Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Nanogel-antimiR-31 conjugates affect colon cancer cells behaviour†
C. Dispenza *ab,
M. A. Sabatinoa,
A. Ajovalasita,
L. A. Dittaa,
M. Ragusac,
M. Purrelloc,
V. Costad,
A. Conigliaroe and
R. Alessandroef
*ab,
M. A. Sabatinoa,
A. Ajovalasita,
L. A. Dittaa,
M. Ragusac,
M. Purrelloc,
V. Costad,
A. Conigliaroe and
R. Alessandroef
aDipartimento dell'Innovazione Industriale e Digitale (DIID), Università degli Studi di Palermo, Viale delle Scienze 6, 90128 Palermo, Italy. E-mail: clelia.dispenza@unipa.it
bIstituto di Biofisica (IBF), Consiglio Nazionale delle Ricerche, Via Ugo La Malfa 153, 90146 Palermo, Italy
cDipartimento di Scienze Biomediche e Biotecnologiche, Unità di BioMedicina Molecolare, Genomica e dei Sistemi Complessi (BMGS), Università di Catania, Via S Sofia 87, 95123 Catania, Italy
dPiattaforma Tecnologica per l'Ingegneria Tissutale, Teranostica ed Oncologia, Istituto Ortopedico Rizzoli, Palermo, Italy
eDipartimento di Biopatologia e Biotecnologie Mediche, Sezione di Biologia e Genetica, Università degli Studi di Palermo, Via Divisi 83, 90133, Palermo, Italy
fIstituto di Biomedicina ed Immunologia Molecolare (IBIM), Consiglio Nazionale delle Ricerche, Via Ugo La Malfa 153, 90146 Palermo, Italy
First published on 9th November 2017
Abstract
Soft and flexible nanogels, produced by electron beam (e-beam) irradiation of poly(N-vinyl pyrrolidone) and acrylic acid, were evaluated as delivery devices of the inhibitor of miR-31, a small RNA molecule with an important role in colorectal cancer (CRC) progression. The nanogel carriers developed possess both carboxyl and primary amino groups; the former were activated to react with the primary amino group present in the purposely-functionalised AntimiR-31. Very high conjugation reaction yields were attained, as well as a remarkable colloidal and storage stability of the conjugates. The ability of these nanoconstructs to be internalized by cells and the specific interaction of conjugated AntimiR with its biological target, without being detached from the nanogel, was demonstrated in vitro. These results are a strong encouragement to further proceed in the pre-clinical evaluation of the therapeutic effects of these formulations in CRC.
Introduction
MiRNAs are small single strand non-coding RNA molecules with a critical role in several biological functions. The known mechanism of action is through interaction with messenger RNA (mRNA), which results in its degradation/destabilization or translation inhibition.1–3 Binding of miRNAs to mRNAs occurs through base pairing of miRNA nucleotides 2–8 at the 5′ region (the seed) with the mRNA (most frequently, its 3′ UTR). This allows one miRNA to recognize a number of mRNAs. Furthermore, a specific mRNA may interact with many miRNAs: therefore, the regulatory effects of miRNAs can be exerted through a combination of concurring mechanisms. Recent studies have demonstrated that the alteration of miRNA expression may cause or be associated with the progression of several neoplastic and degenerative diseases. Therefore, therapies targeting miRNA dysregulation are very promising. The therapeutic strategy can be either to augment the miRNA activity by administering exogenous miRNA, or to silence its activity with miRNA inhibitors (AntimiRs) (i.e., genetically engineered single-strand oligodeoxynucleotides (ODNs) that can either bind to miRNA or to the mRNA sequence that binds miRNA).4Dysregulation of miRNAs expression in tumors has been intensively investigated in the last decade.5–7 Cancer cells with abnormal miRNA expression may undergo restless proliferation, evade growth suppression, resist cell death, activate invasion and metastasis, and induce angiogenesis. Since miRNAs may function as either oncogenes or tumor suppressors, their specific functions must be clearly identified. Once clarified, the advantage of using miRNA or miRNA-inhibitor (antimiR) therapeutics is based on its ability to concurrently target multiple pathway effectors involved in tumorigenesis. As with previous RNA-based therapies, the translation of these approaches to the clinic is subjected to the development of efficacious and affordable delivery strategies that can ensure spatial–temporal control of the therapeutic oligonucleotides at the target cells, overcoming biological barriers. Similarly to siRNAs, due to their polyanionic nature, miRNAs and antimiRs display reduced cellular uptake and poor intracellular trafficking, where sequestration in endosomes is one of the major limitations.8 Another obstacle is the limited stability of RNAs and RNA's inhibitors in body fluids, mainly caused by the ubiquitous presence of nucleases.9
This study is testing the delivery platform based on covalent conjugation of AntimiRs to soft and flexible nanogels. In particular, we focused our interest on miR-31 in colon rectal cancer model, because of its well-known role in colorectal cancer (CRC) progression. Indeed, it has been demonstrated that miR-31 expression is increased in CRC cells originated from metastatic foci and human primary CRC tissues with lymph node metastases.10 In addition, the high-level expression of miR-31 has been significantly associated with a poor prognosis and a more aggressive cancer phenotype in patients with CRC (p < 0.05). The stable over-expression of miR-31 in CRC cells has been also demonstrated to be sufficient to promote cell proliferation, invasion, and migration in vitro, and facilitated tumor growth and metastasis in vivo. Furthermore, miR-31 plays important roles in colon cancer development by targeting E2F2 (transcription factor 2) which, in turn, represses the expression of survivin and plays a crucial role in the control of cell cycle.10 In addition, recent studies observed that miR-31 is involved in tumor therapy resistance: in fact, the suppression of miR-31 increased the sensitivity of colon cancer cells to 5-FU (5-fluorouracil) and affected cell migration and invasion.3,6 These data imply that manipulation of miR-31 levels may have a useful clinical application in colon cancer therapies.
Nanogels (NGs) are an emerging class of nanocarriers characterized by a water-swollen nanosized three-dimensional network.11–13 The synthetized nanogels are characterized by a lightly crosslinked structure, based on poly (N-vinyl pyrrolidone), and multiple functionalities, including carboxylic and primary amino groups. Beside the very soft and conformable 3D structure, their manufacturing process via e-beam irradiation is particularly appealing, since it is performed in the typical irradiation conditions applied for sterilization purposes. Furthermore, the synthesis does not require catalysis, organic solvents or surfactants and yields to nanogels with hydrodynamic size variable from ca. 20 nm to ca. 130 nm and narrow size distribution (PDI < 0.3).13–18 The radiation chemistry of nanogel formation has been recently discussed in ref. 13.
Absence of toxicity, proliferative, immunogenic, and thrombogenic responses of the “empty” nanocarriers has been demonstrated “in vitro”.19 It must be said that the produced NGs and PVP are both not biodegradable. Recently, biodistribution and clearance studies on mice model have shown that these soft polymer nanoparticles can bypass glomerular filtration and be eliminated with the urines within 24 h (submitted manuscript).
The functional groups present in the nanogels have been used for the conjugation of several active components: therapeutic molecules such as doxorubicin20,21 or insulin;19 targeting moieties like folic acid21 or antibodies;22 and chelating groups that can entrap RMI-agents.23 These results prompted us to test the same type of nanocarriers for AntimiR-31 delivery.
To the best of our knowledge, there is only one documented example of AntimiR loaded onto a nanogel.24 As commonly done for other therapeutic genes, the AntimiR is loaded by electrostatic complexation, exploiting the mutual attraction between the cationic groups of the NGs matrix and the negatives charges of the AntimiR. Complexation causes the network to collapse into a dense nanostructure and to entrap the agent with high loading efficiency.24,25 The main limitation of this approach is that the biological effect is deemed to matrix degradation and AntimiR release. This can complicate the temporal control of its action. Besides, it is known that cationic nanoparticles preferentially absorb plasma proteins, which may cause RES activation and relatively fast elimination.26
To overcome this problem, we pursued a different strategy consisting in the conjugation of the AntimiR-31 using the terminal amino-group present in the ODN to form a stable amide bond with one of the carboxyl groups of the nanogel. Preliminary studies using a “non-functional” ODN have shown that the covalent bond to the nanogel does not prevent the ODN binding ability to the complementary sequence, probably due to the flexibility of the particle and its link to the ODN.27 This result was never demonstrated on a cellular model and provides motivation for the present investigation. Moreover, the ability of NG-AntimiR-31 to permeate cell membrane was investigated in the metastatic SW620 colon cancer cell line, which expresses high levels of miR-31.28 Confocal analysis was applied to test the ability of the NG-AntimiR 31 to permeate cell membrane. Moreover, the biological activity of NG-AntimiR 31 was investigated by the analysis of miR-31 amount and its target gene E2F, involved in cancer cell survival.29
Another important aspect investigated was the stability upon storage of the formulation (both as aqueous dispersion and as freeze-dried solid) and the possibility of reconstitution of the dispersion from a dry powder.
Experimental
Materials
AntimiR-31 (5′-AGCUAUGCCAGGAUCUUGCCU-3′), labeled with a fluorescent probe and modified to have a primary amino group, was purchased from Biolegio BV (5′-Label = fluorescein amidite (6-FAM); 3′-Label = amino-modified C6, MW 6689.22 Da). Dimethyl sulfoxide (DMSO), 1-ethyl-3-[3-dimethylaminopropyl] carbodiimide hydrochloride (EDC), N-hydroxysulfosuccinimide sodium salt (Sulfo-NHS), trishydroxymethylaminomethane hydrochloride (TRIS HCl) and ethylenediaminetetraacetic acid (EDTA) were all purchased from Sigma Aldrich and used as received.Synthesis of the nanogels
Two poly(N-vinyl pyrrolidone)-co-acrylic acid nanogel systems (NGs) were synthetized by e-beam irradiation starting from poly(N-vinyl pyrrolidone) (PVP k60 Aldrich, Mw = 0.4 MDa supplied as 45 wt% aqueous solution) and acrylic acid (AA, Aldrich). The rationale behind this approach and the details on the synthetic methodology can be found in ref. 13 and 17, respectively. Briefly, irradiation of poly(N-vinyl pryrrolidone)/acrylic acid solution at 12.5 μM of PVP and 0.9 mM of AA were irradiated at the Institute of Nuclear Chemistry and Technology (ICHTJ) in Warsaw (Poland), using the linear accelerator Electronika 10/10. Irradiation was carried out at an average beam current of 0.45 mA, pulse length of 4.5 μs and using two different pulse repetition rates of 400 Hz and 300 Hz for NG1 and NG2, respectively. The average molecular weight of the two variants produced was determined by Zimm plot analysis of static light scattering (SLS) data, the average numbers of carboxyl groups and primary amino groups per nanoparticle were estimated via colorimetric and fluorimetric methods, respectively.18,21 The results are reported in Table 1.| System | Mw, (MDa) | –COOH/NG (mol/mol) | –NH2/NG (mol/mol) |
|---|---|---|---|
| NG1 | 3.8 ± 0.20 | 50 | 38 |
| NG2 | 4.5 ± 0.12 | 67 | 83 |
Preparation of NG-AntimiR-31 conjugates
Grafting of AntimiR-31 on NGs was carried out following an EDC/Sulfo-NHS protocol.30Firstly, a given volume of NG aqueous dispersion was mixed with EDC and Sulfo-NHS aqueous solution (isotonic phosphate buffered saline pH 7.4) for 30 minutes, then given volumes of AntimiR-31 solution (100 μM) were added, corresponding to various AntimiR-31/NG carboxyl groups ratios (namely, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40, 1
40, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 and 1
20 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10).
10).
The reaction was conducted for further 24 hours under gentle stirring at room temperature. As a control, NGs were also subjected to the above-described EDC/Sulfo-NHS protocol with the exception that AntimiR was not added to the reactor.
AntimiR-conjugated NG systems were then purified through prolonged dialysis (7 days) against milli-Q water at room temperature, using 12 kDa cut-off membrane to remove unreacted reagents. Both synthesis and purification of NG-AntimiR-31 conjugates were conducted under dark conditions. The final concentration of NG-AntimiR-31 nanogels was 3 mg mL−1. The conjugation degree, defined as the average number of AntimiR-31 molecules per nanoparticle, was estimated by UV-vis absorption spectroscopy on the NG-AntimiR-31 variants at λMax Abs = 494 nm (Jasco V 670 spectrophotometer at room temperature, scan-speed 100 nm min−1, band width 1 nm). Samples of NG-AntimiR-31 were also centrifuged at 2500 rpm for 30 min from 100 kDa cut-off centrifuge-filters (Thermo Scientific IEC CL10 Centrifuge) and the eluted fractions were analyzed by UV-vis absorption spectroscopy to ascertain that no AntimiR was released. The three variants of AntimiR-31 conjugates corresponding to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40, 1
40, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 and 1
20 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 AntimiR-31/NG carboxyl groups ratios were named after NG1(or 2)-AntimiR-31-HC, NG1(or 2)-AntimiR-31-IC and NG1(or 2)-AntimiR-31-LC, respectively.
10 AntimiR-31/NG carboxyl groups ratios were named after NG1(or 2)-AntimiR-31-HC, NG1(or 2)-AntimiR-31-IC and NG1(or 2)-AntimiR-31-LC, respectively.
Physico-chemical characterizations of NG-AntimiR-31 conjugates
The hydrodynamic dimensions of AntimiR-conjugated NG variants were measured by dynamic light scattering (DLS) using a Brookhaven Instruments BI200-SM goniometer, equipped with a 50 mW He–Ne laser tuned at λ= 632.8 nm. Measurements were carried out at 25 ± 0.1 °C and at 90° scattering angle, after dilution and filtration with 0.45 μm pore size syringe filters to eliminate impurities introduced with the dialysis. Preliminary tests made on the non-filtrated, “as synthetized” nanogels showed that 0.45 μm as well as 0.22 μm filtration has no effect neither on the light scattering intensity nor on the particle size distribution. DLS data were analyzed according to the method of cumulants to determine the average hydrodynamic diameter and the width of the distribution that is a measure of polydispersity.15,31 ζ-Potential measurements were acquired at 25 °C using a Malvern Zetasizer Nano-ZS (Malvern instrument Ltd., Malvern, UK), equipped with a He–Ne laser at a power of 4.0 mW. For systems characterized by a monomodal ζ-potential distribution, the mean ζ-potential and the relative distribution width are reported. Conversely, for systems that show a more heterogeneous surface charge density (i.e., multimodal ζ-potential distribution), mean ζ-potential values (mean), relative widths (std. dev.) and relative abundance (area %) of the different modes are reported.Morphological analysis
Transmission electron microscopy was performed by JEOL, JEM- 2100 LaB6 on the purified NG1(2)-AntimiR-31 dispersions deposited onto carbon-coated cupper grids after being air-dried for 48 h.Stability studies
The eventual release of AntimiR-31 from NG after conjugation was investigated upon incubation at 37 °C for 24 h with (i) isotonic PBS buffer; and (ii) PBS buffer at pH 9, while gently stirring. After incubation, samples were filtered with 100 kDa cut-off centrifuge-filters (Thermo Scientific IEC CL10 Centrifuge) and the eluted fractions were analyzed by UV-vis absorption and emission spectroscopies.The colloidal stability of NG-AntimiR-31 conjugates in fetal bovine serum (FBS, 10% v/v), at 37 °C, was tested after 24 hours incubation. DLS measurements were then performed at 25 ± 0.1 °C and at 90° scattering angle, before and after dilution with isotonic PBS to the final concentration of 0.025 wt% to minimize FBS absorption at the laser wavelength, without filtration.
The colloidal stability upon storage at 4 °C both in water and in isotonic PBS buffer was investigated by performing DLS analyses. NG-AntimiR-31 conjugates dispersed in water were tested every two weeks for three months, while systems dispersed in isotonic PBS were tested after 20 days of storage.
Nanogel conjugates were also freeze-dried, stored in glass vials up to three months at room temperature, redispersed in PBS buffer, and tested for particle size distribution.
Biological evaluations
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per cm2 and transfected with 15 pmol mL−1 of miR-31 inhibitor (Biolegio BV) or scrambled negative control (cat. number: 4464058, Life Technologies), 24 hours after transfection the medium was collected and the cells processed for following assays.
000 cells per cm2 and transfected with 15 pmol mL−1 of miR-31 inhibitor (Biolegio BV) or scrambled negative control (cat. number: 4464058, Life Technologies), 24 hours after transfection the medium was collected and the cells processed for following assays.| Gene | Primer forward | Primer reverse |
|---|---|---|
| E2F | CGCCATCCAGGAAAAGGTGT | ACATCGATCGGGCCTTGTTT |
| RHO A | GAAAACCGGTGAATCTGCGC | AGAACACATCTGTTTGCGGA |
| B-ACTIN | ATCAAGATCATTGCTCCTCCTGA | CTGCTTGCTGATCCACATCTG |
Results and discussion
Synthesis, physico-chemical and morphological characterization of AntimiR-31-conjugated nanogels
Poly(N-vinyl pyrrolidone)-co-acrylic acid nanogel systems were selected as potential nanocarriers of AntimiR-31. These nanogels possess both carboxyl groups, mainly due to the grafted acrylic acid, and amino groups deriving from radiation- induced double-cleavage of the pyrrolidone rings.18 Two nanogel variants with different average hydrodynamic size and ratio between carboxyl groups and primary amino groups were synthetized varying the pulse frequency of the e-beam accelerator. NG1 system consists of smaller nanogels (average hydrodynamic size of about 40 nm) with a prevalence of carboxyl groups that grant their surface a slightly negative net electric charge density. NG2 has larger average hydrodynamic size, ca. 60 nm, a prevalence of amino groups and a broader distribution of surface electric charge density, with a larger fraction of nanoparticles that result globally non-charged.Being the amino groups present at the backbone of the polymeric network and the carboxyl groups on short side branches, we decided to bind AntimiR-31 to the nanogels through the carboxyl groups of the nanogel. For the purpose, the ODN was synthetised to have a primary amino group at the 3′ terminus (Fig. 1).
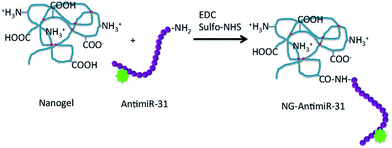 | ||
| Fig. 1 Scheme of the conjugation reaction between nanogel and AntimiR. The red circles represent crosslinking points on PVP chain. | ||
Since, in principle, the carboxyl groups of the nanogel could react with the amino groups of the same nanogel (intra-molecular crosslinking) or of another nanogel (inter-molecular crosslinking), a preliminary experiment was carried out charging the reactor with the nanogel and with EDC and sulfo-NHS used to activate the carboxyl groups, without addition of ODN.
After purification by dialysis, the systems were subjected to DLS analysis to check if aggregation or contraction of particle size had occurred. The particle size distribution of both NG1 and NG2 nanogels after reaction resulted substantially unmodified. The lack of mutual reactivity between primary amino groups and carboxyl groups of nanogels is likely due to their sterical hindrance.
For the conjugation of the ODN to the nanogel, the molar ratio between AntimiR-31 and carboxyl groups was kept lower than the stoichiometric ratio in order to have only a few ODNs per nanoparticle, leaving most of the carboxyl groups unreacted, thus able to contribute to colloidal stability of the system and/or to be used for further conjugation reactions. The conjugation reactions were carried out starting from the same weight fraction of nanogels in the reactor feed, varying the concentration of AntimiR-31. The reaction yield is higher for NG1 than for NG2 and augments with the increase of ODN concentration in the reactor, as shown in Table 3. Despite the slightly higher number of carboxyl groups per nanogel of the NG2 system (see Table 1), they are less densely distributed since the particles are bigger; this results in a lower conjugation efficiency.
| Sample | AntimiR/COOH | NG1 | NG2 | ||
|---|---|---|---|---|---|
| Average AntimiR-31/NG | Reaction yield% | Average AntimiR-31/NG | Reaction yield% | ||
| NG-AntimiR-LC | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40 40 |
0.7 | 79.7 | 0.5 | 45.0 |
| NG-AntimiR-IC | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 20 |
1.6 | 80.1 | 1.0 | 51.5 |
| NG-AntimiR-HC | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 10 |
3.5 | 87.5 | 2.5 | 62.5 |
The estimation of the degree of conjugation was performed by analyzing the various NG-AntimiR-31 conjugates by UV-vis absorption spectroscopy. The molar ratio of ODN to nanogel increases at the increase of the ODN in the reactor, from one AntimiR-31 every two nanogels for the NG-AntimiR-LC, to three or four ODN per nanogel for the NG-AntimiR-HC. The reactions conditions that did not ensure grafting of minimum one ODN per nanogel (LC-systems) were not further considered.
The four AntimiR-NGs systems with degree of conjugation higher than one were further characterized for their hydrodynamic size and surface charge density. Data are reported in Table 4. No aggregation can be appreciated; only a slight increase in size and polydispersity, probably due to the increase of the branching degree after conjugation. The surface charge density of the nanogels is only very little modified upon conjugation. NG1 systems, that were initially anionic and fairly monodisperse in terms of surface charge density, kept this feature; likewise, the NG2 systems that were instead more polydisperse.
| Sample | Hydrodynamic diameter, nm | ζ-Potential |
|---|---|---|
| Mean (mV); area (%); St. Dev. (mV) | ||
| NG1 | 40.0 ± 15.1 | −25.2; 100; 5.7 |
| NG1-AntimiR-IC | 58.4 ± 31.7 | −22.2; 100; 7.9 |
| NG1-AntimiR-HC | 47.1 ± 19.2 | −38.1; 100; 10.0 |
| Peak 1 | Peak 2 | Peak 3 | ||
|---|---|---|---|---|
| NG2 | 60.1 ± 16.0 | −50; 2.2; 5.59 | −3.56; 81.7; 13.2 | +30; 14.3; 8.16 |
| NG2-AntimiR-IC | 63.2 ± 25.1 | −73.3; 2.5; 3.90 | −11.1; 84.5; 13.6 | +23.3; 12.9; 6.47 |
| NG2-AntimiR-HC | 63.6 ± 25.3 | −40; 19.0; 7.53 | −3.89; 71.7; 12.0 | +27.6; 9.3; 6.46 |
The morphological analysis carried out by TEM microscopy did not reveal significant differences among the various samples in good agreement with DLS data. Single particles as well as loose aggregates were visible on the hollow carbon-coated copper stab, the aggregation being the result of the receding water droplet during the drying process. A representative TEM micrograph is shown in Fig. 2, which refers to the NG1-AntimiR-HC system.
Stability studies
To test the strength of the link of AntimiR to nanogel and rule out that the bond between the nanogel and the ODN was purely electrostatic in its nature, samples of the NG1-AntimiR-HC system were incubated at 37 °C for 24 h with isotonic PBS and with PBS at pH 9. After incubation, samples were centrifuge filtered and the eluted solutions were analyzed by both UV-vis absorption and emission spectroscopies to check the presence of signals from the FAM emissive label of the AntimiR. No signals were detected (see Fig. S1 of ESI†). Therefore, neither the relatively high salt concentration of isotonic PBS nor the alkaline pH, that would disrupt eventual electrostatic complexes formed, triggered the release of the ODN, thus supporting the covalent attachment of the AntimiR to the nanogel through an amide bond.The colloidal stability of NG1-AntimiR-HC upon incubation for 24 h at 37 °C in FBS (10% v/v) was investigated. The hydrodynamic dimensions of the system did not change, suggesting that no irreversible aggregation phenomena occur.
The colloidal stability of the formulations upon storage at 4 °C was investigated, in water and isotonic PBS buffer, measuring the hydrodynamic diameters as function of the time. Data are reported in Table 5. An increase in average hydrodynamic size occurs when the nature of the dispersing medium is changed from water to isotonic buffer for the AntimiR-conjugated systems only. No aggregation occurs upon storage at 4 °C for three months in each of the two media. This quite remarkable colloidal stability is due to the presence of ionizable groups, to the small density mismatch between the water-swollen nanogel and dispersing/swelling medium, and to the presence of loose chains stretching out in the medium.
| System – dispersing medium | NG1-AntimiR-31 | NG2-AntimiR-31 | ||
|---|---|---|---|---|
| t = 0 | t = 3 month | t = 0 | t = 3 month | |
| NG-AntimiR-IC – water | 58.0 ± 31.7 | 61.0 ± 31.2 | 63.2 ± 25.1 | 63.4 ± 26.5 |
| NG-AntimiR-HC – water | 47.1 ± 19.2 | 49.4 ± 23.1 | 63.6 ± 25.3 | 77.0 ± 25.3 |
| t = 0 | t = 20 days | t = 0 | t = 20 days | |
|---|---|---|---|---|
| NG-AntimiR-IC – PBS | 82.0 ± 21.1 | 83.2 ± 28.1 | 79.7 ± 24.8 | 76.2 ± 23.6 |
| NG-AntimiR-HC – PBS | 72.2 ± 22.1 | 71.4 ± 25.3 | 79.8 ± 24.8 | 82.6 ± 26.6 |
The slight increase of average particle size in the presence of saline solutions of the AntimiR conjugates could be due to the displacement of “internal” interactions between the strongly anionic AntimiR and the cationic groups present in the nanogels by the small ions of the saline solution. This effect resembles the anti-polyelectrolyte effect of zwitterion polymers.32
Table 6 reports data from DLS analysis of nanogel-conjugates, soon after freeze-drying, after one month and after three months storage at room temperature. Data indicate that no aggregation occurs if the dispersion is reconstituted immediately after freeze-drying or after one-month storage, while some degree of aggregation occurs after three-months, probably because the storage was not carried out in completely moisture-free conditions.
| Sample | Before freeze-drying | Soon after freeze-drying | 1 month after freeze-drying | 3 month after freeze-drying |
|---|---|---|---|---|
| NG1 – water | 40.0 ± 15.1 | 40.4 ± 18.1 | 42.2 ± 20.3 | 63.3 ± 26.2 |
| NG1-AntimiR-IC – water | 58.0 ± 31.7 | 73.0 ± 23.5 | 80.1 ± 28.2 | 123.2 ± 34.3 |
Biological effects of NG-AntimiR-31
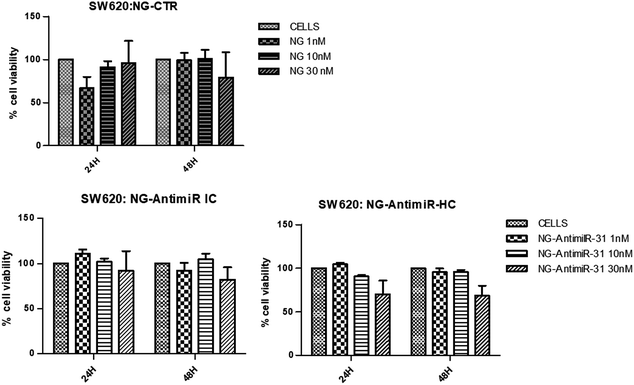
The aim of the biological evaluations performed is a preliminary investigation of in vitro effects of NG-AntimiR-31 on colon cancer cells, in the prospect of developing a hypothetical new therapeutic approach for colon cancer progression. In particular, we studied the cytotoxicity of AntimiR-31 conjugated to NGs and its ability to modulate the expression of miR-31 target genes on a suitable colon cancer cell line model.In consideration of the higher yields of conjugation reaction of ODN to NG1 compared to NG2, conjugates of NG1 were selected for the biological evaluation. To investigate their biocompatibility, we performed a cell viability assay on SW620 cells treated with different concentrations (1 nM, 10 nM and 30 nM) of bare NGs or conjugated with AntimiR-31 (both IC and HC). As shown in Fig. 3, SW cells viability was evaluated 24 and 48 hours after NGs treatments. While NG-AntimiR-31-IC treatments did not reveal any significant toxicity, the higher concentration of NG-AntimiR-31-HC reduced cell viability in SW620 cells.
No toxicity was revealed by transfecting SW620 cells with 10 nM or 30 nM of AntimiR-31 (Fig. S2 of ESI†). Further experiments, aimed to investigate the biological properties of NG-AntimiR-31, were then performed using NG-AntimiR-IC.
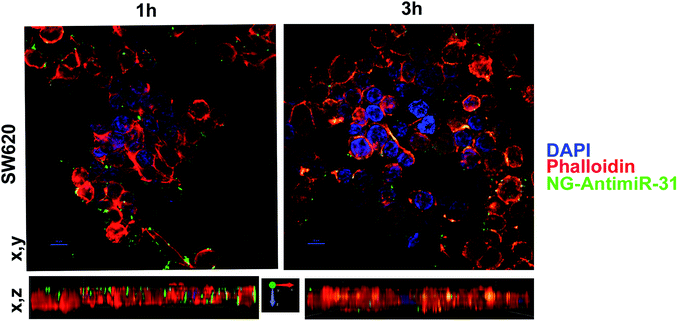
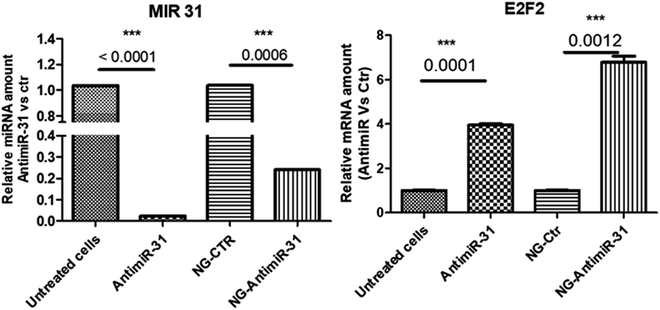
Uptake assay followed by confocal analysis allowed to demonstrate the ability of NG-AntimiR-31 to permeate SW cells. As shown in Fig. 4, NG-AntimiR-31, mainly localized on cell surface one hour after incubation (green dots), penetrated SW-cells after three hours (orange dots). Subsequently, we explored the biological activity of the transported AntimiR-31. As shown in Fig. 5, the treatment of cell line with 30 nM of NG-AntimiR-IC suppressed miR-31 levels with efficiency comparable to that obtained by AntimiR-31 transfection with Attractene. This data confirms that the conjugation of the oligonucleotide to our nanogels did not affect the annealing between exogenous AntimiR-31 and endogenous miR-31.
To further evaluate the biological functions of NG-AntimiR-31, we analyzed the transcriptional levels of miR-31 target (obtained by miRTarBase) E2F.33 As shown in Fig. 5 the effects induced by NG-AntimiR-31 treatment are in line with those obtained after transfection of AntimiR-31. These data demonstrated that NG conjugation did not affect AntimiR-31 activity.
Conclusions
Two poly(N-vinyl pyrrolidone)-based nanogel systems, with different average hydrodynamic size and concentration of carboxyl and primary amino groups, were synthetized by e-beam irradiation and covalently conjugated to AntimiR-31. Our interest in AntimiR-31 comes from the many evidences about the role of miR-31 in cancer progression and tumor therapy resistance. The NG-AntimiR-31 conjugates showed no evidence of AntimiR-31 release over time, at body temperature, both in physiologic conditions and alkaline solution, and no aggregation in blood serum. Furthermore, they showed good colloidal stability upon storage at 4 °C for 3 months and full redispersibility soon after freeze-drying and also after one month storage at 4 °C. The biological effects of NG-AntimiR-31 conjugates were investigated in a syngenic in vitro model. Cell viability evaluation in SW620 cells, treated with increasing concentrations of NG-AntimiR conjugates (up to 30 nM), showed that no concentration-dependent toxicity is induced 24 h and 48 h after the treatment. Internalization into colon cancer cells of the designed NG-AntimiR nanoconstruct was appreciable already after 3 hours of treatment. Moreover, NG-AntimiR-31 conjugates showed ability to vehicle AntimiR-31 in SW620 cells regulating the expression of miR-31 and its target genes, thus demonstrating that the covalent link of the ODN to these soft nanogels does not affect the AntimiR-31 functionality in vitro. Future work will focus on understanding the in vivo performance of NG-AntimiR-31 formulations, with the prospect of overcoming some of the limits of the current pharmacological therapies for aggressive colon cancer.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was supported by HIPPOCRATES - PON02_00355_2964193 and IAEA CRP F22064 Nanosized delivery systems for radiopharmaceuticals. The Authors wish to thank Prof. Grazyna Przybytniak and the Centre for Radiation Research and Technology, Institute of Nuclear Chemistry and Technology, Warsaw, Poland for e-beam irradiation and Prof. Gaetano Giammona, STEBICEF, Università degli Studi di Palermo, for Z-potential measurements. TEM experimental data were provided by Centro Grandi Apparecchiature e UniNetLab e Universita' di Palermo, funded by P. O. R. Sicilia 2000 e 2006, Misura 3.15, Quota Regionale. C. Dispenza designed the nanocarrier and the experiments, analysed the data and wrote the manuscript, which was reviewed by all authors. M. A. Sabatino and L. A. Ditta synthesized and characterised the non targeted NGs. M. A. Sabatino developed the conjugation protocols. A. Ajovalasit synthetized and characterised the AntimiR-NG conjugates. M. Ragusa and Purrello chose the target (miR-31) and designed the targeting agent. V. Costa, A. Conigliaro and R. Alessandro helped with the experimental design, confocal and data analysis and manuscript writing.References
- M. Wery, M. Kwapisz and A. Morillon, Wiley Interdiscip. Rev.: Syst. Biol. Med., 2011, 3(6), 728–738 CrossRef CAS PubMed.
- B. Yan, Z. H. Wang and J. T. Guo, Genomics, 2012, 99(2), 76–80 CrossRef CAS PubMed.
- M. D. Jansson and A. H. Lund, Mol. Oncol., 2012, 6(6), 590–610 CrossRef CAS PubMed.
- J. Krutzfeldt, N. Rajewsky, R. Braich, K. G. Rajeev, T. Tuschl, M. Manoharan and M. Stoffel, Nature, 2005, 438(7068), 685–689 CrossRef PubMed.
- G. A. Calin, C. D. Dumitru, M. Shimizu, R. Bichi, S. Zupo, E. Noch, H. Aldler, S. Rattan, M. Keating, K. Rai, L. Rassenti, T. Kipps, M. Negrini, F. Bullrich and C. M. Croce, Proc. Natl. Acad. Sci. U. S. A., 2002, 99(24), 15524–15529 CrossRef CAS PubMed.
- C. M. Croce, Nat. Rev. Genet., 2009, 10(10), 704–714 CrossRef CAS PubMed.
- J. Lu, G. Getz, E. A. Miska, E. Alvarez-Saavedra, J. Lamb, D. Peck, A. Sweet-Cordero, B. L. Ebert, R. H. Mak, A. A. Ferrando, J. R. Downing, T. Jacks, H. R. Horvitz and T. R. Golub, Nature, 2005, 435(7043), 834–838 CrossRef CAS PubMed.
- G. Tiram, A. Scomparin, P. Ofek and R. Satchi-Fainaro, J. Biomed. Nanotechnol., 2014, 10(1), 50–66 CrossRef CAS PubMed.
- Y. Zhang, Z. Wang and R. A. Gemeinhart, J. Controlled Release, 2013, 172(3), 962–974 CrossRef CAS PubMed.
- T. Mitamura, H. Watari, L. Wang, H. Kanno, M. K. Hassan, M. Miyazaki, Y. Katoh, T. Kimura, M. Tanino, H. Nishihara, S. Tanaka and N. Sakuragi, Oncogenesis, 2013, 25(2), e40 CrossRef PubMed.
- A. V. Kabanov and S. V. Vinogradov, Angew. Chem., Int. Ed. Engl., 2009, 48(30), 5418–5429 CrossRef CAS PubMed.
- R. T. Chacko, J. Ventura, J. Zhuang and S. Thayumanavan, Adv. Drug Delivery Rev., 2012, 64(9), 836–851 CrossRef CAS PubMed.
- C. Dispenza, G. Spadaro and M. Jonsson, Top. Curr. Chem., 2016, 374(5), 69 CrossRef CAS PubMed.
- C. Dispenza, N. Grimaldi, M. A. Sabatino, I. L. Soroka and M. Jonsson, J. Nanosci. Nanotechnol., 2015, 15(5), 3445–3467 CrossRef CAS PubMed.
- C. Dispenza, M. A. Sabatino, N. Grimaldi, D. Bulone, M. L. Bondì, M. P. Casaletto, S. Rigogliuso, G. Adamo and G. Ghersi, Biomacromolecules, 2012, 13, 1805–1817 CrossRef CAS PubMed.
- M. A. Sabatino, D. Bulone, M. Veres, A. Spinella, G. Spadaro and C. Dispenza, Polymer, 2013, 53, 54–64 CrossRef.
- N. Grimaldi, M. A. Sabatino, G. Przybytniak, I. Kaluska, M. L. Bondì, D. Bulone, S. Alessi, G. Spadaro and C. Dispenza, Radiat. Phys. Chem., 2014, 94, 76–79 CrossRef CAS.
- C. Dispenza, M. A. Sabatino, N. Grimaldi, M. R. Mangione, M. Walo, E. Murugan and M. Jonsson, RSC Adv., 2016, 6(4), 2582–2591 RSC.
- P. Picone, L. A. Ditta, M. A. Sabatino, V. Militello, P. L. San Biagio, M. L. Di Giacinto, L. Cristaldi, D. Nuzzo, C. Dispenza, D. Giacomazza and M. Di Carlo, Biomaterials, 2016, 80, 179–194 CrossRef CAS PubMed.
- G. Adamo, N. Grimaldi, S. Campora, M. A. Sabatino, C. Dispenza and G. Ghersi, Chem. Eng. Trans., 2014, 38, 457–462 Search PubMed.
- G. Adamo, N. Grimaldi, S. Campora, D. Bulone, M. L. Bondi, M. Al-Sheikhly, M. A. Sabatino, C. Dispenza and G. Ghersi, Molecules, 2016, 21, 1594 CrossRef PubMed.
- G. Adamo, N. Grimaldi, M. A. Sabatino, M. Walo, C. Dispenza and G. Ghersi, Biol. Chem., 2017, 398, 277–287 CrossRef CAS PubMed.
- S. Pardo, C. Dispenza, M. A. Sabatino, N. Grimaldi, A. Ajovalasit, L. Ditta and M. Jonsson, ECR, 2014 DOI:10.1594/ecr2014/C-0556.
- H. Zou, Z. Wang and M. Feng, J. Controlled Release, 2015, 214, 121–133 CrossRef CAS PubMed.
- J. Yu, H. Zhao, L. Ye, H. Yang, S. Ku, N. Yang and N. Xiao, J. Mater. Chem., 2009, 19, 1265–1270 RSC.
- F. Alexis, E. Pridgen, L. K. Molnar and O. C. Farokhzad, Mol. Pharm., 2008, 5, 505–515 CrossRef CAS PubMed.
- C. Dispenza, G. Adamo, N. Grimaldi, D. Bulone, M. L. Bondì, S. Rigogliuso and G. Ghersi, J. Appl. Polym. Sci., 2014, 131, 239774–239780 CrossRef.
- M. H. Yang, J. Yu, N. Chen, X. Y. Wang, X. Y. Liu, S. Wang and Y. Q. Ding, PLoS ONE, 2013, 8(12), e85353, DOI:10.1371/journal.pone.0085353.
- A. Lo Dico, V. Costa, C. Martelli, C. Diceglie, F. Rajata, A. Rizzo, C. Mancone, M. Tripodi, L. Ottobrini, R. Alessandro and A. Conigliaro, Theranostics, 2016, 6, 1105–1118 CrossRef PubMed.
- G. T. Hermanson, Bioconjugate Techniques, Academic Press, New York, 2nd edn., 2008 Search PubMed.
- D. E. Koppel, J. Chem. Phys., 1972, 57, 4814–4820 CrossRef CAS.
- F. Wang, J. F. Yang and J. Zhao, Polym. Int., 2015, 64(8), 999–1005 CrossRef CAS.
- T. Chen, L. Q. Yao, Q. Shi, Z. Ren, L. C. Ye, J. M. Xu, P. H. Zhou and Y. S. Zhong, Cancer Biol. Ther., 2014, 15(5), 516–523 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra09797b |
| This journal is © The Royal Society of Chemistry 2017 |