proliferation, angiogenesis, tumor invasion, metastasis, and inhibition of apoptosis.2,3 Over-expression of EGFR is also crucial for the evolution of malignant tumors, such as glioma cell, kidney, lung, breast, prostate and pancreatic cancer.4–6 The activation of EGFR phosphorylates its downstream signalling molecules PI3K and Akt, which are involved in cellular migration, apoptosis and mitogen-activated protein kinase signalling, Ras/Raf/MEK/ERK, as implicated in cell proliferation.7,8 It is well known that inhibition of EGFR kinase-mediated signal is a well-established strategy for the treatment of advanced stage non-small cell lung cancer (NSCLC) and also correlated well with the proliferation and migration of aggressive triple negative breast cancer, MDA-MB-231.9,10Studies have shown that triple-negative breast cancer was considered another potential indication for EGFR kinase inhibitors because they are not amenable to endocrine or HER-2 targeted therapies, while they highly-express EGFR kinase.10,11 Some literatures have suggested a crucial role of EGFR in breast cancer proliferation and migration. However, inhibition of EGFR by gefitinib or erlotinib showed low efficacy in arresting tumor cell growth at the cell line level and minimum clinical activity.7,12 Such extraordinary phenomenon always attracts us to explore the underlying mechanism and discover new potent EGFR inhibitors that are naturally derived chemicals as breast cancer therapeutics.
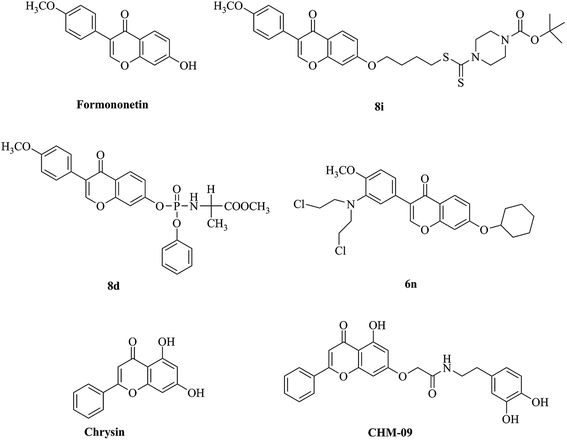
Natural products are a major source of drug discovery and cancer therapy. Formononetin (Fig. 1) is a bioactive isoflavone found in diverse Chinese medicine herbals, such as Amorpha fruticose,13Glycyrrhiza,13,14Trifolium pretense,13,14Astragalus membranaceus,15 which belonging to the family of Leguminosae. It has attracted broad attention because of multiple pharmacological activities, such as anti-bacterial,16,17 anti-cancer,17 anti-oxidant,18 anti-diabetic19,20 and anti-hyperlipidemia.21 Currently, many formononetin analogues (Fig. 1) were designed as anticancer agents and explored the biological mechanism against various human cancer cell lines. As reported that formononetin can induce cell apoptosis via PI3K/AKT signaling pathway in human nasopharyngeal and cervical carcinoma cells.22 Formononetin–dithiocarbamate hybrids 8i could inhibit growth and migration of PC-3 cells via MAPK/Wnt signaling pathway and cause cell cycle arrest in G1 phase.23 While its 7-phosphoramidate derivative 8d and nitrogen-mustard derivative 6n could induce cell cycle arrest in G2/M phase and cell apoptosis.24,25 In addition, Manupati et al. reported a chrysin analog CHM-09 which potentiated the mesenchymal–epithelial transition of breast cancer stem cells through inhibiting EGFR.10 In the light of previous studies, we reported here the designation of novel formononetin derivatives as potent anti-cancer agents through targeting EGFR and further investigation of their underlying biological mechanisms.
Results and discussion
Chemistry
In this study, we hypothesize that the design of synthetic derivatives of formononetin against EGFR with lapatinib as positive control may improve its anticancer efficacy. Before synthesis, a molecular screening process was performed by computer assist drug design (CADD) method. Molecular docking was conducted to calculate the binding energy (CDOCKER INTERACTION ENERGY in CDOCKER protocol in Discovery Studio 3.5) between compounds and EGFR complex structure (1XKK.pdb, downloaded from the PDB). Finally, 22 compounds were picked to continue assays subsequently and their binding energy were shown in Table S1 (ESI†).In view of the binding energy, 4v is the best one among them. For better understanding of the potential of 4v, we further examined the interaction of 4v with EGFR (PDB code 1XKK) with lapatinib as the positive control. All the amino acid residues which had interactions with lapatinib were exhibited and the results were depicted in Fig. 2A. In the binding model, lapatinib bound well to EGFR mainly via three hydrogen bonds with ARG 803, ASP 800 and PHE856, a π-cation bond with LYS 745 and a π–π bond with PHE856. In the binding model of 4v with EGFR (Fig. 2C), there formed a hydrogen bond with MET 793, a π-cation bond with LYS 745, a π-anion bond with ASP 800, a π–π bond with PHE856 and a π-sigma bond with MET766 which are the main contributors for the effective combination between them. The 3D models of the interaction between lapatinib, 4v and EGFR were also depicted in Fig. 2B and D. For Fig. 2E, we further compared the structure and target combine similarity of 4v and lapatinib. The result demonstrated that 1c might be a potential EGFR inhibitor.
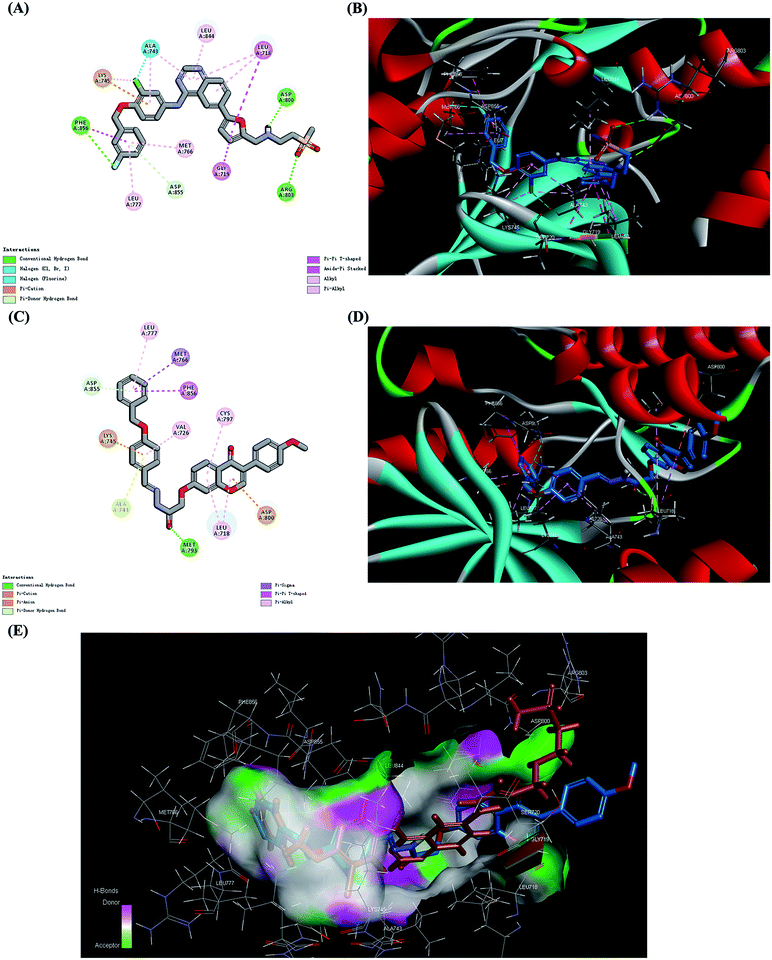 | ||
| Fig. 2 Molecular docking analysis of 4v and lapatinib, showing proposed binding modes with EGFR (PDB code 1XKK). (A) Interactions of lapatinib with the amino acid residues at the binding pocket of EGFR (carbon atom, gray; oxygen atom, red; hydrogen atom, white; nitrogen atom, purple; chlorine atom, bright green; fluorine atom, light green; sulphur atom, orange). (B) Binding pose of lapatinib in the protein surface of EGFR (carbon atom, blue; oxygen atom, red; hydrogen atom, white; nitrogen atom, purple; chlorine atom, bright green; fluorine atom, sulphur atom, orange). Hydrogen bonds were displayed as green dashed lines. (C) Interactions of 4v with the amino acid residues at the binding pocket of EGFR (carbon atom, gray; oxygen atom, red; hydrogen atom, white; nitrogen atom, purple; sulphur atom, orange). (D) Binding pose of 4v in the protein surface of EGFR (carbon atom, blue; oxygen atom, red; hydrogen atom, white; nitrogen atom, purple; sulphur atom, orange). Hydrogen bond was displayed as green dashed lines. (E) 3D mode of the interaction between the compounds (lapatinib, brown; 4v, blue) and the EGFR binding site. The protein is represented by molecular surface. | ||
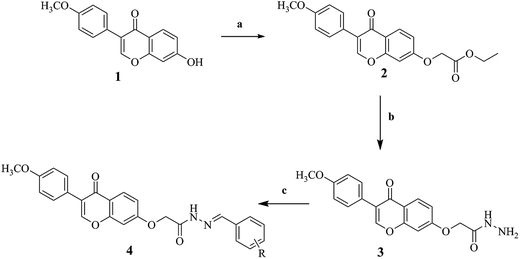
The synthetic routes of novel formononetin derivatives are outlined in Scheme 1, and structure of derivatives 4a–4v are shown in Table 1. These compounds were obtained by three steps which were elucidated in the Experimental section. All of them are first reported and characterized by 1H NMR, 13C NMR, elemental analysis, melting test and mass spectroscopy, and results are in full accordance with their depicted structures.
| Compound | R1 | R2 | R3 | R4 | R5 |
|---|---|---|---|---|---|
| 4a | H | H | H | H | H |
| 4b | F | H | H | H | H |
| 4c | H | F | H | H | H |
| 4d | H | H | F | H | H |
| 4e | H | F | F | H | H |
| 4f | Cl | H | H | H | H |
| 4g | H | Cl | H | H | H |
| 4h | H | H | Cl | H | H |
| 4i | Cl | Cl | H | H | H |
| 4j | Cl | H | H | H | Cl |
| 4k | H | Br | H | H | H |
| 4l | H | OH | H | H | H |
| 4m | H | H | OH | H | H |
| 4n | H | H | NH2 | H | H |
| 4o | CF3 | H | H | H | H |
| 4p | H | H | CF3 | H | H |
| 4q | H | H | CH3 | H | H |
| 4r | OCH3 | H | H | H | H |
| 4s | H | H | OCH3 | H | H |
| 4t | H | OCH3 | OCH3 | H | H |
| 4u | H | H | CH(CH3)2 | H | H |
| 4v | H | H | OC7H7 | H | H |
Biological studies
In order to screen out the optimal active compounds from all synthesized formononetin derivatives, a comparative study was carried out to evaluated their in vitro EGFR inhibition activities. As shown in Table 2, we clearly found that most of the tested compounds presented stronger EGFR inhibitory activity compared to formononetin itself. Especially, compound 4v was the most potent inhibitor of EGFR (IC50 = 14.5 nM), which was at least 10-fold stronger than formononetin (IC50 > 150 nM) and almost comparable to gefitinib (IC50 = 6.2 nM) and lapatinib (IC50 = 5.6 nM). Among them, compound 4n also showed potent EGFR inhibitory activity with an IC50 value of 20.6 nM. Generally, electron-donating group improved the enzymatic inhibitory activity more significantly than the electron-withdrawing group, especially when the substitution appeared at the R3-position of benzene ring of formononetin skeleton.| Compound | EGFR kinase | |
|---|---|---|
| % inhibition at 150 nM | IC50 (nM) | |
| 4a | 54.7 | 120.4 |
| 4b | 51.6 | 131.6 |
| 4c | 33.5 | >150 |
| 4d | 70.3 | 52.6 |
| 4e | 68.4 | 66.2 |
| 4f | 52.3 | 142.2 |
| 4g | 50.2 | 146.3 |
| 4h | 72.8 | 30.7 |
| 4i | 60.1 | 98.4 |
| 4j | 58.5 | 105.8 |
| 4k | 56.2 | 112.5 |
| 4l | 50.9 | 139.5 |
| 4m | 73.5 | 33.1 |
| 4n | 75.7 | 20.6 |
| 4o | 43.1 | >150 |
| 4p | 66.8 | 48.9 |
| 4q | 70.4 | 30.4 |
| 4r | 62.9 | 56.4 |
| 4s | 68.3 | 40.6 |
| 4t | 80.6 | 47.8 |
| 4u | 76.9 | 35.5 |
| 4v | 87.3 | 14.5 |
| Formononetin | 26.4 | >150 |
| Gefitinib | 91.5 | 6.2 |
| Lapatinib | 94.2 | 5.6 |
Then we further evaluated their anti-proliferation activities against four cancer cell lines, human breast cancer cells (MCF-7, MDA-MB-231), human lung carcinoma cells (H460, H1650), as well as two non-cancer cell lines, normal human liver cells (L02) and African green monkey kidney cell (VERO) by the cell based MTT assay. The results are summarized in Table 3 and expressed by the half maximal inhibitory concentration (IC50) values. As shown in Table 3, all compounds showed good anti-proliferation activity against the cancer cells, and most of them even better than formononetin itself with the standard of IC50 values. To be specific, most compounds showed better anti-proliferation activity against MDA-MB-231 than MCF-7, which might because of the high expression of EGFR in the former. For MDA-MB-231 cell line, 4h (IC50 = 8.30 ± 1.23), 4m (IC50 = 9.35 ± 1.02 μM), 4n (IC50 = 9.25 ± 1.05 μM), 4t (IC50 = 8.42 ± 1.21 μM) and 4v (IC50 = 5.44 ± 1.28 μM) showed relatively better anti-proliferation activities than other compounds and their inhibitory efficacy against MDA-MB-231 were double stronger than against MCF-7 cell line (4h, IC50 = 17.8 ± 1.64 μM, 4m, IC50 = 20.9 ± 2.24 μM, 4n, IC50 = 19.5 ± 1.73 μM, 4t, IC50 = 12.3 ± 1.11 μM and 4v, IC50 = 11.5 ± 1.52 μM). Combined with the data of two human lung cancer cell lines, we clearly found that 4h and 4v showed much stronger inhibition effect than others, especially for 4v (MCF-7, IC50 = 11.5 ± 1.52 μM, MDA-MB-231, IC50 = 5.44 ± 1.28 μM, H460, IC50 = 6.36 ± 1.55 μM, H1650, IC50 = 7.26 ± 1.02 μM), which effect is almost comparable to that of lapatinib (MCF-7, IC50 = 7.1 ± 2.81 μM, MDA-MB-231, IC50 = 2.48 ± 1.04 μM, H460, IC50 = 1.22 ± 0.35 μM, H1650, IC50 = 1.64 ± 0.98 μM) and much better than the inhibitory effect of gefitinib against breast cancer cell lines (MCF-7, IC50 = 13.5 ± 3.08 μM, MDA-MB-231, IC50 = 12.5 ± 2.13 μM). Generally speaking, for comprehensive consideration anti-proliferation activities and the impact on log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P (Table S2, ESI†), 4v possessing 4-benzyloxy can be considered the optimal one in all newly synthetic compounds, and appeared to be a more efficient inhibitor of MDA-MB-231 cell viability (IC50 = 5.44 ± 1.28 μM) when compared with formononetin (IC50 = 26.6 ± 2.84 μM).
P (Table S2, ESI†), 4v possessing 4-benzyloxy can be considered the optimal one in all newly synthetic compounds, and appeared to be a more efficient inhibitor of MDA-MB-231 cell viability (IC50 = 5.44 ± 1.28 μM) when compared with formononetin (IC50 = 26.6 ± 2.84 μM).
| Compound | IC50 (μM) | |||||
|---|---|---|---|---|---|---|
| MCF-7 | MDA-231 | H460 | H1650 | L02 | VERO | |
| 4a | 23.7 ± 1.87 | 15.1 ± 4.13 | 21.5 ± 2.04 | 11.9 ± 1.18 | 43.5 ± 3.64 | 25.3 ± 1.74 |
| 4b | 24.5 ± 1.98 | 20.7 ± 1.62 | 18.5 ± 2.33 | 38.6 ± 3.56 | >100 | >100 |
| 4c | 42.6 ± 0.25 | 33.6 ± 1.83 | 16.2 ± 2.05 | 17.8 ± 2.13 | >100 | >100 |
| 4d | 21.5 ± 0.91 | 18.3 ± 1.03 | 16.5 ± 1.22 | 49.6 ± 3.65 | 65.8 ± 4.62 | 62.5 ± 2.01 |
| 4e | 30.7 ± 1.42 | 27.8 ± 0.75 | 22.1 ± 2.87 | 33.4 ± 3.59 | >100 | >100 |
| 4f | 23.9 ± 1.12 | 17.9 ± 2.25 | 24.3 ± 1.46 | 13.9 ± 1.44 | >100 | 41.6 ± 2.48 |
| 4g | 41.5 ± 2.01 | 19.5 ± 1.52 | 16.5 ± 5.49 | 12.9 ± 2.58 | >100 | >100 |
| 4h | 17.8 ± 1.64 | 8.30 ± 1.23 | 8.02 ± 1.02 | 9.45 ± 1.08 | >100 | 29.3 ± 1.94 |
| 4i | 25.4 ± 1.73 | 16.3 ± 1.24 | 58.1 ± 3.54 | 19.7 ± 2.27 | >100 | 57.9 ± 1.42 |
| 4j | 25.7 ± 1.53 | 18.7 ± 1.02 | 19.6 ± 1.55 | 29.7 ± 2.59 | >100 | 78.8 ± 2.23 |
| 4k | 21.5 ± 0.63 | 23.2 ± 2.19 | 22.7 ± 2.28 | 10.8 ± 1.77 | >100 | 36.5 ± 3.14 |
| 4l | 21.1 ± 2.76 | 16.1 ± 2.75 | 16.3 ± 1.99 | 12.4 ± 1.68 | >100 | >100 |
| 4m | 20.9 ± 2.24 | 9.35 ± 1.02 | 14.2 ± 1.69 | 22.5 ± 2.68 | >100 | >100 |
| 4n | 19.5 ± 1.73 | 9.25 ± 1.05 | 18.2 ± 2.24 | 15.6 ± 2.07 | >100 | 67.3 ± 1.52 |
| 4o | 31.3 ± 1.93 | 33.8 ± 2.15 | 42.5 ± 6.42 | 10.1 ± 1.46 | >100 | >100 |
| 4p | 24.6 ± 2.52 | 12.3 ± 1.49 | 15.5 ± 1.95 | 19.3 ± 2.04 | >100 | >100 |
| 4q | 17.2 ± 1.93 | 15.4 ± 1.03 | 16.7 ± 2.49 | 8.79 ± 1.08 | >100 | >100 |
| 4r | 15.9 ± 3.42 | 13.1 ± 1.84 | 24.6 ± 3.39 | 40.2 ± 3.88 | >100 | 55.7 ± 2.05 |
| 4s | 29.1 ± 1.27 | 10.1 ± 1.55 | 21.58 ± 2.09 | 15.3 ± 2.31 | >100 | >100 |
| 4t | 12.3 ± 1.11 | 8.42 ± 1.21 | 18.6 ± 2.23 | 24.3 ± 1.89 | >100 | >100 |
| 4u | 25.9 ± 1.94 | 13.2 ± 4.02 | 14.3 ± 1.84 | 12.2 ± 1.45 | >100 | >100 |
| 4v | 11.5 ± 1.52 | 5.44 ± 1.28 | 6.36 ± 1.55 | 7.26 ± 1.02 | >100 | 95.2 ± 4.72 |
| Formononetin | 36.4 ± 3.01 | 26.6 ± 2.84 | 27.8 ± 2.22 | 29.5 ± 3.25 | 82.6 ± 5.73 | 89.5 ± 5.82 |
| Gefitinib | 13.5 ± 3.08 | 12.5 ± 2.13 | 3.02 ± 0.89 | 2.64 ± 0.98 | 35.2 ± 3.08 | 38.6 ± 2.56 |
| Lapatinib | 7.10 ± 2.81 | 2.48 ± 1.04 | 1.22 ± 0.35 | 1.64 ± 0.98 | 42.5 ± 2.59 | 35.6 ± 2.25 |
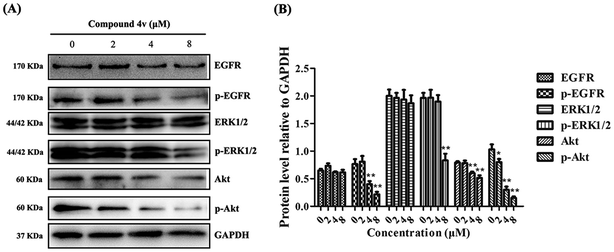
EGFR activation has been associated with multiple biological processes in cancer progression, such as cell proliferation, migration and metastasis through different signaling pathways.26 To investigate the effect of 4v on signal transduction mechanism, we performed immunoblot analysis of EGFR and its downstream signaling pathways: extracellular signal regulated kinase (ERK) and Akt. As shown in Fig. 3, MDA-MB-231 cells that express high EGFR were treated with increasing concentrations of 4v and demonstrated a dose-dependent decrease in phosphorylation of EGFR, ERK and Akt.
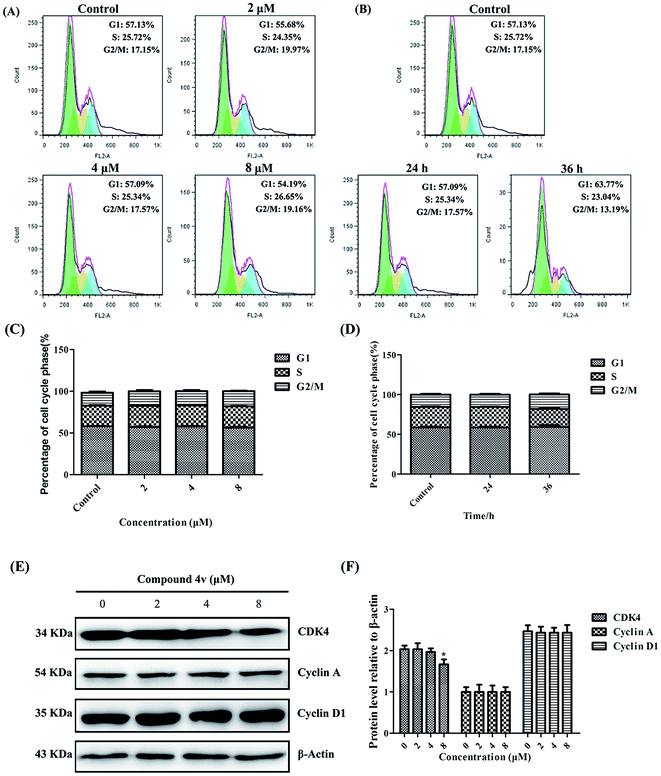
To further substantiate the anti-proliferative effect of 4v on MDA-MB-231 cells, we evaluated its impact on the cell cycle arrest. As illustrated in Fig. 4A–D, 4v treated cells demonstrated no obvious cell cycle arrest versus the control groups. Then, we performed immunoblot analysis of cell cycle related proteins, Cyclin A, Cyclin D1 and CDK4. And the results shown in Fig. 4E and F indicated that the expression of the cell cycle related proteins in MDA-MB-231 cells did not increase after treated with compound 4v.
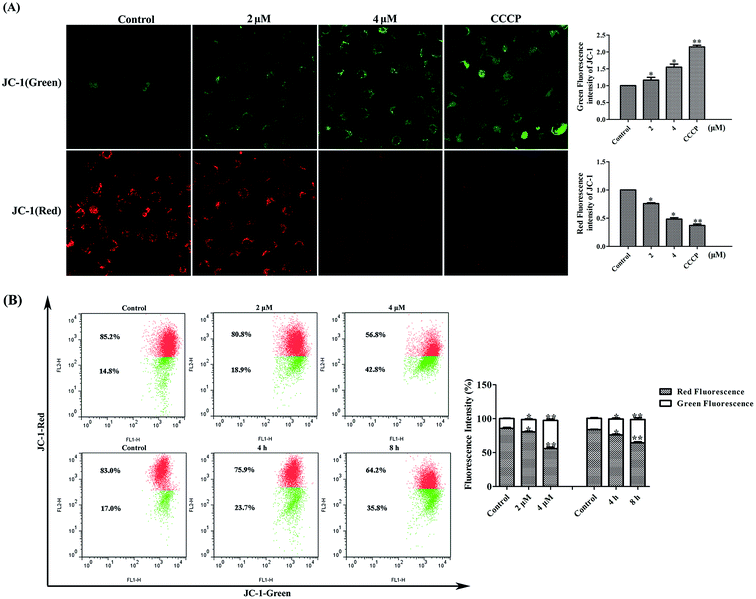
The transmembrane potential (ΔΨm) is an important parameter for mitochondrial functionality and its dissipation means the involvement of mitochondria in early apoptosis.27 As shown in Fig. 5, microscopy revealed that in untreated MDA-MB-231 cells, well-polarized mitochondria were marked by red fluorescent staining. After treated with 2 or 4 μM of 4v, this pattern was replaced by green fluorescence in MDA-MB-231 cells. CCCP was used as the positive control here. Additionally, flow cytometry results also showed that untreated cells with well-polarized, red-emitting mitochondria localized in the upper region of the plot. Cells exposed to 4v for 12 hours underwent a progressive loss of red fluorescence, indicated by a downward in 4v treated groups. The ratio of cells with high membrane potential decreased from 85.2% to 80.8% after exposed to 2 μM 4v for 12 hours and even decreased to 56.8% in 4 μM 4v treated group. Likewise, the loss of mitochondrial potential in MDA-MB-231 cells caused by 4v also showed time-dependent manner.
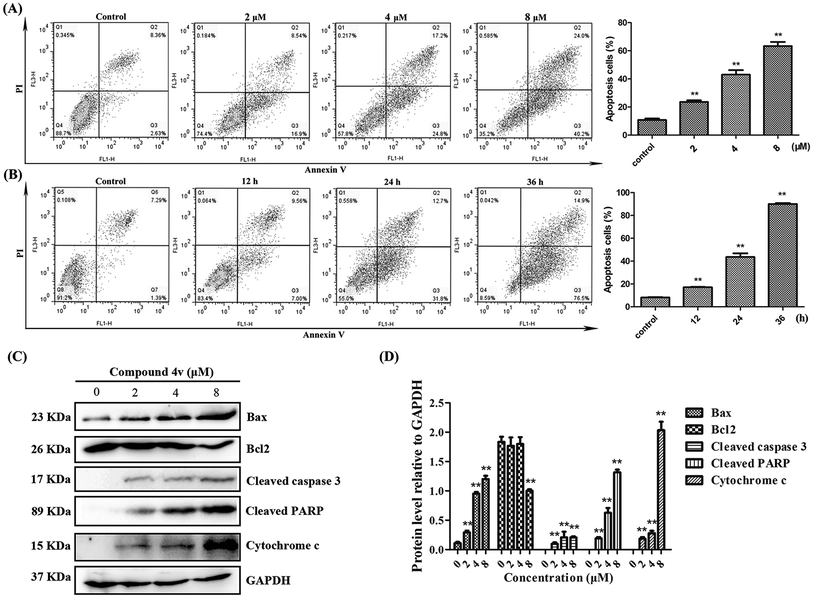
Based on the above results, we also evaluated the apoptotic populations of MDA-MB-231 cells treated with 4v using flow cytometry analysis. As shown in Fig. 6A, cells treated with increasing concentrations of 4v and demonstrated a dose-dependent increase in apoptotic populations. In addition, cells treated with 4v for increasing time also demonstrated a time-dependent manner in apoptotic populations and a significant high percentage (approximately 90.4%) of apoptotic cells were observed in 36 hours-treated cells compared to vehicle control (Fig. 6B). This observation was further substantiated by concomitant increased expression of pro-apoptotic proteins, Bax, cytochrome c, cleaved caspase-3, cleaved PARP and decreased expression of anti-apoptotic Bcl2 after exposure to 4v (Fig. 6C).
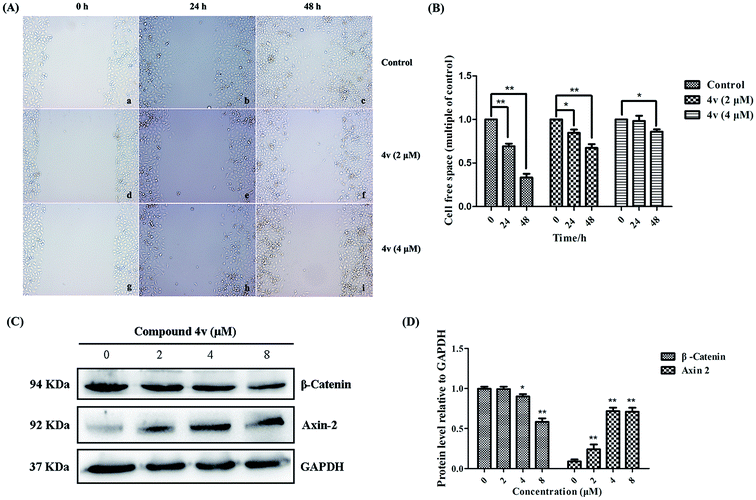
Tumor cell migration has been identified as one hallmark of malignant tumor progression, particularly for the establishment of lethal secondary metastases at distant organs. To investigate whether 4v could inhibit the migratory capacity of MDA-MB-231 cells, the conventional in vitro wound healing assay was also conducted. As shown in Fig. 7A and B, the control group cells gradually occupied the cell-free space within the channel over time and the cells almost fill the channel after 48 hours. Whereas, cells migration capacity of 4v-treated group was greatly inhibited and the inhibition was dose-dependent. The Wnt signaling pathway is crucial for multiple cellular processes including cell proliferation and migration. Once activated, β-catenin disassembles from complex comprised of Axin, APC and GSK3β and translocates into nuclear. Then it interacts with TCF4 which promotes the downstream target gene transcription. On the basis of the wound healing assay, we subsequently investigated the influence of 4v on Wnt signaling pathway in MDA-MB-231 cells. Western bolt analysis results shown in Fig. 7C indicated that treatment of MDA-MB-231 cells with 4v resulted in the decrease expression of β-catenin and the increase expression of Axin-2 in dose-dependent manners. These results collectively suggested that 4v might inhibit migration of MDA-MB-231 cells via Wnt signaling pathway.
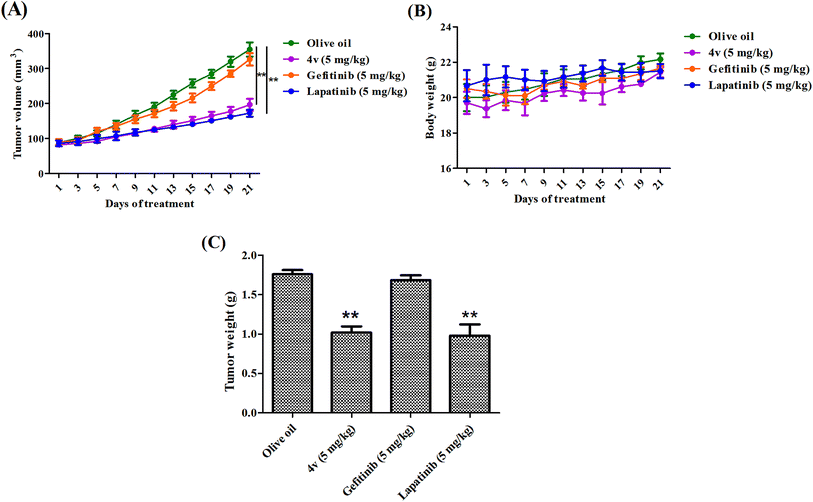
To evaluate the anti-tumor effects of 4vin vivo, we subcutaneously inoculated MDA-MB-231 cells in to the right flank of female nude mice. Two weeks later, all the mice got visible tumors. The mice were randomized into 3 groups, vehicle (olive oil), 4v (5 mg kg−1) and positive control (gefitinib, 5 mg kg−1; lapatinib, 5 mg kg−1). As shown in Fig. 8A, the administration of 4v greatly inhibited the tumor growth compared with the vehicle group and the inhibition effect was similar to that of lapatinib, while the traditional EGFR inhibitor gefitinib showed extremely low therapeutic efficiency on MDA-MB-231 cells. After the last time of treatment on day 21, the average weight of tumors from 4v-treated mice was nearly 2-fold less than the olive oil treated group (Fig. 8C). Meanwhile, 4v has no significant influence on the body weight of the mice (Fig. 8B).
Experimental
General information and materials
Formononetin (purity ≥ 99%), ethyl bromoacetate, hydrazine hydrate, a series of benzaldehyde with different substituents, acetic acid, acetone, ethanol, etc. According to experimental conditions, all reagents used were of analytical reagent grade (AR) except ethanol which was purified and purchased from Nanjing Chemical Reagent Co. Ltd. (Nanjing, China). Gefitinib (purity ≥ 99%) and lapatinib (purity ≥ 99%) were purchased from J&K Scientific Ltd. (Shanghai, China). 1H NMR and 13C NMR spectra of all intermediate and final products were recorded on a DRX 600 spectrometer and DMSO-d6 as solvent. Chemical shifts (d) for 1H NMR and 13C NMR spectra were reported in ppm (d). The ESI-MS spectra were obtained on a Mariner Biospectrometry Workstation (ESI-TOF) mass spectrometer. Melting points of products were determined using a XT4 MP Apparatus (Taike Corp., Beijing, China) and uncorrected. TLC was carried out on the glass-backed silica gel sheets (silica gel 60 Å GF254) and visualized in UV light (254 nm).Cell culture reagents were purchased from Gibco Life Technology (Grand Island, NY, USA). MTT, PMSF, and RIPA lysis buffer (#P0013B) were purchased from Beyotime Institute of Biotechnology (Haimen, China). Cell Cycle Detection Kit and Annexin V-FITC/PI apoptosis detection kit were purchased from KeyGEN Biotech Co. Ltd (Nanjing, China). Mitochondrial membrane potential assay kit with JC-1 was purchased from Beyotime Biotech Co. Ltd (Haimen, China). BCA protein assay kit (#23227) was purchased from Pierce (Rockford, IL, USA). PVDF membranes were purchased from Biosharp (Hefei, China). Anti-Axin-2 (20540-1-AP) was purchased from Proteintech (Wuhan, China); anti-cleaved PARP-1 (#sc-56196) was purchased from Santa Cruz Biotechnology Inc. (Dallas, Texas, USA). Anti-EGFR (WL0682a), anti-ERK (WL01864), anti-p-ERK (WLP1512), anti-Akt (WL0003b), anti-p-Akt (WLP001a), anti-CDK4 (WL02143), anti-Cyclin A (WL01753), anti-Cyclin D1 (WL01435a), anti-β-Actin (WL0002c), anti-Bax (WL01637), anti-Bcl2 (WL01556), anti-cleaved caspase-3 (WL01992), anti-cytochrome c (WL01571) and anti-β-catenin (WL0962a) were purchased from Wanlei Biotech Co. Ltd (Shenyang, China). GAPDH antibody (#E1A7021) and secondary antibody (anti-mouse or anti-rabbit IgG) (#E1WP319 or #E1WP318) were purchased from Enogene (Nanjing, China). ECL Kit (#34077) were purchased from Thermo Scientific (USA).
General procedure for preparation of compound 2
A mixture of formononetin (0.04 mol, 11.44 g) and potassium carbonate (0.08 mol, 22.48 g) were stirred in acetone under the condition of reflux at 60 °C for 3 hours to get dissolved and clarified. Meanwhile, proportionable ethyl bromoacetate (0.04 mol, 4.9 g) was prepared which dissolved in appropriate acetone and added to the system dropwise to react another 10 hours. And then reaction system was filtered in hot to obtain filtrate. When the filtrate tended to be cooling, a large amount of compound 2 separated out and washed with petroleum ether, 1% sodium hydroxide and distilled water successively. The product was dried in the oven weighing 9.97 g (87%). White powder, yield 95%; mp: 188.5–189.3 °C; 1H NMR (300 MHz, CDCl3) δ 8.24 (d, J = 8.9 Hz, 1H), 7.92 (s, 1H), 7.53–7.46 (m, 2H), 7.03 (dd, J = 8.9, 2.4 Hz, 1H), 7.00–6.94 (m, 2H), 6.84 (d, J = 2.4 Hz, 1H), 4.73 (s, 2H), 4.31 (q, J = 7.1 Hz, 2H), 3.84 (s, 3H), 1.25 (dd, J = 8.4, 5.6 Hz, 3H). ESI-TOF, calcd for C19H16O6 ([M + Na]+) 363.33, found 363.09. Anal. calcd for C19H16O6: C, 67.06; H, 4.74; O, 28.21. Found: C, 66.87; H, 4.92; O, 28.29.General procedure for preparation of compound 3
Compound 2 (0.025 mol, 8.85 g) was added to a clean round-bottom flask which contained 30–40 mL of ethanol. The reaction mixture was stirred under the condition of reflux at 80 °C. Hydrazine hydrate (0.075 mol, 3.9 g) was added and stirred for a further 10 hours with the reaction system getting clarified gradually. While stirring, the product was checked with TLC continually to choose the right point in time to stop the reaction. After the system temperature cooling to room temperature, the compound 3 which is white was isolated by filtration when precipitated. The product was dried in the oven weighing 7.45 g (84%). White powder, yield 87%; mp: 200.4–201.7 °C; 1H NMR (300 MHz, DMSO) δ 9.32 (s, 1H), 7.78 (d, J = 70.9 Hz, 1H), 7.18 (d, J = 8.2 Hz, 2H), 7.07–6.68 (m, 3H), 6.43 (d, J = 34.4 Hz, 2H), 4.42 (s, 2H), 4.31 (s, 2H), 3.71 (s, 3H). ESI-TOF, calcd for C17H14N2O5 ([M + Na]+) 349.31, found 349.09. Anal. calcd for C17H14N2O5: C, 62.57; H, 4.32; N, 8.59; O, 24.52. Found: C, 61.94; H, 4.41; N, 8.66; O, 24.59.General procedure for preparation of compounds 4a–4v
Compounds 4a–4v were synthesized by reacting the compound 3 (0.0008 mol) with the different substituted ethyl benzaldehyde (0.0008 mol) in a Schiff base condensation reaction. The reaction was stirred at 80 °C for 8 hours in the presence of a bit of glacial acetic acid (4–8 drops) in ethanol. The compounds 4a–4v were obtained by filtering after precipitation and all structure shown in Table 1.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 8.32–8.00 (m, 1H, Ar-H), 7.90–7.62 (m, 3H, N-NH, Ar-H), 7.44 (d, J = 3.6 Hz, 3H, Ar-H), 7.27–7.11 (m, 2H, Ar-H), 7.03 (t, J = 8.6 Hz, 1H, Ar-H), 6.85 (d, J = 8.1 Hz, 2H, Ar-H), 6.60–6.24 (m, 2H, Ar-H), 5.09–4.62 (m, 2H, –O–CH2–), 3.72 (s, 3H, –OCH3). 13C NMR (75 MHz, DMSO) δ 169.43 (s, 14C), 164.73 (s, 18C), 159.64 (s, 11C), 158.04 (s, 6C), 157.08 (s, 16C), 148.40 (s, 9C), 144.35 (s, 19C), 134.56 (s, 20C), 134.32 (s, 23C), 130.68 (s, 2C), 130.46 (s, 4C), 129.32 (s, 21C, 25C), 129.29 (s, 22C, 24C), 127.62 (s, 13C), 127.40 (s, 3C), 126.75 (s, 8C), 119.13 (s, 15C), 114.32 (s, 1C, 5C), 105.83 (s, 12C), 102.47 (s, 10C), 66.88 (s, 17C), 55.45 (s, 7C). ESI-TOF, calcd for C25H20N2O5 ([M + Na]+) 451.1372, found 451.1351. Anal. calcd for C25H20N2O5: C, 70.09; H, 4.71; N, 6.54; O, 18.67. Found: C, 69.58; H, 4.75; N, 6.61; O, 18.74.
N), 8.32–8.00 (m, 1H, Ar-H), 7.90–7.62 (m, 3H, N-NH, Ar-H), 7.44 (d, J = 3.6 Hz, 3H, Ar-H), 7.27–7.11 (m, 2H, Ar-H), 7.03 (t, J = 8.6 Hz, 1H, Ar-H), 6.85 (d, J = 8.1 Hz, 2H, Ar-H), 6.60–6.24 (m, 2H, Ar-H), 5.09–4.62 (m, 2H, –O–CH2–), 3.72 (s, 3H, –OCH3). 13C NMR (75 MHz, DMSO) δ 169.43 (s, 14C), 164.73 (s, 18C), 159.64 (s, 11C), 158.04 (s, 6C), 157.08 (s, 16C), 148.40 (s, 9C), 144.35 (s, 19C), 134.56 (s, 20C), 134.32 (s, 23C), 130.68 (s, 2C), 130.46 (s, 4C), 129.32 (s, 21C, 25C), 129.29 (s, 22C, 24C), 127.62 (s, 13C), 127.40 (s, 3C), 126.75 (s, 8C), 119.13 (s, 15C), 114.32 (s, 1C, 5C), 105.83 (s, 12C), 102.47 (s, 10C), 66.88 (s, 17C), 55.45 (s, 7C). ESI-TOF, calcd for C25H20N2O5 ([M + Na]+) 451.1372, found 451.1351. Anal. calcd for C25H20N2O5: C, 70.09; H, 4.71; N, 6.54; O, 18.67. Found: C, 69.58; H, 4.75; N, 6.61; O, 18.74.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 8.62–8.19 (m, 1H, Ar-H), 7.91 (dt, J = 15.0, 7.6 Hz, 1H, Ar-H), 7.76 (s, 1H, N-NH), 7.55–7.39 (m, 1H, Ar-H), 7.35–7.22 (m, 2H, Ar-H), 7.21–7.13 (m, 2H, Ar-H), 7.02 (t, J = 9.1 Hz, 1H, Ar-H), 6.84 (d, J = 8.0 Hz, 2H, Ar-H), 6.62–6.22 (m, 2H, Ar-H), 5.15–4.60 (m, 2H, –O–CH2–), 3.71 (s, 3H, –OCH3). 13C NMR (75 MHz, DMSO) δ 169.54 (s, 14C), 164.86 (s, 18C), 161.96 (s, 11C), 160.31 (s, 6C), 157.05 (s, 25C), 149.08 (s, 16C), 145.26 (s, 9C), 141.19 (s, 19C), 137.13 (s, 23C), 132.38 (d, J = 8.5 Hz, 21C), 126.90 (d, J = 15.3 Hz, 2C, 4C), 125.39 (s, 13C), 122.11 (s, 3C), 121.97 (s, 22C), 119.14 (s, 15C), 116.40 (s, 8C, 20C), 116.53 (s, 24C), 114.31 (s, 1C, 5C), 105.81 (s, 12C), 102.48 (s, 10C), 65.12 (s, 17C), 55.45 (s, 7C). ESI-TOF, calcd for C25H19FN2O5 ([M + Na]+) 469.1278, found 469.1263. Anal. calcd for C25H19FN2O5: C, 67.26; H, 4.29; F, 4.26; N, 6.28; O, 17.92. Found: C, 67.14; H, 4.34; F, 4.31; N, 6.35; O, 18.02.
N), 8.62–8.19 (m, 1H, Ar-H), 7.91 (dt, J = 15.0, 7.6 Hz, 1H, Ar-H), 7.76 (s, 1H, N-NH), 7.55–7.39 (m, 1H, Ar-H), 7.35–7.22 (m, 2H, Ar-H), 7.21–7.13 (m, 2H, Ar-H), 7.02 (t, J = 9.1 Hz, 1H, Ar-H), 6.84 (d, J = 8.0 Hz, 2H, Ar-H), 6.62–6.22 (m, 2H, Ar-H), 5.15–4.60 (m, 2H, –O–CH2–), 3.71 (s, 3H, –OCH3). 13C NMR (75 MHz, DMSO) δ 169.54 (s, 14C), 164.86 (s, 18C), 161.96 (s, 11C), 160.31 (s, 6C), 157.05 (s, 25C), 149.08 (s, 16C), 145.26 (s, 9C), 141.19 (s, 19C), 137.13 (s, 23C), 132.38 (d, J = 8.5 Hz, 21C), 126.90 (d, J = 15.3 Hz, 2C, 4C), 125.39 (s, 13C), 122.11 (s, 3C), 121.97 (s, 22C), 119.14 (s, 15C), 116.40 (s, 8C, 20C), 116.53 (s, 24C), 114.31 (s, 1C, 5C), 105.81 (s, 12C), 102.48 (s, 10C), 65.12 (s, 17C), 55.45 (s, 7C). ESI-TOF, calcd for C25H19FN2O5 ([M + Na]+) 469.1278, found 469.1263. Anal. calcd for C25H19FN2O5: C, 67.26; H, 4.29; F, 4.26; N, 6.28; O, 17.92. Found: C, 67.14; H, 4.34; F, 4.31; N, 6.35; O, 18.02.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 8.39–7.98 (m, 1H, Ar-H), 7.72 (s, 1H, N-NH), 7.63–7.37 (m, 3H, Ar-H), 7.31–7.11 (m, 3H, Ar-H), 7.08–6.92 (m, 1H, Ar-H), 6.84 (d, J = 7.9 Hz, 2H, Ar-H), 6.46 (t, J = 21.5 Hz, 2H, Ar-H), 5.15–4.60 (m, 2H, –O–CH2–), 3.71 (s, 3H, –OCH3). ESI-TOF, calcd for C25H19FN2O5 ([M + Na]+) 469.1278, found 469.1263. Anal. calcd for C25H19FN2O5: C, 67.26; H, 4.29; F, 4.26; N, 6.28; O, 17.92. Found: C, 67.14; H, 4.34; F, 4.31; N, 6.35; O, 18.02.
N), 8.39–7.98 (m, 1H, Ar-H), 7.72 (s, 1H, N-NH), 7.63–7.37 (m, 3H, Ar-H), 7.31–7.11 (m, 3H, Ar-H), 7.08–6.92 (m, 1H, Ar-H), 6.84 (d, J = 7.9 Hz, 2H, Ar-H), 6.46 (t, J = 21.5 Hz, 2H, Ar-H), 5.15–4.60 (m, 2H, –O–CH2–), 3.71 (s, 3H, –OCH3). ESI-TOF, calcd for C25H19FN2O5 ([M + Na]+) 469.1278, found 469.1263. Anal. calcd for C25H19FN2O5: C, 67.26; H, 4.29; F, 4.26; N, 6.28; O, 17.92. Found: C, 67.14; H, 4.34; F, 4.31; N, 6.35; O, 18.02.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 8.35–7.96 (m, 1H, Ar-H), 7.89 (s, 1H, N-NH), 7.75 (dt, J = 10.2, 5.3 Hz, 2H, Ar-H), 7.25 (ddd, J = 21.7, 13.2, 6.1 Hz, 4H, Ar-H), 7.00 (s, 1H, Ar-H), 6.83 (s, 2H, Ar-H), 6.50 (d, J = 16.2 Hz, 2H, Ar-H), 5.1–4.55 (m, 2H, –O–CH2–), 3.70 (s, 3H, –OCH3). ESI-TOF, calcd for C25H19FN2O5 ([M + Na]+) 469.1278, found 469.1263. Anal. calcd for C25H19FN2O5: C, 67.26; H, 4.29; F, 4.26; N, 6.28; O, 17.92. Found: C, 67.14; H, 4.34; F, 4.31; N, 6.35; O, 18.02.
N), 8.35–7.96 (m, 1H, Ar-H), 7.89 (s, 1H, N-NH), 7.75 (dt, J = 10.2, 5.3 Hz, 2H, Ar-H), 7.25 (ddd, J = 21.7, 13.2, 6.1 Hz, 4H, Ar-H), 7.00 (s, 1H, Ar-H), 6.83 (s, 2H, Ar-H), 6.50 (d, J = 16.2 Hz, 2H, Ar-H), 5.1–4.55 (m, 2H, –O–CH2–), 3.70 (s, 3H, –OCH3). ESI-TOF, calcd for C25H19FN2O5 ([M + Na]+) 469.1278, found 469.1263. Anal. calcd for C25H19FN2O5: C, 67.26; H, 4.29; F, 4.26; N, 6.28; O, 17.92. Found: C, 67.14; H, 4.34; F, 4.31; N, 6.35; O, 18.02.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 8.34–7.94 (m, 1H, Ar-H), 7.90–7.64 (m, 2H, N-NH, Ar-H), 7.64–7.40 (m, 2H, Ar-H), 7.19 (d, J = 8.6 Hz, 2H, Ar-H), 7.01 (t, J = 8.6 Hz, 1H, Ar-H), 6.84 (d, J = 7.0 Hz, 2H, Ar-H), 6.45 (t, J = 20.6 Hz, 2H, Ar-H), 5.15–4.60 (m, 2H, –O–CH2–), 3.71 (s, 3H, –OCH3). ESI-TOF, calcd for C25H19F2N2O5 ([M + Na]+) 487.1184, found 487.1173. Anal. calcd for C25H19FN2O5: C, 64.66; H, 3.91; F, 8.18; N, 6.03; O, 17.22. Found: C, 64.58; H, 3.99; F, 8.23; N, 6.11; O, 17.31.
N), 8.34–7.94 (m, 1H, Ar-H), 7.90–7.64 (m, 2H, N-NH, Ar-H), 7.64–7.40 (m, 2H, Ar-H), 7.19 (d, J = 8.6 Hz, 2H, Ar-H), 7.01 (t, J = 8.6 Hz, 1H, Ar-H), 6.84 (d, J = 7.0 Hz, 2H, Ar-H), 6.45 (t, J = 20.6 Hz, 2H, Ar-H), 5.15–4.60 (m, 2H, –O–CH2–), 3.71 (s, 3H, –OCH3). ESI-TOF, calcd for C25H19F2N2O5 ([M + Na]+) 487.1184, found 487.1173. Anal. calcd for C25H19FN2O5: C, 64.66; H, 3.91; F, 8.18; N, 6.03; O, 17.22. Found: C, 64.58; H, 3.99; F, 8.23; N, 6.11; O, 17.31.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 8.78–8.32 (m, 1H, Ar-H), 8.00 (m, 2H, Ar-H), 7.78 (s, 1H, N-NH), 7.57–7.37 (m, 2H, Ar-H), 7.21 (dd, J = 7.0, 1.4 Hz, 2H, Ar-H), 7.09–6.95 (m, 1H, Ar-H), 6.86 (d, J = 8.5 Hz, 2H, Ar-H), 6.59–6.31 (m, 2H, Ar-H), 5.20–4.48 (m, 2H, –O–CH2–), 3.73 (s, 3H, –OCH3). ESI-TOF, calcd for C25H19ClN2O5 ([M + Na]+) 485.0982, found 485.0974. Anal. calcd for C25H19ClN2O5: C, 64.87; H, 4.14; Cl, 7.66; N, 6.05; O, 17.28. Found: C, 64.69; H, 4.27; Cl, 7.74; N, 6.12; O, 17.32.
N), 8.78–8.32 (m, 1H, Ar-H), 8.00 (m, 2H, Ar-H), 7.78 (s, 1H, N-NH), 7.57–7.37 (m, 2H, Ar-H), 7.21 (dd, J = 7.0, 1.4 Hz, 2H, Ar-H), 7.09–6.95 (m, 1H, Ar-H), 6.86 (d, J = 8.5 Hz, 2H, Ar-H), 6.59–6.31 (m, 2H, Ar-H), 5.20–4.48 (m, 2H, –O–CH2–), 3.73 (s, 3H, –OCH3). ESI-TOF, calcd for C25H19ClN2O5 ([M + Na]+) 485.0982, found 485.0974. Anal. calcd for C25H19ClN2O5: C, 64.87; H, 4.14; Cl, 7.66; N, 6.05; O, 17.28. Found: C, 64.69; H, 4.27; Cl, 7.74; N, 6.12; O, 17.32.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 8.35–7.95 (m, 1H, Ar-H), 7.94 (s, 1H, N-NH), 7.85–7.72 (m, 1H, Ar-H), 7.68 (d, J = 3.2 Hz, 1H, Ar-H), 7.48 (dd, J = 13.8, 7.9 Hz, 2H, Ar-H), 7.21 (s, 2H, Ar-H), 7.04 (s, 1H, Ar-H), 6.87 (d, J = 22.1 Hz, 2H, Ar-H), 6.58–6.27 (m, 2H, Ar-H), 5.20–4.60 (m, 2H, –O–CH2–), 3.71 (s, 3H, –OCH3). ESI-TOF, calcd for C25H19ClN2O5 ([M + Na]+) 485.0982, found 485.0974. Anal. calcd for C25H19ClN2O5: C, 64.87; H, 4.14; Cl, 7.66; N, 6.05; O, 17.28. Found: C, 64.69; H, 4.27; Cl, 7.74; N, 6.12; O, 17.32.
N), 8.35–7.95 (m, 1H, Ar-H), 7.94 (s, 1H, N-NH), 7.85–7.72 (m, 1H, Ar-H), 7.68 (d, J = 3.2 Hz, 1H, Ar-H), 7.48 (dd, J = 13.8, 7.9 Hz, 2H, Ar-H), 7.21 (s, 2H, Ar-H), 7.04 (s, 1H, Ar-H), 6.87 (d, J = 22.1 Hz, 2H, Ar-H), 6.58–6.27 (m, 2H, Ar-H), 5.20–4.60 (m, 2H, –O–CH2–), 3.71 (s, 3H, –OCH3). ESI-TOF, calcd for C25H19ClN2O5 ([M + Na]+) 485.0982, found 485.0974. Anal. calcd for C25H19ClN2O5: C, 64.87; H, 4.14; Cl, 7.66; N, 6.05; O, 17.28. Found: C, 64.69; H, 4.27; Cl, 7.74; N, 6.12; O, 17.32.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 8.35–8.10 (m, 1H, Ar-H), 7.74 (dd, J = 8.5, 4.8 Hz, 2H, Ar-H), 7.51 (dd, J = 8.4, 5.0 Hz, 2H, Ar-H), 7.21 (d, J = 8.6 Hz, 2H, Ar-H), 7.08–6.75 (m, 7.5 Hz, 3H, Ar-H), 6.59–6.27 (m, 2H, Ar-H), 5.15–4.55 (m, 2H, –O–CH2–), 3.72 (s, 3H, –OCH3). ESI-TOF, calcd for C25H19ClN2O5 ([M + Na]+) 485.0982, found 485.0979. Anal. calcd for C25H19ClN2O5: C, 64.87; H, 4.14; Cl, 7.66; N, 6.05; O, 17.28. Found: C, 64.69; H, 4.27; Cl, 7.74; N, 6.12; O, 17.32.
N), 8.35–8.10 (m, 1H, Ar-H), 7.74 (dd, J = 8.5, 4.8 Hz, 2H, Ar-H), 7.51 (dd, J = 8.4, 5.0 Hz, 2H, Ar-H), 7.21 (d, J = 8.6 Hz, 2H, Ar-H), 7.08–6.75 (m, 7.5 Hz, 3H, Ar-H), 6.59–6.27 (m, 2H, Ar-H), 5.15–4.55 (m, 2H, –O–CH2–), 3.72 (s, 3H, –OCH3). ESI-TOF, calcd for C25H19ClN2O5 ([M + Na]+) 485.0982, found 485.0979. Anal. calcd for C25H19ClN2O5: C, 64.87; H, 4.14; Cl, 7.66; N, 6.05; O, 17.28. Found: C, 64.69; H, 4.27; Cl, 7.74; N, 6.12; O, 17.32.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 8.80–8.35 (m, 1H, Ar-H), 8.04–7.87 (m, 1H, Ar-H), 7.86–7.59 (m, 2H, N-NH, Ar-H), 7.43 (td, J = 7.9, 4.1 Hz, 1H, Ar-H), 7.19 (d, J = 8.4 Hz, 2H, Ar-H), 7.07–6.94 (m, 1H, Ar-H), 6.84 (d, J = 8.2 Hz, 2H, Ar-H), 6.46 (t, J = 23.2 Hz, 2H, Ar-H), 5.15–4.60 (m, 2H, –O–CH2–), 3.71 (s, 3H, –OCH3). ESI-TOF, calcd for C25H18Cl2N2O5 ([M + Na]+) 519.0593, found 519.0582. Anal. calcd for C25H18Cl2N2O5: C, 60.38; H, 3.65; Cl, 14.26; N, 5.63; O, 16.08. Found: C, 60.29; H, 3.69; Cl, 14.30; N, 5.74; O, 16.15.
N), 8.80–8.35 (m, 1H, Ar-H), 8.04–7.87 (m, 1H, Ar-H), 7.86–7.59 (m, 2H, N-NH, Ar-H), 7.43 (td, J = 7.9, 4.1 Hz, 1H, Ar-H), 7.19 (d, J = 8.4 Hz, 2H, Ar-H), 7.07–6.94 (m, 1H, Ar-H), 6.84 (d, J = 8.2 Hz, 2H, Ar-H), 6.46 (t, J = 23.2 Hz, 2H, Ar-H), 5.15–4.60 (m, 2H, –O–CH2–), 3.71 (s, 3H, –OCH3). ESI-TOF, calcd for C25H18Cl2N2O5 ([M + Na]+) 519.0593, found 519.0582. Anal. calcd for C25H18Cl2N2O5: C, 60.38; H, 3.65; Cl, 14.26; N, 5.63; O, 16.08. Found: C, 60.29; H, 3.69; Cl, 14.30; N, 5.74; O, 16.15.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 8.55–8.22 (m, 1H, Ar-H), 7.95–7.60 (m, 1H, N-NH), 7.55 (d, J = 7.9 Hz, 2H, Ar-H), 7.48–7.35 (m, 1H, Ar-H), 7.18 (d, J = 8.0 Hz, 2H, Ar-H), 7.10–6.75 (m, 3H, Ar-H), 6.50 (d, J = 29.2 Hz, 2H, Ar-H), 5.05–4.60 (m, 2H, –O–CH2–), 3.70 (s, 3H, –OCH3). 13C NMR (75 MHz, DMSO) δ 169.71 (s, 14C), 165.00 (s, 18C), 159.57 (s, 11C), 158.11 (s, 6C), 157.12 (s, 16C), 143.84 (s, 9C), 139.31 (s, 19C), 134.43 (s, 21C, 25C), 134.39 (s, 20C), 131.59 (s, 13C), 129.93 (s, 2C, 4C), 129.87 (s, 23C), 129.54 (s, 22C, 24C), 128.12 (s, 8C), 126.77 (s, 3C), 119.14 (s, 15C), 114.31 (s, 1C, 5C), 105.72 (s, 12C), 102.33 (s, 10C), 66.86 (s, 17C), 55.45 (s, 7C). ESI-TOF, calcd for C25H18Cl2N2O5 ([M + Na]+) 519.0593, found 519.0582. Anal. calcd for C25H18Cl2N2O5: C, 60.38; H, 3.65; Cl, 14.26; N, 5.63; O, 16.08. Found: C, 60.29; H, 3.69; Cl, 14.30; N, 5.74; O, 16.15.
N), 8.55–8.22 (m, 1H, Ar-H), 7.95–7.60 (m, 1H, N-NH), 7.55 (d, J = 7.9 Hz, 2H, Ar-H), 7.48–7.35 (m, 1H, Ar-H), 7.18 (d, J = 8.0 Hz, 2H, Ar-H), 7.10–6.75 (m, 3H, Ar-H), 6.50 (d, J = 29.2 Hz, 2H, Ar-H), 5.05–4.60 (m, 2H, –O–CH2–), 3.70 (s, 3H, –OCH3). 13C NMR (75 MHz, DMSO) δ 169.71 (s, 14C), 165.00 (s, 18C), 159.57 (s, 11C), 158.11 (s, 6C), 157.12 (s, 16C), 143.84 (s, 9C), 139.31 (s, 19C), 134.43 (s, 21C, 25C), 134.39 (s, 20C), 131.59 (s, 13C), 129.93 (s, 2C, 4C), 129.87 (s, 23C), 129.54 (s, 22C, 24C), 128.12 (s, 8C), 126.77 (s, 3C), 119.14 (s, 15C), 114.31 (s, 1C, 5C), 105.72 (s, 12C), 102.33 (s, 10C), 66.86 (s, 17C), 55.45 (s, 7C). ESI-TOF, calcd for C25H18Cl2N2O5 ([M + Na]+) 519.0593, found 519.0582. Anal. calcd for C25H18Cl2N2O5: C, 60.38; H, 3.65; Cl, 14.26; N, 5.63; O, 16.08. Found: C, 60.29; H, 3.69; Cl, 14.30; N, 5.74; O, 16.15.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 8.30–7.95 (m, 1H, Ar-H), 7.90 (d, J = 9.7 Hz, 1H, Ar-H), 7.69 (d, J = 7.7 Hz, 1H, Ar-H), 7.59 (d, J = 6.6 Hz, 1H, Ar-H), 7.39 (dd, J = 14.5, 6.7 Hz, 1H, Ar-H), 7.19 (d, J = 8.4 Hz, 2H, Ar-H), 6.99 (d, J = 8.1 Hz, 1H, Ar-H), 6.84 (s, 2H, Ar-H), 6.56–6.22 (m, 2H, Ar-H), 5.15–4.59 (m, 2H, –O–CH2–), 3.70 (s, 3H, –OCH3). ESI-TOF, calcd for C25H19BrN2O5 ([M + Na]+) 529.0477, found 529.0438. Anal. calcd for C25H19BrN2O5: C, 59.19; H, 3.77; Br, 15.75; N, 5.52; O, 15.77. Found: C, 59.10; H, 3.83; Br, 15.82; N, 5.59; O, 15.80.
N), 8.30–7.95 (m, 1H, Ar-H), 7.90 (d, J = 9.7 Hz, 1H, Ar-H), 7.69 (d, J = 7.7 Hz, 1H, Ar-H), 7.59 (d, J = 6.6 Hz, 1H, Ar-H), 7.39 (dd, J = 14.5, 6.7 Hz, 1H, Ar-H), 7.19 (d, J = 8.4 Hz, 2H, Ar-H), 6.99 (d, J = 8.1 Hz, 1H, Ar-H), 6.84 (s, 2H, Ar-H), 6.56–6.22 (m, 2H, Ar-H), 5.15–4.59 (m, 2H, –O–CH2–), 3.70 (s, 3H, –OCH3). ESI-TOF, calcd for C25H19BrN2O5 ([M + Na]+) 529.0477, found 529.0438. Anal. calcd for C25H19BrN2O5: C, 59.19; H, 3.77; Br, 15.75; N, 5.52; O, 15.77. Found: C, 59.10; H, 3.83; Br, 15.82; N, 5.59; O, 15.80.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 8.25–7.90 (m, 1H, Ar-H), 7.68 (s, 1H, N-NH), 7.28–6.95 (m, 6H, Ar-H), 6.83 (d, J = 8.1 Hz, 3H, Ar-H), 6.52 (d, J = 25.9 Hz, 2H, Ar-H), 5.15–4.60 (m, 2H, –O–CH2–), 3.72 (s, 3H, –OCH3). 13C NMR (100 MHz, DMSO) δ 169.31 (s, 14C), 164.66 (s, 18C), 158.14 (s, 14C), 158.10 (s, 6C), 157.08 (s, 24C), 148.44 (s, 9C), 144.54 (s, 19C), 135.83 (s, 20C), 135.66 (s, 22C), 130.35 (s, 2C, 4C), 128.09 (s, 13C), 126.74 (s, 3C), 119.33 (s, 8C), 118.85 (s, 21C), 118.02 (s, 23C), 117.76 (s, 14C), 114.32 (s, 25C, 1C, 5C), 113.23 (s), 113.12 (s), 105.81 (s, 12C), 102.82 (s, 10C), 66.87 (s, 17C), 55.45 (s, 7C). ESI-TOF, calcd for C25H20N2O6 ([M + Na]+) 467.1321, found 467.1314. Anal. calcd for C25H20N2O6: C, 67.56; H, 4.54; N, 6.30; O, 21.60. Found: C, 67.47; H, 4.61; N, 6.39; O, 21.65.
N), 8.25–7.90 (m, 1H, Ar-H), 7.68 (s, 1H, N-NH), 7.28–6.95 (m, 6H, Ar-H), 6.83 (d, J = 8.1 Hz, 3H, Ar-H), 6.52 (d, J = 25.9 Hz, 2H, Ar-H), 5.15–4.60 (m, 2H, –O–CH2–), 3.72 (s, 3H, –OCH3). 13C NMR (100 MHz, DMSO) δ 169.31 (s, 14C), 164.66 (s, 18C), 158.14 (s, 14C), 158.10 (s, 6C), 157.08 (s, 24C), 148.44 (s, 9C), 144.54 (s, 19C), 135.83 (s, 20C), 135.66 (s, 22C), 130.35 (s, 2C, 4C), 128.09 (s, 13C), 126.74 (s, 3C), 119.33 (s, 8C), 118.85 (s, 21C), 118.02 (s, 23C), 117.76 (s, 14C), 114.32 (s, 25C, 1C, 5C), 113.23 (s), 113.12 (s), 105.81 (s, 12C), 102.82 (s, 10C), 66.87 (s, 17C), 55.45 (s, 7C). ESI-TOF, calcd for C25H20N2O6 ([M + Na]+) 467.1321, found 467.1314. Anal. calcd for C25H20N2O6: C, 67.56; H, 4.54; N, 6.30; O, 21.60. Found: C, 67.47; H, 4.61; N, 6.39; O, 21.65.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 10.01 (s, 1H, –OH), 8.25–7.85 (m, 1H, Ar-H), 7.76 (s, 1H, N-NH), 7.51 (d, J = 8.4 Hz, 2H, Ar-H), 7.19 (dd, J = 8.8, 2.6 Hz, 2H, Ar-H), 7.01 (t, J = 8.5 Hz, 1H, Ar-H), 6.89–6.73 (m, 4H, Ar-H), 6.54–6.25 (m, 2H, Ar-H), 5.10–4.55 (m, 2H, –O–CH2–), 3.70 (s, 3H, –OCH3). ESI-TOF, calcd for C25H20N2O6 ([M + Na]+) 467.1321, found 467.1314. Anal. calcd for C25H20N2O6: C, 67.56; H, 4.54; N, 6.30; O, 21.60. Found: C, 67.47; H, 4.61; N, 6.39; O, 21.65.
N), 10.01 (s, 1H, –OH), 8.25–7.85 (m, 1H, Ar-H), 7.76 (s, 1H, N-NH), 7.51 (d, J = 8.4 Hz, 2H, Ar-H), 7.19 (dd, J = 8.8, 2.6 Hz, 2H, Ar-H), 7.01 (t, J = 8.5 Hz, 1H, Ar-H), 6.89–6.73 (m, 4H, Ar-H), 6.54–6.25 (m, 2H, Ar-H), 5.10–4.55 (m, 2H, –O–CH2–), 3.70 (s, 3H, –OCH3). ESI-TOF, calcd for C25H20N2O6 ([M + Na]+) 467.1321, found 467.1314. Anal. calcd for C25H20N2O6: C, 67.56; H, 4.54; N, 6.30; O, 21.60. Found: C, 67.47; H, 4.61; N, 6.39; O, 21.65.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 8.14–7.78 (m, 1H, Ar-H), 7.75 (s, 1H, N-NH), 7.34 (d, J = 8.5 Hz, 2H, Ar-H), 7.17 (dt, J = 14.1, 7.0 Hz, 2H, Ar-H), 7.00 (t, J = 8.3 Hz, 1H, Ar-H), 6.82 (t, J = 10.8 Hz, 2H, Ar-H), 6.56 (d, J = 8.0 Hz, 2H, Ar-H), 6.47–6.28 (m, 2H, Ar-H), 5.61 (d, J = 9.2 Hz, 2H, –NH2), 5.55–4.51 (m, 2H, –O–CH2–), 3.70 (s, 3H, –OCH3). ESI-TOF, calcd for C25H21N3O5 ([M + Na]+) 466.1481, found 466.1473. Anal. calcd for C25H21N3O5: C, 67.71; H, 4.77; N, 9.48; O, 18.04. Found: C, 67.65; H, 4.82; N, 9.53; O, 18.14.
N), 8.14–7.78 (m, 1H, Ar-H), 7.75 (s, 1H, N-NH), 7.34 (d, J = 8.5 Hz, 2H, Ar-H), 7.17 (dt, J = 14.1, 7.0 Hz, 2H, Ar-H), 7.00 (t, J = 8.3 Hz, 1H, Ar-H), 6.82 (t, J = 10.8 Hz, 2H, Ar-H), 6.56 (d, J = 8.0 Hz, 2H, Ar-H), 6.47–6.28 (m, 2H, Ar-H), 5.61 (d, J = 9.2 Hz, 2H, –NH2), 5.55–4.51 (m, 2H, –O–CH2–), 3.70 (s, 3H, –OCH3). ESI-TOF, calcd for C25H21N3O5 ([M + Na]+) 466.1481, found 466.1473. Anal. calcd for C25H21N3O5: C, 67.71; H, 4.77; N, 9.48; O, 18.04. Found: C, 67.65; H, 4.82; N, 9.53; O, 18.14.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 8.75–8.32 (m, 1H, Ar-H), 8.19 (dd, J = 16.1, 7.6 Hz, 1H, Ar-H), 7.85–7.52 (m, 4H, N-NH, Ar-H), 7.19 (d, J = 7.3 Hz, 2H, Ar-H), 7.09–6.91 (m, 1H, Ar-H), 6.84 (d, J = 7.5 Hz, 2H, Ar-H), 6.62–6.22 (m, 2H, Ar-H), 5.21–4.60 (m, 2H, –O–CH2–), 3.70 (s, 3H, –OCH3). ESI-TOF, calcd for C26H19F3N2O5 ([M + Na]+) 519.1246, found 519.1209. Anal. calcd for C26H19F3N2O5: C, 62.90; H, 3.86; F, 11.48; N, 5.64; O, 16.11. Found: C, 62.82; H, 3.94; F, 11.53; N, 5.72; O, 16.24.
N), 8.75–8.32 (m, 1H, Ar-H), 8.19 (dd, J = 16.1, 7.6 Hz, 1H, Ar-H), 7.85–7.52 (m, 4H, N-NH, Ar-H), 7.19 (d, J = 7.3 Hz, 2H, Ar-H), 7.09–6.91 (m, 1H, Ar-H), 6.84 (d, J = 7.5 Hz, 2H, Ar-H), 6.62–6.22 (m, 2H, Ar-H), 5.21–4.60 (m, 2H, –O–CH2–), 3.70 (s, 3H, –OCH3). ESI-TOF, calcd for C26H19F3N2O5 ([M + Na]+) 519.1246, found 519.1209. Anal. calcd for C26H19F3N2O5: C, 62.90; H, 3.86; F, 11.48; N, 5.64; O, 16.11. Found: C, 62.82; H, 3.94; F, 11.53; N, 5.72; O, 16.24.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 8.43–8.05 (m, 1H, Ar-H), 7.92 (d, J = 8.1 Hz, 2H, Ar-H), 7.78 (t, J = 7.2 Hz, 3H, N-NH, Ar-H), 7.19 (dd, J = 8.7, 2.1 Hz, 2H, Ar-H), 7.02 (t, J = 8.7 Hz, 1H, Ar-H), 6.84 (d, J = 8.3 Hz, 2H, Ar-H), 6.61–6.29 (m, 2H, Ar-H), 5.17–4.61 (m, 2H, –O–CH2–), 3.71 (s, 3H, –OCH3). 13C NMR (75 MHz, DMSO) δ 169.73 (s, 14C), 165.08 (s, 18C), 159.79–159.10 (m, 11C), 158.05 (s, 6C), 157.08 (s, 16C), 146.63 (s, 9C), 142.67 (s, 19C), 138.56 (s, 20C), 138.39 (s, 23C), 130.18 (s, 2C, 4C), 128.20 (s, 21C, 25C), 128.01 (s, 13C), 126.74 (s, 22C, 24C), 126.16 (d, J = 4.7 Hz, 3C), 125.46 (s,
N), 8.43–8.05 (m, 1H, Ar-H), 7.92 (d, J = 8.1 Hz, 2H, Ar-H), 7.78 (t, J = 7.2 Hz, 3H, N-NH, Ar-H), 7.19 (dd, J = 8.7, 2.1 Hz, 2H, Ar-H), 7.02 (t, J = 8.7 Hz, 1H, Ar-H), 6.84 (d, J = 8.3 Hz, 2H, Ar-H), 6.61–6.29 (m, 2H, Ar-H), 5.17–4.61 (m, 2H, –O–CH2–), 3.71 (s, 3H, –OCH3). 13C NMR (75 MHz, DMSO) δ 169.73 (s, 14C), 165.08 (s, 18C), 159.79–159.10 (m, 11C), 158.05 (s, 6C), 157.08 (s, 16C), 146.63 (s, 9C), 142.67 (s, 19C), 138.56 (s, 20C), 138.39 (s, 23C), 130.18 (s, 2C, 4C), 128.20 (s, 21C, 25C), 128.01 (s, 13C), 126.74 (s, 22C, 24C), 126.16 (d, J = 4.7 Hz, 3C), 125.46 (s, ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) F3), 123.66 (s, 8C), 119.13 (s, 15C), 114.31 (s, 1C, 5C), 105.79 (s, 12C), 102.54 (s, 10C), 66.84 (s, 17C), 55.44 (s, 7C). ESI-TOF, calcd for C26H19F3N2O5 ([M + Na]+) 519.1246, found 519.1209. Anal. calcd for C26H19F3N2O5: C, 62.90; H, 3.86; F, 11.48; N, 5.64; O, 16.11. Found: C, 62.82; H, 3.94; F, 11.53; N, 5.72; O, 16.24.
F3), 123.66 (s, 8C), 119.13 (s, 15C), 114.31 (s, 1C, 5C), 105.79 (s, 12C), 102.54 (s, 10C), 66.84 (s, 17C), 55.44 (s, 7C). ESI-TOF, calcd for C26H19F3N2O5 ([M + Na]+) 519.1246, found 519.1209. Anal. calcd for C26H19F3N2O5: C, 62.90; H, 3.86; F, 11.48; N, 5.64; O, 16.11. Found: C, 62.82; H, 3.94; F, 11.53; N, 5.72; O, 16.24.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 8.32–7.9 (m, 1H, Ar-H), 7.74 (s, 1H, N-NH), 7.58 (d, J = 7.8 Hz, 2H, Ar-H), 7.35–7.10 (m, 4H, Ar-H), 7.01 (t, J = 8.6 Hz, 1H, Ar-H), 6.84 (d, J = 7.6 Hz, 2H, Ar-H), 6.45 (dd, J = 28.6, 17.3 Hz, 2H, Ar-H), 5.17–4.51 (m, 2H, –O–CH2–), 3.71 (s, 3H, –OCH3), 2.32 (s, 3H, –CH3). ESI-TOF, calcd for C26H22N2O5 ([M + Na]+) 465.1529, found 465.1513. Anal. calcd for C26H22N2O5: C, 70.58; H, 5.01; N, 6.33; O, 18.08. Found: C, 70.14; H, 5.16; N, 6.42; O, 18.14.
N), 8.32–7.9 (m, 1H, Ar-H), 7.74 (s, 1H, N-NH), 7.58 (d, J = 7.8 Hz, 2H, Ar-H), 7.35–7.10 (m, 4H, Ar-H), 7.01 (t, J = 8.6 Hz, 1H, Ar-H), 6.84 (d, J = 7.6 Hz, 2H, Ar-H), 6.45 (dd, J = 28.6, 17.3 Hz, 2H, Ar-H), 5.17–4.51 (m, 2H, –O–CH2–), 3.71 (s, 3H, –OCH3), 2.32 (s, 3H, –CH3). ESI-TOF, calcd for C26H22N2O5 ([M + Na]+) 465.1529, found 465.1513. Anal. calcd for C26H22N2O5: C, 70.58; H, 5.01; N, 6.33; O, 18.08. Found: C, 70.14; H, 5.16; N, 6.42; O, 18.14.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 8.71–8.32 (m, 1H, Ar-H), 7.84 (ddd, J = 16.7, 7.7, 1.5 Hz, 2H, N-NH, Ar-H), 7.47–7.34 (m, 1H, Ar-H), 7.21 (dd, J = 8.8, 2.4 Hz, 2H, Ar-H), 7.09 (t, J = 9.1 Hz, 1H, Ar-H), 7.02 (dt, J = 10.7, 8.2 Hz, 2H, Ar-H), 6.86 (d, J = 8.0 Hz, 2H, Ar-H), 6.60–6.27 (m, 2H, Ar-H), 5.15–4.55 (m, 2H, –O–CH2–), 3.86 (d, J = 3.2 Hz, 3H, –OCH3), 3.72 (s, 3H, –OCH3). ESI-TOF, calcd for C26H22N2O6 ([M + Na]+) 481.1478, found 481.1463. Anal. calcd for C26H22N2O6: C, 68.11; H, 4.84; N, 6.11; O, 20.94. Found: C, 68.02; H, 4.92; N, 6.23; O, 20.98.
N), 8.71–8.32 (m, 1H, Ar-H), 7.84 (ddd, J = 16.7, 7.7, 1.5 Hz, 2H, N-NH, Ar-H), 7.47–7.34 (m, 1H, Ar-H), 7.21 (dd, J = 8.8, 2.4 Hz, 2H, Ar-H), 7.09 (t, J = 9.1 Hz, 1H, Ar-H), 7.02 (dt, J = 10.7, 8.2 Hz, 2H, Ar-H), 6.86 (d, J = 8.0 Hz, 2H, Ar-H), 6.60–6.27 (m, 2H, Ar-H), 5.15–4.55 (m, 2H, –O–CH2–), 3.86 (d, J = 3.2 Hz, 3H, –OCH3), 3.72 (s, 3H, –OCH3). ESI-TOF, calcd for C26H22N2O6 ([M + Na]+) 481.1478, found 481.1463. Anal. calcd for C26H22N2O6: C, 68.11; H, 4.84; N, 6.11; O, 20.94. Found: C, 68.02; H, 4.92; N, 6.23; O, 20.98.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 8.29–7.95 (m, 1H, Ar-H), 7.75–7.56 (m, 2H, Ar-H), 7.21 (d, J = 7.5 Hz, 2H, Ar-H), 7.01 (dd, J = 8.7, 6.8 Hz, 3H, Ar-H), 6.85 (s, 2H), 6.63–6.22 (m, 2H, Ar-H), 5.12–4.59 (m, 2H, –O–CH2–), 3.80 (d, J = 2.8 Hz, 3H, –OCH3), 3.72 (s, 3H, –OCH3). ESI-TOF, calcd for C26H22N2O6 ([M + Na]+) 481.1478, found 481.1463. Anal. calcd for C26H22N2O6: C, 68.11; H, 4.84; N, 6.11; O, 20.94. Found: C, 68.02; H, 4.92; N, 6.23; O, 20.98.
N), 8.29–7.95 (m, 1H, Ar-H), 7.75–7.56 (m, 2H, Ar-H), 7.21 (d, J = 7.5 Hz, 2H, Ar-H), 7.01 (dd, J = 8.7, 6.8 Hz, 3H, Ar-H), 6.85 (s, 2H), 6.63–6.22 (m, 2H, Ar-H), 5.12–4.59 (m, 2H, –O–CH2–), 3.80 (d, J = 2.8 Hz, 3H, –OCH3), 3.72 (s, 3H, –OCH3). ESI-TOF, calcd for C26H22N2O6 ([M + Na]+) 481.1478, found 481.1463. Anal. calcd for C26H22N2O6: C, 68.11; H, 4.84; N, 6.11; O, 20.94. Found: C, 68.02; H, 4.92; N, 6.23; O, 20.98.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 8.28–7.90 (m, 1H, Ar-H), 7.73 (s, 1H, Ar-H), 7.32 (dd, J = 9.1, 1.6 Hz, 1H, Ar-H), 7.21 (dd, J = 8.4, 2.6 Hz, 3H, Ar-H), 7.03 (dd, J = 16.7, 8.3 Hz, 2H, Ar-H), 6.86 (d, J = 6.8 Hz, 2H, Ar-H), 6.59–6.31 (m, 2H, Ar-H), 5.18–4.60 (m, 2H, –O–CH2–), 3.81 (dd, J = 8.5, 6.6 Hz, 6H, –OCH3), 3.72 (s, 3H, –OCH3). ESI-TOF, calcd for C27H24N2O7 ([M + Na]+) 511.1584, found 511.1558. Anal. calcd for C27H24N2O7: C, 66.39; H, 4.95; N, 5.73; O, 22.93. Found: C, 66.24; H, 5.03; N, 5.86; O, 23.06.
N), 8.28–7.90 (m, 1H, Ar-H), 7.73 (s, 1H, Ar-H), 7.32 (dd, J = 9.1, 1.6 Hz, 1H, Ar-H), 7.21 (dd, J = 8.4, 2.6 Hz, 3H, Ar-H), 7.03 (dd, J = 16.7, 8.3 Hz, 2H, Ar-H), 6.86 (d, J = 6.8 Hz, 2H, Ar-H), 6.59–6.31 (m, 2H, Ar-H), 5.18–4.60 (m, 2H, –O–CH2–), 3.81 (dd, J = 8.5, 6.6 Hz, 6H, –OCH3), 3.72 (s, 3H, –OCH3). ESI-TOF, calcd for C27H24N2O7 ([M + Na]+) 511.1584, found 511.1558. Anal. calcd for C27H24N2O7: C, 66.39; H, 4.95; N, 5.73; O, 22.93. Found: C, 66.24; H, 5.03; N, 5.86; O, 23.06.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 8.31–7.95 (m, 1H, Ar-H), 7.75 (s, 1H, N-NH), 7.61 (d, J = 8.2 Hz, 2H, Ar-H), 7.30 (dd, J = 8.0, 5.1 Hz, 2H, Ar-H), 7.19 (dd, J = 8.8, 2.5 Hz, 2H, Ar-H), 7.02 (t, J = 8.6 Hz, 1H, Ar-H), 6.84 (d, J = 8.1 Hz, 2H, Ar-H), 6.56–6.30 (m, 2H, Ar-H), 5.12–4.55 (m, 2H, –O–CH2–), 3.71 (s, 3H, –OCH3), 2.90 (m, 1H,
N), 8.31–7.95 (m, 1H, Ar-H), 7.75 (s, 1H, N-NH), 7.61 (d, J = 8.2 Hz, 2H, Ar-H), 7.30 (dd, J = 8.0, 5.1 Hz, 2H, Ar-H), 7.19 (dd, J = 8.8, 2.5 Hz, 2H, Ar-H), 7.02 (t, J = 8.6 Hz, 1H, Ar-H), 6.84 (d, J = 8.1 Hz, 2H, Ar-H), 6.56–6.30 (m, 2H, Ar-H), 5.12–4.55 (m, 2H, –O–CH2–), 3.71 (s, 3H, –OCH3), 2.90 (m, 1H, 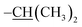 ), 1.20 (dd, J = 6.9, 1.8 Hz, 6H,
), 1.20 (dd, J = 6.9, 1.8 Hz, 6H, 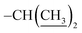 ). ESI-TOF, calcd for C28H26N2O5 ([M + Na]+) 493.1842, found 493.1837. Anal. calcd for C28H26N2O5: C, 71.48; H, 5.57; N, 5.95; O, 17.00. Found: C, 71.31; H, 5.62; N, 5.995; O, 17.12.
). ESI-TOF, calcd for C28H26N2O5 ([M + Na]+) 493.1842, found 493.1837. Anal. calcd for C28H26N2O5: C, 71.48; H, 5.57; N, 5.95; O, 17.00. Found: C, 71.31; H, 5.62; N, 5.995; O, 17.12.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 8.30–7.90 (m, 1H, Ar-H), 7.80 (d, J = 8.4 Hz, 1H, N-NH), 7.63 (d, J = 8.3 Hz, 2H, Ar-H), 7.38 (ddd, J = 19.5, 13.1, 6.8 Hz, 5H, Ar-H), 7.19 (d, J = 8.0 Hz, 2H, Ar-H), 7.05 (dd, J = 15.8, 7.5 Hz, 3H, Ar-H), 6.85 (s, 2H, Ar-H), 6.58–6.22 (m, 2H, Ar-H), 5.14 (s, 2H, Ph-CH2), 5.10–4.55 (m, 2H, –O–CH2–), 3.70 (s, 3H, –OCH3). 13C NMR (75 MHz, DMSO) δ 169.17 (s, 14C), 164.43 (s, 18C), 160.48 (s, 11C), 160.26 (s, 23C), 148.21 (s, 6C), 144.14 (s, 16C), 137.21 (s, 9C), 129.24 (s, 19C), 128.99 (s, 27C), 128.94 (s, 2C, 4C, 21C, 25C), 128.42 (s, 29C, 31C), 128.25 (s, 13C), 127.31 (s, 28C, 32C), 127.23 (s, 31C), 115.62 (s, 20C, 3C), 114.32 (s, 1C, 5C, 22C, 24C), 105.80 (s, 8C, 12C), 102.86 (s, 15C), 102.45 (s, 10C), 69.81 (s, 26C), 66.89 (s, 17C), 55.45 (s, 7C). ESI-TOF, calcd for C32H26N2O6 ([M + Na]+) 557.1791, found 557.1805. Anal. calcd for C32H26N2O6: C, 71.90; H, 4.90; N, 5.24; O, 17.96. Found: C, 71.83; H, 4.98; N, 5.31; O, 18.03.
N), 8.30–7.90 (m, 1H, Ar-H), 7.80 (d, J = 8.4 Hz, 1H, N-NH), 7.63 (d, J = 8.3 Hz, 2H, Ar-H), 7.38 (ddd, J = 19.5, 13.1, 6.8 Hz, 5H, Ar-H), 7.19 (d, J = 8.0 Hz, 2H, Ar-H), 7.05 (dd, J = 15.8, 7.5 Hz, 3H, Ar-H), 6.85 (s, 2H, Ar-H), 6.58–6.22 (m, 2H, Ar-H), 5.14 (s, 2H, Ph-CH2), 5.10–4.55 (m, 2H, –O–CH2–), 3.70 (s, 3H, –OCH3). 13C NMR (75 MHz, DMSO) δ 169.17 (s, 14C), 164.43 (s, 18C), 160.48 (s, 11C), 160.26 (s, 23C), 148.21 (s, 6C), 144.14 (s, 16C), 137.21 (s, 9C), 129.24 (s, 19C), 128.99 (s, 27C), 128.94 (s, 2C, 4C, 21C, 25C), 128.42 (s, 29C, 31C), 128.25 (s, 13C), 127.31 (s, 28C, 32C), 127.23 (s, 31C), 115.62 (s, 20C, 3C), 114.32 (s, 1C, 5C, 22C, 24C), 105.80 (s, 8C, 12C), 102.86 (s, 15C), 102.45 (s, 10C), 69.81 (s, 26C), 66.89 (s, 17C), 55.45 (s, 7C). ESI-TOF, calcd for C32H26N2O6 ([M + Na]+) 557.1791, found 557.1805. Anal. calcd for C32H26N2O6: C, 71.90; H, 4.90; N, 5.24; O, 17.96. Found: C, 71.83; H, 4.98; N, 5.31; O, 18.03.
Cell lines and culture conditions
The cell lines used in this study were MCF-7, MDA-MB-231, H460, H1650, L02 and VERO. They were obtained from State Key Laboratory of Pharmaceutical Biotechnology, Nanjing University. MCF-7, MDA-MB-231, L02 and VERO cells were maintained in DMEM/high glucose supplemented with 10% FBS, L-glutamine, and penicillin/streptomycin and incubated at 37 °C in humidified atmosphere containing 5% CO2. H460 and H1650 cells were maintained in RPMI.1640 medium supplemented with 10% FBS and penicillin/streptomycin and incubated at 37 °C in humidified atmosphere containing 5% CO2.Animals feeding
Female nude mice (5–7 weeks old) were obtained from Model Animal Research Center of Nanjing University (Nanjing, China). Briefly, mice were fed with free access to pellet food and water in plastic cages at 21 ± 2 °C and kept on a 12 hours light–dark cycle. All animal procedures were performed in accordance with the Guidelines for Care and Use of Laboratory Animals of Nanjing University and approved by the Animal Ethics Committee of the Ministry of Science and Technology of China (2006). All efforts were made to minimize animal suffering and to reduce the number of animals used.EGFR kinase phosphorylation assay
The assays were carried out as described previously.3,28 Phosphorylation assays were performed in a final volume of 20 μL containing 8 mM MOPS (pH 7.0), 0.2 mM EDTA, 10 mM MnCl2, 200 μM substrate peptide, 0.25 mM DTT, 0.1 mg mL−1 BSA, 10 ng of wild-type EGFR kinase (catalogue no. 40187, BPS Bioscience), 10 mM magnesium acetate, 100 μM γ-[32P] ATP, and inhibitors or DMSO control (1.25% v/v). For IC50 curves with the wild-type enzyme, the following concentrations of the compounds were tested in triplicate: 150, 100, 50, 25, 15, 10, 7.5, 5, and 2.5 nM. Reactions were started by the addition of the magnesium acetate/ATP mixture. After 30 minutes incubation at 30 °C, 5 μL of each reaction was spotted on phosphocellulose P81 paper (Whatman). The P81 paper was then washed 5 times with 50 mM phosphoric acid for 15 minutes, dried, and exposed to a phosphorimager screen (Storm, GE Healthcare), which was scanned and densitometrically analyzed the next day. The sequence of the substrate peptide was derived from phospholipase C-γ1. The IC50 values were calculated using nonlinear regression with normalized dose response fit using Prism GraphPad software.Anti-proliferation assay
The anti-proliferation activity of the prepared compounds against four cancer cell lines, MCF-7, MDA-MB-231, H460, H1650 and two normal cell lines, L02 and VERO were evaluated as described elsewhere.29 Four kinds of cancer cell lines and two normal cell lines were plated in 96-well plates at the appropriate density (2.0 × 103 cells per well) with maintained in DMEM (or 1640 medium) with 10% FBS and cultivated for 12 hours at 37 °C, 5% CO2 atmosphere. All cells treated with different concentrations (0, 0.1, 1, 10 and 100 μM) of each compound with formononetin and lapatinib as positive references. After 24 hours, 20 μL PBS containing of MTT solution (2.5 mg mL−1) was added to each well and incubated for a further 4 hours. Then 150 μL of DMSO was added to each well to dissolve purple crystal. The plates were shaken to ensure the completion solubility for 10 minutes at room temperature. The values of absorbance were recorded on ELISA reader (ELx800, BioTek, USA) at a wavelength of 570 nm. All experiments were performed in three replicate wells for each concentration of all compounds including formononetin, and all assay experiments were carried out three times. The IC50 values defined as the concentrations that cause a 50% loss of cell viability were calculated by Origin 7.5 and exhibited in Table 3.Apoptosis analysis by flow cytometry
Cell apoptosis assay of compound 4v for MDA-MB-231 cell was carried out by Annexin V/PI assays and monitored by flow cytometry. Briefly, 5 × 103 cells per well were seeded in 6-well plates for 12 hours and then treated with 4v (0, 2, 4 and 8 μM) for 24 hours or treated with 4 μM 4v for 0, 12, 24 and 36 hours. Next, cells were collected and washed twice with PBS and then centrifuged to obtain sedimentation. 500 μL 1× binding buffer (10 mM HEPES, PH 7.4, 140 mM NaOH, 2.5 mM CaCl2) was added to each EP tube to make cells resuspended and stained with 5 μL of Annexin V-FITC and 5 μL of PI for 30 minutes at room temperature in the dark. Apoptotic cells were quantified using a BD Accuri C6 Flowjo Cytometer (BD, USA). Statistical analysis was done using Flowjo 7.6.1 software.Mitochondrial membrane potential staining
JC-1 is an ideal fluorescent probe which is widely used in the detection of mitochondrial membrane potential (ΔΨm). According to the requirements, MDA-MB-231 cells were cultured in 6-well plates (5 × 103 cells per well) for 12 hours at 37 °C, 5% CO2 atmosphere and then treated with compound 4v (0, 2 and 4 μM) for 12 hours and with positive control drug, CCCP in another well for 30 minutes at 37 °C. Cell in all wells were collected and rinsed with PBS. Then 500 μL JC-1 staining solution (5×) was added to each EP tube and incubated at 37 °C for 20 minutes. Finally, cells were washed twice with JC-1 staining solution (1×) and then resuspended to detect by BD Accuri C6 Flowjo Cytometer and Olympus confocal microscope, and data was analyzed using FV-10-ASW 1.7 viewer.Cell cycle arrested analysis by flow cytometry
After incubation in 6-well plates (5 × 103 cells per well), MDA-MB-231 cells were treated with compound 4v (0, 2, 4 and 8 μM) for 24 hours or treated with 4 μM 4v for 0, 24 and 36 hours. After that, cells were collected and washed twice with PBS and then fixed in 70% ethanol at 4 °C for at least 2 hours. Next, cells were resuspended in PBS with 0.1 mg mL−1 RNase A and 5 mg mL−1 propidium iodide (PI) in the dark condition. The measure was performed by BD Accuri C6 Flowjo Cytometer (BD, USA). The date of cells in the G1, S and G2/M phases of the cell cycle were counted using the Flowjo 7.6.1 software excluding cell debris.Wound-healing assay
MDA-MB-231 cells were seeded in a tissue culture 6-well plate at an initial density of 2 × 105 cells per cm2 overnight. A micropipette tip was used to create a wound in the monolayer by scraping after cells seeding and incubation for 24 hours. Identical cell-free space was observed by phase-contrast microscopy (NIKON, Japan) and digital images were taken by cells at 0, 24 and 48 hours. Subsequently, the NIH Image J image analysis software was used to outline the wound areas and analysis quantifies the reduced cell-free space, n = 3.Western blot analysis
Total proteins were extracted from MDA-MB-231 cells which were treated with compound 4v at different concentrations (0, 2, 4 and 8 μM). Cells obtained were lysed in RIPA lysis buffer containing protease inhibitor phenylmethanesulfonyl fluoride (PMSF, PMSF![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) RIPA = 1
RIPA = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100) on ice for 20–30 minutes. Protein content was measured using a Pierce BCA protein assay kit. Protein samples were separated by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred onto polyvinylidene difluoride (PVDF) membranes which were blocked with 5% non-fat-milk for 1 hour. All membranes were separated and incubated with primary antibodies including EGFR (rabbit polyclonal IgG, 1
100) on ice for 20–30 minutes. Protein content was measured using a Pierce BCA protein assay kit. Protein samples were separated by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred onto polyvinylidene difluoride (PVDF) membranes which were blocked with 5% non-fat-milk for 1 hour. All membranes were separated and incubated with primary antibodies including EGFR (rabbit polyclonal IgG, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 dilution), p-EGFR (rabbit polyclonal IgG, 1
1000 dilution), p-EGFR (rabbit polyclonal IgG, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 dilution), ERK (rabbit polyclonal IgG, 1
1000 dilution), ERK (rabbit polyclonal IgG, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 dilution), p-ERK (rabbit polyclonal IgG, 1
500 dilution), p-ERK (rabbit polyclonal IgG, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 dilution), Akt (rabbit polyclonal IgG, 1
500 dilution), Akt (rabbit polyclonal IgG, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 dilution), p-Akt (rabbit polyclonal IgG, 1
500 dilution), p-Akt (rabbit polyclonal IgG, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 dilution), GAPDH (rabbit polyclonal IgG, 1
500 dilution), GAPDH (rabbit polyclonal IgG, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 dilution), CDK4 (rabbit polyclonal IgG, 1
1000 dilution), CDK4 (rabbit polyclonal IgG, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 dilution), Cyclin A (rabbit polyclonal IgG, 1
500 dilution), Cyclin A (rabbit polyclonal IgG, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 dilution), Cyclin D1 (rabbit polyclonal IgG, 1
500 dilution), Cyclin D1 (rabbit polyclonal IgG, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 dilution), β-actin (rabbit polyclonal IgG, 1
500 dilution), β-actin (rabbit polyclonal IgG, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 dilution), Bax (rabbit polyclonal IgG, 1
500 dilution), Bax (rabbit polyclonal IgG, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 dilution), Bcl2 (rabbit polyclonal IgG, 1
500 dilution), Bcl2 (rabbit polyclonal IgG, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 dilution), cleaved caspase-3 (rabbit polyclonal IgG, 1
500 dilution), cleaved caspase-3 (rabbit polyclonal IgG, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 dilution), cleaved-PARP (mouse monoclonal IgG, 1
500 dilution), cleaved-PARP (mouse monoclonal IgG, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600 dilution), cytochrome c (rabbit polyclonal IgG, 1
600 dilution), cytochrome c (rabbit polyclonal IgG, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 dilution), β-catenin (rabbit polyclonal IgG, 1
500 dilution), β-catenin (rabbit polyclonal IgG, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 dilution) and Axin-2 (rabbit polyclonal IgG, 1
500 dilution) and Axin-2 (rabbit polyclonal IgG, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 dilution) at 4 °C shaking overnight. After the membrane was washed three times with tris-buffered saline buffer, HRP-conjugated secondary antibody (diluted 1
1000 dilution) at 4 °C shaking overnight. After the membrane was washed three times with tris-buffered saline buffer, HRP-conjugated secondary antibody (diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50
50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) was used at room temperature. Detection was performed by an enhanced chemiluminescent reagent (Thermo Fisher Scientific, USA) according to the manufacturer's instructions. Bands were then recorded by a digital camera (Tanon 5200, Tanon, China). Finally, the results were analyzed with Image J Software (National Institutes of Health, BetheSEMa, Maryland, USA), and all the targeted protein were normalized to GAPDH or β-actin.
000) was used at room temperature. Detection was performed by an enhanced chemiluminescent reagent (Thermo Fisher Scientific, USA) according to the manufacturer's instructions. Bands were then recorded by a digital camera (Tanon 5200, Tanon, China). Finally, the results were analyzed with Image J Software (National Institutes of Health, BetheSEMa, Maryland, USA), and all the targeted protein were normalized to GAPDH or β-actin.
Transplantation of MDA-MB-231 cells into nude mice
Cultured MDA-MB-231 cells were washed with and resuspended in ice-cold PBS. Portions of the suspension (6 × 106 cells in 0.1 mL) were injected into the right flanks of female nu/nu mice. Two weeks after the injection, the mice bearing tumors (an average size of 90 mm3) were distributed into 3 groups (n = 8 mice per group) according to tumor volumes. 4v (5 mg kg−1) and lapatinib (5 mg kg−1) were dissolved in olive oil and administered once every two days for 11 times by intraperitoneal injection. Vehicle (olive oil)-treated group was included as control. Body weight and tumor volumes were measured and recorded every two days. Long diameter (L) and short diameter (S) of a tumor were measured with a vernier caliper and the tumor volume was calculated using the following formula: L × (S)2/2. Twenty-one days after injection, the mice were killed and tumors were separated. Tumor weight was measured and tumor sections were fixed in paraformaldehyde. The rest of the sections were frozen in liquid nitrogen and stored at −80 °C.Data and statistical analysis
All experiments were repeated at least three times. Data are presented as mean ± S.E.M. and statistical comparisons between the treated and untreated groups were performed using one-way ANOVA using GraphPad PRISM5 (Graphpad Inc., La Jolla, USA). P-Values of <0.05 were considered statistically significant; P-values of <0.01 were considered statistically very significant.Conclusion
In this study, a series of novel formononetin derivatives targeting EGFR kinase were designed, synthesized and evaluated for their potential anti-proliferation activities against human breast and lung cancer cell lines. Among them, compound 4v (IC50 = 14.5 nM) showed potent EGFR inhibitory activity which was almost comparable to that of gefitinib (IC50 = 6.2 nM) and lapatinib (IC50 = 5.6 nM). For in vitro anti-proliferation activities, 4v exhibited potent inhibitory activity against MDA-MB-231 cell line (IC50 = 5.44 ± 1.28 μM) which was also comparable to lapatinib (IC50 = 2.48 ± 1.04 μM) but much better than gefitinib (IC50 = 12.5 ± 2.13 μM) and formononetin (IC50 = 26.6 ± 2.84 μM) itself. The structure–activity relationship (SAR) analyzed by molecular docking showing that 4v could bind well to the active site of EGFR as lapatinib. Flow cytometry analysis and mitochondrial membrane potential staining results demonstrated that 4v could effectively cause loss of mitochondrial membrane potential and then induce apoptosis on MDA-MB-231 cells, but had no significant influence on cell cycle distribution. Western blot results further confirmed that 4v inhibited MDA-MB-231 cell proliferation by blocking EGFR downstream Ras/Raf/MEK/ERK signaling pathway and induced MDA-MB-231 cell apoptosis by regulating EGFR/PI3K/Akt/Bad pathway. In addition, 4v could also effectively inhibit migration of MDA-MB-231 cells involving to Wnt signaling pathway. The preliminary in vivo anti-tumor results demonstrated the effectiveness of 4v in tumor chemotherapy in mice and indicated its possibility as a new EGFR inhibitor in curing MDA-MB-231 malignant tumor.Conflicts of interest
The authors declare no conflict of interest.Abbreviations
| CADD | Computer assist drug design |
| CCCP | Carbonylcyanidem-chlorophen-ylhydrazone |
| EGF | Epidermal growth factor |
| EGFR | Epidermal growth factor receptor |
| ESI-MS | Electrospray ionization-mass spectrometry |
| HER2 | Human epidermal growth factor receptor-2 |
| HEPES | 4-(2-Hydroxyethyl)-1-piperazineethanesulfonic acid |
| MAPK | Mitogen-activated protein kinase |
| MTT | 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide |
| NSCLC | Non-small-cell carcinoma |
| PARP | Poly-ADP ribose polymerase |
| PI | Propidium iodide |
| SAR | Structure–activity relationship |
| TLC | Thin layer chromatography |
| UV | Ultraviolet |
Acknowledgements
The authors are grateful to the Program for Changjiang Scholars and Innovative Research Team in University (IRT_14R27), the National Natural Science Foundation of China (31470384, 31171161, and 31670298), and the Fundamental Research Funds for the Central Universities (020814380002).Notes and references
- A. Olayioye, R. M. Neve and N. E. Hynes, EMBO J., 2000, 17, 3159–3167 CrossRef PubMed.
- I. Okamoto, FEBS J., 2010, 277, 309–315 CrossRef CAS PubMed.
- M. M. Hamed, S. S. Darwish, J. Herrmann, A. H. Abadi and M. Engel, J. Med. Chem., 2017, 60, 2853–2868 CrossRef CAS PubMed.
- Y. Yamanaka, H. Friess, M. Kobrin, M. Buchler, H. Beger and M. Korc, Anticancer Res., 1992, 13, 565–569 Search PubMed.
- J. M. Rae and M. E. Lippman, Breast Cancer Res. Treat., 2004, 83, 99–107 CrossRef CAS PubMed.
- R. E. Sobol, R. W. Astarita, C. Hofeditz, H. Masui, R. Fairshter, I. Royston and J. Mendelsohn, J. Natl. Cancer Inst., 1987, 79, 403–407 CAS.
- S. Balakrishnan, S. Mukherjee, S. Das, F. A. Bhat, P. Raja Singh, C. R. Patra and J. Arunakaran, Cell Biochem. Funct., 2017, 35, 217–231 CrossRef CAS PubMed.
- K. Takasawa, A. Takasawa, M. Osanai, T. Aoyama, Y. Ono, T. Kono, Y. Hirohashi, M. Murata and N. Sawada, Cancer Lett., 2017, 403, 66–73 CrossRef CAS PubMed.
- A. Sgambato, F. Casaluce, P. Maione, A. Rossi, E. Rossi, A. Napolitano, G. Palazzlol, M. A. Bareschion, C. Schettino, P. C. Sacco, F. Ciadiello and C. Gridelli, Curr. Med. Chem., 2012, 19, 3337–3352 CrossRef CAS PubMed.
- K. Manupati, N. R. Dhoke, T. Debnath, R. Yeeravalli, K. Goguloth, S. Saeidpour, U. C. De, S. Debnath and A. Das, FEBS J., 2017, 284, 1830–1854 CrossRef CAS PubMed.
- M. J. Duffy, P. M. Mcgowan and J. Crown, Int. J. Cancer, 2012, 131, 2471–2477 CrossRef CAS PubMed.
- D. Zhang, A. M. Tari, U. Akar, B. K. Arun, T. A. LaFortune, R. Nieves-Alicea, G. N. Hortobagyi and N. T. Ueno, Mol. Cancer Ther., 2010, 9, 3090–3099 CrossRef CAS PubMed.
- O. Lapčík, M. Hill, I. Černý, J. Lachman, N. Al-Maharik, H. Adlercreutz and R. Hampl, Plant Sci., 1999, 148, 111–119 CrossRef.
- L. Hutabarat, H. Greenfield and M. Mulholland, J. Chromatogr. A, 2000, 886, 55–63 CrossRef CAS PubMed.
- X. Y. Wu, H. Xu, Z. F. Wu, C. Chen, J. Y. Liu, G. N. Wu, X. Q. Yao, F. K. Liu, G. Li and L. Shen, Oncotarget, 2015, 6, 44563–44578 CrossRef PubMed.
- X. Yu, W. Wang and M. Yang, Food Chem., 2007, 104, 715–720 CrossRef CAS.
- T. Fukai, A. Marumo, K. Kaitou, T. Kanda, S. Terada and T. Nomura, Life Sci., 2002, 71, 1449–1463 CrossRef CAS PubMed.
- Y. Ungar, O. F. Osundahunsi and E. Shimoni, J. Agric. Food Chem., 2003, 51, 4394–4399 CrossRef CAS PubMed.
- S. Sato, J. Takeo, C. Aoyama and H. Kawahara, Bioorg. Med. Chem., 2007, 15, 3445–3449 CrossRef CAS PubMed.
- Z. N. Ji, W. Zhao, G. Liao, R. Choi, C. K. Lo, T. T. X. Dong and K. W. K. Tsim, Gynecol. Endocrinol., 2006, 22, 578–584 CrossRef CAS PubMed.
- S. Medjakovic and A. Jungbauer, J. Steroid Biochem., 2008, 108, 171–177 CrossRef CAS PubMed.
- Y. M. Jin, T. M. Xu, Y. H. Zhao, Y. C. Wang and M. H. Cui, Tumor Biol., 2014, 35, 2279–2284 CrossRef CAS PubMed.
- D. J. Fu, L. Zhang, J. Song, R. W. Mao, R. H. Zhao, Y. C. Liu, Y. H. Hou, J. H. Li, J. J. Yang, C. Y. Jin, P. Li, X. L. Zi, H. M. Liu, S. Y. Zhang and Y. B. Zhang, Eur. J. Med. Chem., 2016, 127, 87–99 CrossRef PubMed.
- Y. Q. Li, F. Yang, L. Wang, Z. Cao, T. J. Han, Z. A. Duan, Z. Li and W. J. Zhao, Eur. J. Med. Chem., 2016, 112, 196–208 CrossRef CAS PubMed.
- J. Ren, H. J. Xu, H. Cheng, W. Q. Xin, X. Chen and K. Hu, Eur. J. Med. Chem., 2012, 54, 175–187 CrossRef CAS PubMed.
- P. V. M. D. Alan, Cancer, 2005, 103, 2435–2446 CrossRef PubMed.
- X. J. Zhu, Y. Shi, J. Peng, C. S. Guo, N. N. Shan, P. Qin, X. B. Ji and M. Hou, Blood, 2009, 114, 5362–5367 CrossRef CAS PubMed.
- Y. G. Zheng, J. Su, C. Y. Gao, P. Jiang, L. An, Y. S. Xue, J. Gao and Y. Liu, Eur. J. Med. Chem., 2017, 130, 393–405 CrossRef CAS PubMed.
- H. Y. Lin, Z. K. Li, L. F. Bai, S. K. Baloch, F. Wang, H. Y. Qiu, X. Wang, J. L. Qi, R. W. Yang, X. M. Wang and Y. H. Yang, Biochem. Pharmacol., 2015, 96, 93–106 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra09825a |
| ‡ These two authors contribute equally to this article. |
| This journal is © The Royal Society of Chemistry 2017 |