Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
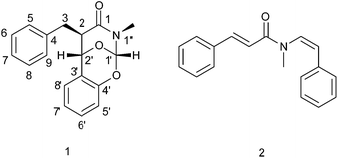
Clausoxamine, an alkaloid possessing a 1,3-oxazine-4-one ring from the seeds of Clausena lansium and the anti-obesity effect of lansiumamide B†
Li Huang‡
a,
Dan Li‡a,
Yun-Shao Xua,
Zhe-Ling Fenga,
Fan-Chen Menga,
Qing-Wen Zhanga,
Li-She Ganb and
Li-Gen Lin *a
*a
aState Key Laboratory of Quality Research in Chinese Medicine, Institute of Chinese Medical Sciences, University of Macau, Macao 999078, People's Republic of China. E-mail: ligenl@umac.mo
bCollege of Pharmaceutical Sciences, Zhejiang University, 866 Yuhangtang Road, Hangzhou 310058, People's Republic of China
First published on 4th October 2017
Abstract
A novel alkaloid, clausoxamine (1), which possesses an unprecedented 1,3-oxazinan-4-one ring, was isolated from the seeds of Clausena lansium, along with six known analogues. The structure and absolute configuration of 1 were established on the basis of detailed NMR and HRESIMS analysis, as well as comparison of its experimental and TD-DFT calculated ECD spectra. A proposed biosynthetic pathway was predicted for compound 1. The major alkaloid, lansiumamide B, was found to reduce fat mass gain and improve insulin sensitivity on high fat diet treated mice, which might be a potential anti-obesity and insulin sensitizing agent.
Introduction
Obesity is approaching epidemic proportions worldwide, resulting in higher risks of insulin resistance, hyperlipidemia, and cardiovascular diseases.1,2 Over nutrition often leads to expansion of visceral adipose tissue, elevated triglycerides and low-density lipoprotein (LDL), and decreased high-density lipoprotein (HDL), which eventually causes type 2 diabetes. The US Food and Drug Administration approved anti-obesity drugs, either appetite suppressors or pancreatic lipase inhibitors, always have unpleasant adverse effects.3 Alternatively, natural products have attracted considerable attention in the discovery and development of new anti-obesity drugs due to their safety and efficacy.Clausena lansium Skeels (Rutaceae) is widely cultivated in southern China and Southeast Asia, and the edible fruit, called wampee, is highly aromatic and pleasantly sweet. In traditional medicines, the leaves and roots of C. lansium have been used for treatment of cough, asthma, dermatological disease, and gastro-intestinal diseases. The seeds have been used to treat acute and chronic gastro-intestinal inflammation and ulcers.4 Extensive phytochemical investigations on different parts of this plant have revealed the presences of amide alkaloids, carbazole alkaloids, and coumarins with anti-inflammation, anti-oxidation, and anti-cancer properties.5–9 In our research for new anti-obesity natural products, one new amide alkaloids with an unprecedented skeleton was identified from the seeds of C. lansium, together with six known analogues. Compound 1 possessed an unusual 1,3-oxazine-4-one ring. Herein, the isolation, structure characterization, and proposed biosynthetic pathways of the new compound, as well as anti-obesity effect of lansiumamide B on high-fat diet treated mice, are presented.
Results and discussion
The EtOAc-soluble fraction of a 95% ethanol extract of C. lansium was purified by column chromatography over silica gel, Sephadex LH-20 and preparative HPLC to afford seven amide alkaloids, including one new compound (1, Fig. 1) and six known analogues.Compound 1 was isolated as a white amorphous powder, and its molecular formula was designated as C18H17NO3 according to the protonated ion peak at m/z 296.1261 [M + H]+ (calcd for C18H18NO3, 296.1287) in its HRESIMS, requiring eleven degrees of unsaturation. The IR spectrum exhibited absorptions for an amide group (1658 cm−1) and a phenyl group (1491 and 1459 cm−1). The 13C NMR and DEPT spectra (Table 1) revealed the presence of 18 carbon resonances encompassing one methyl group, one methylene group, 12 methine groups (including three sp3 carbons and nine aryl carbons), and 4 quaternary carbons (including three aryl carbons and one carbonyl group). The 1H NMR data of 1 (Table 1), with the aid of the HSQC data, displayed signals ascribed to one ortho-disubstituted benzene moiety [δH 6.91 (dd, J = 8.2, 1.1 Hz)/δC 117.2; δH 7.24 (m)/δC 130.0; δH 6.98 (td, J = 7.5, 1.1 Hz)/δC 122.5; δH 6.88 (dd, J = 7.7, 1.6 Hz)/δC 126.4], one monosubstituted benzene moiety [δH 7.34–7.39 (4H, m)/δC 129.5 (2C), 129.1 (2C); δH 7.27 (ddd, J = 8.4, 6.9, 1.7 Hz)/δC 127.1], three sp3 methines [δH 6.55 (s)/δC 98.2; δH 4.82 (s)/δC 70.1; δH 2.81 (m)/δC 50.8], one sp3 methylene [δH 3.06 (dd, J = 13.8, 5.5 Hz), 2.99 (dd, J = 13.8, 10.2 Hz)/δC 35.9], and one methyl [δH 2.82 (3H, s)/δC 30.2]. The aforementioned functionalities accounted for nine degrees of unsaturation, the remaining ones required for two additional rings in the structure of 1.
| Position | δC | δH |
|---|---|---|
| 1 | 168.1 C | |
| 2 | 50.8 CH | 2.81, m |
| 3 | 35.9 CH2 | 3.06, dd, 13.8, 5.5; 2.99, dd, 13.8, 10.2 |
| 4 | 138.8 C | |
| 5 | 129.5 CH | 7.34–7.39, 1H, m |
| 6 | 129.1 CH | 7.34–7.39, 1H, m |
| 7 | 127.1 CH | 7.27, ddd, 8.4, 6.9, 1.7 |
| 8 | 129.1 CH | 7.34–7.39, 1H, m |
| 9 | 129.5 CH | 7.34–7.39, 1H, m |
| 1′ | 98.2 CH | 6.55, s |
| 2′ | 70.1 CH | 4.82, s |
| 3′ | 122.8 C | |
| 4′ | 147.5 C | |
| 5′ | 117.2 CH | 6.91, dd, 8.2, 1.1 |
| 6′ | 130.0 CH | 7.24, m |
| 7′ | 122.5 CH | 6.98, td, 7.5, 1.1 |
| 8′ | 126.4 CH | 6.88, dd, 7.7, 1.6 |
| N–CH3 | 30.2 CH3 | 2.82, s |
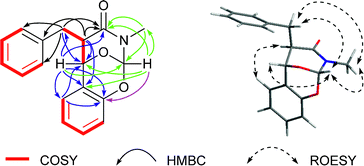
Next, 2D NMR experiments were carried out to construct the planar structure of 1. The 1H–1H COSY spectrum suggested the presence of three spin systems of H2-3/H-2/H-2′, H-5/H-6/H-7/H-8/H-9, and H-5′/H-6′/H-7′/H-8′ (Fig. 2). The HMBC correlations between H-2 and C-1, C-3 and C-4; between the methylene (H2-3) and C-1, C-2, C-4 and C-5/9; between H-5/9 and C-3, revealed the presence of a 3-phenylpropanamide moiety in 1 (Fig. 2), which is common in amide alkaloids from the Clausena species. In addition, the HMBC correlations from H-2 to C-3′, from H2-3 to C-2′, from the oxygenated proton (H-2′) to C-1, C-2, C-3, C-3′, C-4′ and C-8′, and from H-8′ to C-2′ and C-4′ (Fig. 2), combined with the corresponding 1H–1H COSY correlations, suggested a benzyl group is linked to the 3-phenylpropanamide at C-2. Further HMBC cross-peaks from the low-field proton (H-1′) to C-1, C-2′, C-3′ and the methyl, from H-2′ to C-1′, and the methyl to C-1 and C-1′ (Fig. 2), constructed a 1,3-oxazinan-4-one ring, possessing a methyl group on the nitrogen. Finally, the HMBC correlation from H-1′ to C-4′, in combination with the molecular formula, indicated the presence of an oxygen bridge between C-1′ and C-4′ to form another ring. Thus, the gross structure of 1 was constructed as shown in Fig. 1.
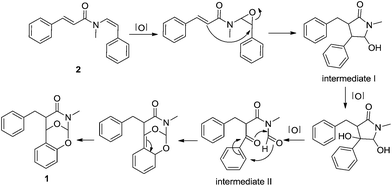
The relative configuration of 1 was inferred on the basis of analysis of ROESY spectrum (Fig. 2). The NOE correlations of H-1′/N–CH3, H-1′/H-2′, H-1′/H-3 and H-3/N–CH3 suggested the cofacial relationship among H-1′, H-2′, N–CH3 and the benzyl group, and the 1,3-oxazinan-4-one ring adopts a stable chair conformation. Additionally, the NOE cross-peaks between H-2 and H-8′ revealed these two protons are spatially close. Moreover, the 1H NMR data gave no coupling constant (J2,2′ ≈ 0 Hz) between H-2 and H-2′, revealing a trans configuration of the two carbons. The absolute configuration of compound 1 was determined by comparison of the experimental ECD spectrum with the theoretical ECD spectrum predicted from quantum chemical TDDFT (time-dependent density functional theory) calculation. In the 190–400 nm region (Fig. 3), both the experimental ECD spectrum and the calculated one showed a positive first Cotton effect and a negative second Cotton effect. Therefore, qualitative analysis of the calculated and experimental ECD spectra allowed the assignments of the absolute configuration of 1 to be 2R, 1′S, 2′S, and 1′′S. Therefore, the structure of clausoxamine (1) was elucidated as shown (Fig. 1). Clausoxamine (1) is the first example of amide alkaloids possessing a 1,3-oxazinan-4-one ring. A proposed biosynthetic pathway for the formation of compound 1 was showed in Scheme 1. Compound 1 might be synthesized from the major amide alkaloid, lansiumamide B (2). The double bond in the 2-phenylethenamine moiety of compound 2 could be oxidized to form epoxide ring, followed by a nucleophilic substitution reaction to form a γ-lactam ring. The intermediate I could be further oxidized to produce intermediate II. The core skeleton of 1 could be constructed by oxa-Diels–Alder reaction. Compound 1 could be formed by further double bond shift.
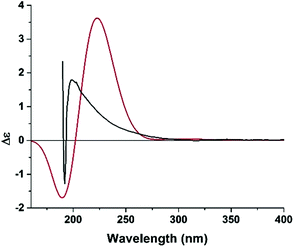 | ||
| Fig. 3 Experimental (black line) and B3LYP/6-311++G(2d,2p)//B3LYP/6-31G(d) calculated (red line) ECD spectra of 1. | ||
Six known compounds, lansiumamide B (2),10 (E)-N-2-phenethyl cinnamide (3),11 lansiumamides C and D (4 and 5),10 ξ-clausenamide (6),12 and SB-204900 (7)13 were isolated. Their structures were identified by comparison of the NMR and MS spectra with the reported data.
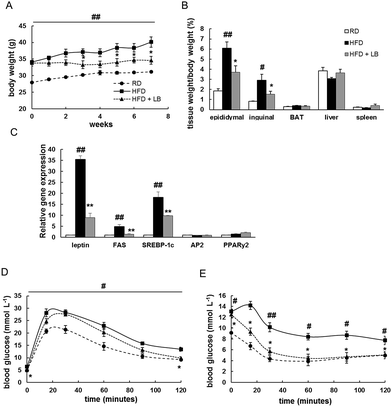
Lansiumamide B (2), the major alkaloid from the seeds of C. lansium, was chosen to evaluate anti-obesity and insulin sensitizing effects on high fat diet (HFD) treated mice. After eight-week HFD treatment, the body weight of mice in HFD group was significantly higher compared to that of regular chow diet (RD) group (Fig. 4A). Then, the HFD treated mice were orally administrated with lansiumamide B (HFD + LB, 20 mg kg−1, dissolved in PEG 400: distilled water = 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4, v/v) or vehicle (PEG 400: distilled water = 6
4, v/v) or vehicle (PEG 400: distilled water = 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4, v/v). From three weeks on, the body weight of mice from HFD + LB group was obviously reduced compared to that from HFD group (Fig. 4A). After 8 weeks treatment, the average weight gain of 13.92 ± 4.30% was observed in HFD group against 2.02 ± 2.82% in HFD + LB group. Analyses of the percentage of different tissue mass by total body weight revealed that the lower body weight of mice in HFD + LB group was mainly due to the reduced visceral (epididymal) and subcutaneous (inguinal) fat tissues, but not liver or kidney (Fig. 4B). Obesity is always accompanied by elevated lipid in serum, resulting in insulin resistance and type 2 diabetes. Therefore, the serum lipid profile was analyzed. As expected, the serum cholesterol and LDL were elevated in HFD-treated mice, but the serum HDL levels were unchanged (Table 2). Lansiumamide B treatment significantly reversed the increases of serum cholesterol and LDL levels (Table 2). Similarly, the cholesterol and LDL were increased, and HDL was decreased in liver from HFD-treated mice, compared with those of RD mice; while, lansiumamide B treatment almost reversed these changes (ESI Table S1†). Furthermore, q-RT-PCR analysis revealed HFD treatment elevated the expressions of adipokine, leptin, and lipogenic markers, FAS (fatty acid synthase) and SREBP-1c (Sterol regulatory element-binding protein 1c), but not adipogenic markers, AP2 (adipocyte protein 2) and PPARγ2 (Peroxisome proliferator-activated receptor γ2); and lansiumamide B treatment significantly suppressed the expressions of leptin, FAS and SREBP-1c (Fig. 4C). Taken together, lansiumamide B prevented HFD-induced obesity through suppressing lipogenesis.
4, v/v). From three weeks on, the body weight of mice from HFD + LB group was obviously reduced compared to that from HFD group (Fig. 4A). After 8 weeks treatment, the average weight gain of 13.92 ± 4.30% was observed in HFD group against 2.02 ± 2.82% in HFD + LB group. Analyses of the percentage of different tissue mass by total body weight revealed that the lower body weight of mice in HFD + LB group was mainly due to the reduced visceral (epididymal) and subcutaneous (inguinal) fat tissues, but not liver or kidney (Fig. 4B). Obesity is always accompanied by elevated lipid in serum, resulting in insulin resistance and type 2 diabetes. Therefore, the serum lipid profile was analyzed. As expected, the serum cholesterol and LDL were elevated in HFD-treated mice, but the serum HDL levels were unchanged (Table 2). Lansiumamide B treatment significantly reversed the increases of serum cholesterol and LDL levels (Table 2). Similarly, the cholesterol and LDL were increased, and HDL was decreased in liver from HFD-treated mice, compared with those of RD mice; while, lansiumamide B treatment almost reversed these changes (ESI Table S1†). Furthermore, q-RT-PCR analysis revealed HFD treatment elevated the expressions of adipokine, leptin, and lipogenic markers, FAS (fatty acid synthase) and SREBP-1c (Sterol regulatory element-binding protein 1c), but not adipogenic markers, AP2 (adipocyte protein 2) and PPARγ2 (Peroxisome proliferator-activated receptor γ2); and lansiumamide B treatment significantly suppressed the expressions of leptin, FAS and SREBP-1c (Fig. 4C). Taken together, lansiumamide B prevented HFD-induced obesity through suppressing lipogenesis.
| RD | HFD | HFD + LB | |
|---|---|---|---|
| a Data are shown as mean ± S.D., n = 6.b p < 0.05 RD vs. HFD group of mice.c p < 0.05, HFD + LB vs. HFD group of mice. | |||
| Cholesterol (mmol L−1) | 4.85 ± 0.90 | 10.11 ± 0.50b | 7.03 ± 0.64c |
| HDL (mmol L−1) | 3.01 ± 0.44 | 3.66 ± 0.41 | 2.85 ± 0.22 |
| LDL (mmol L−1) | 0.76 ± 0.12 | 3.12 ± 0.62b | 1.63 ± 0.28c |
To assess the insulin sensitivity in lansiumamide B treated mice, glucose tolerance tests (GTT) and insulin tolerance tests (ITT) were performed. During GTT, the glucose clearance rate was greatly interrupted in HFD mice, and lansiumamide B treatment obviously improved glucose disposal rate (Fig. 4D). In ITT experiments, the glucose levels of lansiumamide B treated mice were significantly reduced under insulin stimulation, as low as those of RD mice, which indicated lansiumamide B greatly enhanced insulin sensitivity (Fig. 4E) in mice. Collectively, lansiumamide B effectively improved HFD induced insulin resistance.
Conclusions
In conclusion, one novel alkaloid was identified from the seeds of C. lansium. To the best of our knowledge, compound 1 is the first example of alkaloid from the genus Clausena, containing a 1,3-oxazinan-4-one ring. Furthermore, lansiumamide B, a major alkaloid from C. lansium, showed potential anti-obesity effect on HFD treated mice, which might be developed as an anti-obesity agent.Experimental section
General experimental procedures
Optical rotation data were obtained using an Autopol VI polarimeter. UV data were recorded with a Varian Cary 50 spectrophotometer. ECD spectra were recorded with a JASCO J-810 circular dichroism spectrophotometer. IR spectra were recorded on a PerkinElmer spectrum-100 FTIR spectrometer using KBr disks. NMR spectra were recorded on an ASCEND 600 MHz/54 mm NMR spectrometers. The chemical shift (δ) values were given in ppm with TMS as internal standard, and coupling constants (J) were in Hz. ESIMS and HRESIMS spectra were recorded on an LTQ-Orbitrap XL spectrometer. All solvents were analytical grade (Tianjing Chemical Plant, Tianjing, People's Republic of China). Silica gel used for flash chromatography and precoated silica gel GF254 plates used for TLC were produced by Qingdao Haiyang Chemical Co., Ltd. TLC spots were viewed at 254 nm and visualized by spraying with 10% sulfuric acid in alcohol. MCI gel (CHP20P, 75–150 μm, Mitsubishi Chemical Industries Ltd., Tokyo, Japan) and Sephadex LH-20 gel (GE Healthcare Life Sciences, Buckinghamshire, UK) were used for column chromatography (CC). Preparative HPLC was performed on a Shimadzu LC-20AP instrument with a SPD-M20A PDA detector. Chromatographic separation was carried out on a C18 column (19 × 250 mm, 5 μm, Waters, SunFire™), using a gradient solvent system comprised of H2O (A) and CH3CN (B) at a flow rate of 10 mL min−1.Plant material
The seeds of C. lansium were collected from Lincheng, Qinzhou, Guangxi Zhuang Autonomous Region, People's Republic of China, in August 2015, and identified by Professor Jingui Shen from Shanghai Institute of Materia Medica, Chinese Academy of Sciences. A voucher was deposited at the herbarium of the Institute of Chinese Medical Sciences, University of Macau (LL-20151002).Extraction and isolation
The air-dried seeds of C. lansium (2.5 kg) were ground into powder and defatted with petroleum ether (1.5 L) for three times. The residues were then extracted with 95% ethanol under reflux for 3 hours (2 L × 3 times). After evaporation of the collected percolate, the crude extract (120 g) was suspended in 400 mL H2O and extracted with EtOAc (250 mL × 3 times) and n-butanol (250 mL × 3 times), successively. The EtOAc fraction (27.7 g) was subjected to CC over silica gel eluted with petroleum ether/acetone (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 1
1 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v), to yield eight major fractions (A to H). Compound 2 (2.4 g) was crystallized from subfraction C. Fraction D was subjected to CC over silica gel, eluted with CHCl3–EtOAc (10
1, v/v), to yield eight major fractions (A to H). Compound 2 (2.4 g) was crystallized from subfraction C. Fraction D was subjected to CC over silica gel, eluted with CHCl3–EtOAc (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 3
1 to 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) to yield six subfractions (D1 to D6). Fraction D5 was further purified by preparative HPLC, eluting with H2O/CH3CN (1
1, v/v) to yield six subfractions (D1 to D6). Fraction D5 was further purified by preparative HPLC, eluting with H2O/CH3CN (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v), to obtain compound 3 (8.6 mg). Fraction F was purified with Sephadex LH-20, eluting with CHCl3/CH3OH (1
1, v/v), to obtain compound 3 (8.6 mg). Fraction F was purified with Sephadex LH-20, eluting with CHCl3/CH3OH (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) to yield three fractions (F1 to F3). F3 was further separated by preparative HPLC, eluting with H2O/CH3CN (3
1, v/v) to yield three fractions (F1 to F3). F3 was further separated by preparative HPLC, eluting with H2O/CH3CN (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7 to 0
7 to 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v), to obtain compound 7 (9.6 mg) and a mixture of 4/5 (13.2 mg). Fraction G was also purified with Sephadex LH-20, eluting with CHCl3/CH3OH (1
1, v/v), to obtain compound 7 (9.6 mg) and a mixture of 4/5 (13.2 mg). Fraction G was also purified with Sephadex LH-20, eluting with CHCl3/CH3OH (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) to yield five fractions (G1 to G5). Compound 6 (4.2 mg) was obtained from fraction G2 by preparative HPLC, eluting with H2O/CH3CN (1
1, v/v) to yield five fractions (G1 to G5). Compound 6 (4.2 mg) was obtained from fraction G2 by preparative HPLC, eluting with H2O/CH3CN (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 0
1 to 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v). Fraction G was subjected to CC over silica gel, eluted with petroleum ether–acetone (10
1, v/v). Fraction G was subjected to CC over silica gel, eluted with petroleum ether–acetone (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 1
1 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v), to yield six subfractions (G1 to G6). Compound 1 (5.7 mg) was purified from fraction G2 by preparative HPLC, eluding with H2O/CH3CN (1
1, v/v), to yield six subfractions (G1 to G6). Compound 1 (5.7 mg) was purified from fraction G2 by preparative HPLC, eluding with H2O/CH3CN (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v).
1, v/v).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 211 (1.13), 271 (0.22) nm; IR (KBr) vmax 2922, 1658, 1491, 1459, 1254, 1216, 1194, 1033, 977 cm−1; NMR data see Table 1; HRESIMS m/z 296.1261 [M + H]+ (calcd for C18H18NO3, 296.1287).
ε) 211 (1.13), 271 (0.22) nm; IR (KBr) vmax 2922, 1658, 1491, 1459, 1254, 1216, 1194, 1033, 977 cm−1; NMR data see Table 1; HRESIMS m/z 296.1261 [M + H]+ (calcd for C18H18NO3, 296.1287).Computational methods
Conformational analysis of 1 was carried out via Monte Carlo searching using molecular mechanism with MMFF94 force field in the Spartan 08 program.14 The result showed three lowest energy conformers for 1. The conformers were then reoptimized using DFT at the B3LYP/6-31+G(d) level in vacuum in the Gaussian 09 program.15 The B3LYP/6-31+G(d) harmonic vibrational frequencies were further calculated to confirm their stability. The energies, oscillator strengths, and rotational strengths of the first 60 electronic excitations were calculated using the TDDFT methodology at the B3LYP/6-311++G(2d,2p) level in vacuum. The ECD spectra were then simulated by the overlapping Gaussian function (σ = 0.70 eV),16 in which velocity rotatory strengths of the first 20 exited states were adopted. To get the conformationally averaged ECD spectra, the simulated spectra of the lowest energy conformers were averaged according to the Boltzmann distribution theory and their relative Gibbs free energy (ΔG).Animals
All animal experiments were approved by the Animal Ethical and Welfare Committee of University of Macau (No. ICMS-AEC-2014-06). All procedures involved in the animal experiments were carried out in accordance with the approved guidelines and regulations. Male C57BL/6J mice were purchased from the animal facility of Faculty of Health Sciences, University of Macau, and maintained at 22 ± 1 °C under a 12 h light, 12 h dark cycle with a 50% humidity, free access to food and water.Lansiumamide B treatment
Eight-week-old male mice were randomly distributed into three groups, each six mice. One group of mice (RD) were fed with a regular chow-diet (Guangdong Medical Lab Animal Centre, Guangzhou, Guangdong, People's Republic of China). The other two groups of mice were fed with a 45% high fat diet (Trophic Animal Feed High-Tech Co., Nantong, Jiangsu, People's Republic of China) for eight weeks to induce obesity. Then, one group of the high fat diet treated mice (HFD + LB) were orally administrated with 20 mg kg−1 lansiumamide B (dissolved in PEG 400![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) distilled water = 6
distilled water = 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4, v/v) once a day for eight weeks. The other group of the high fat diet treat mice (HFD) and the RD group of mice were orally administrated with the same volume of vehicle solution (PEG 400
4, v/v) once a day for eight weeks. The other group of the high fat diet treat mice (HFD) and the RD group of mice were orally administrated with the same volume of vehicle solution (PEG 400![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) distilled water = 6
distilled water = 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4, v/v) once a day for eight weeks. Body weight was monitored every week. Tissues and serum were collected after 16 hour of fasting and stored at −80 °C for further studies.
4, v/v) once a day for eight weeks. Body weight was monitored every week. Tissues and serum were collected after 16 hour of fasting and stored at −80 °C for further studies.
Analysis of serum lipids
Triglycerides HDL and LDL levels in serum and liver were analysed using commercially available kits (Nanjing Jiancheng, Nanjing, Jiangsu, People's Republic of China), following the manufacture's instruction.q-RT-PCR analysis
Total RNA of epididymal fat was isolated using TRIzol Reagent (Invitrogen, Carlsbad, CA) following the manufacturer's instructions. The cDNA was synthesized from 1 μg RNA using the SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen). Q-PCR was performed on a Bio-Rad qPCR machine using the SYBR Green PCR Master Mix, according to the manufacturer's recommended procedures. The primers were as follows: leptin forward primer 5′-CTATCCAGAAAGTCCAGGATGACA-3′, leptin reverse primer 5′-ATTCTCCAGGTCATTGGCTATCT-3′; adiponectin forward primer 5′-GTTCCTCTTAATCCTGCCCAGTC-3′, adiponectin reverse primer 5′-GATCTTAGTAAAGCGAATGGGTACA-3′; resistin forward primer 5′-GAACTGAGTTGTGTCCTGCTAAGTC-3′, resistin reverse primer 5′-AATTTAAGCCAATGTTCTTTATTGC-3′; FAS forward primer 5′-AGAGATCCCGAGACGCTTCT-3′, FAS reverse primer 5′-GCCTGGTAGGCATTCTGTAGT-3′; SREBP-1c forward primer 5′-GGAGCCATGGATTGCACATT-3′, SREBP-1c reverse primer 5′-CCTGTCTCACCCCCAGCATA-3′; AP2 forward primer 5′-AGTGAAAACTTCGATGATTACATGAA-3′, AP2 reverse primer 5′-GCCTGCCACTTTCCTTGTG-3′; PPARγ2 forward primer 5′-GCCTATGAGCACTTCACAAGAAATT-3′, PPARγ2 reverse primer 5′-TGCGAGTGGTCTTCCATCAC-3′. 18S was used as housekeeping gene: 18S forward primer 5′-AGCCTGCGGCTTAATTTGAC-3′, 18S reverse primer 5′-CAACTAAGAACGGCCATGCA-3′.Glucose tolerance tests and insulin tolerance tests
The glucose tolerance tests (GTT) and insulin tolerance tests (ITT) were performed after six and seven weeks of lansiumamide B treatment, respectively, as described previously.17,18 After 18 hours fasting, the tail blood glucose was measured using OneTouch Ultra blood glucose meter and LifeScan test strips. Then, the mice were received an intraperitoneal injection of glucose solution (Sigma-Aldrich, St. Louis, MO, USA) at a dose of 2.0 g kg−1 body weight. The tail blood glucose was measured at 15, 30, 60 and 120 min after injections. Regarding to ITT, the mice tail blood glucose concentration was measured after 6 hours fasting. The mice were then received an intraperitoneal injection of human insulin (Eli Lilly, Indianapolis, IN, USA) at a dose of 1.0 U kg−1 of body weight. Tail blood glucose concentration was measured at 15, 30, 60, 90 and 120 min after injections.Statistical analysis
Data were presented as mean values and standard errors of the mean. Statistical analyses were performed using one-way ANOVA statistical analysis, Graphpad Prism version 6.0. *p < 0.05 was considered statistically significant.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Financial support by the Science and Technology Development Fund, Macao S. A. R (FDCT 120/2013/A3) and the Research Fund of University of Macau (MYRG2015-00153-ICMS-QRCM, and MYRG2017-00109-ICMS) are gratefully acknowledged. The authors thank Prof. Jingui Shen for identification of plant material.Notes and references
- S. E. Kahn, R. L. Hull and K. M. Utzschneider, Nature, 2006, 444, 840–846 CrossRef CAS PubMed.
- D. M. Muoio and C. B. Newgard, Nat. Rev. Mol. Cell Biol., 2008, 9, 193–205 CrossRef CAS PubMed.
- H. L. Daneschvar, M. D. Aronson and G. W. Smetana, Am. J. Med., 2016, 129, e871–e876 CrossRef PubMed.
- A. C. Adebajo, E. O. Iwalewa, E. M. Obuotor, G. F. Ibikunle, N. O. Omisore, C. O. Adewunmi, O. O. Obaparusi, M. Klaes, G. E. Adetogun, T. J. Schmidt and E. J. Verspohl, J. Ethnopharmacol., 2009, 122, 10–19 CrossRef CAS PubMed.
- D.-Y. Shen, C.-H. Chao, H.-H. Chan, G.-J. Huang, T.-L. Hwang, C.-Y. Lai, K.-H. Lee, T.-D. Thang and T.-S. Wu, Phytochemistry, 2012, 82, 110–117 CrossRef CAS PubMed.
- T. Matsui, C. Ito, H. Furukawa, T. Okada and M. Itoigawa, Inflammation Res., 2013, 62, 333–341 CrossRef CAS PubMed.
- D.-Y. Shen, Y.-Y. Chan, T.-L. Hwang, S.-H. Juang, S.-C. Huang, P.-C. Kuo, T.-D. Thang, E.-J. Lee, A.-G. Damu and T.-S. Wu, J. Nat. Prod., 2014, 77, 1215–1223 CrossRef CAS PubMed.
- K. N. Prasad, H.-H. Xie, J. Hao, B. Yang, S.-X. Qiu, X.-Y. Wei, F. Chen and Y.-M. Jiang, Food Chem., 2010, 118, 62–66 CrossRef CAS.
- H.-Y. Jiang, C.-F. Wang, L. Fang, K. Yang, J.-B. Feng, Z.-F. Geng, J. Xu, Z.-W. Deng, S.-S. Du and H.-B. Yin, Molecules, 2013, 18, 10768–10775 CrossRef CAS PubMed.
- J.-H. Lin, Phytochemistry, 1989, 28, 621–622 CrossRef CAS.
- H.-Y. Yao, Y. Tang and K. Yamamoto, Tetrahedron Lett., 2012, 53, 5094–5098 CrossRef CAS.
- M.-H. Yang, Y.-Y. Chen and L. Huang, Acta Chim. Sin., 1987, 45, 1170–1174 CAS.
- P. H. Milner, N. J. Coates, M. L. Gilpin, S. R. Spear and D. S. Eggleston, J. Nat. Prod., 1996, 59, 400–402 CrossRef CAS.
- Wavefunction Inc, Spartan 08, Irvine, CA Search PubMed.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, B. Mennucci, G. A. Petersson, H. Nakatsuji, M. Caricato, X. Li, H. P. Hratchian, A. F. Izmaylov, J. Bloino, G. Zheng, J. L. Sonnenberg, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, J. A. Montgomery Jr, J. E. Peralta, F. Ogliaro, M. Bearpark, J. J. Heyd, E. Brothers, K. N. Kudin, V. N. Staroverov, R. Kobayashi, J. Normand, K. Raghavachari, A. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, N. Rega, J. M. Millam, M. Klene, J. E. Knox, J. B. Cross, V. Bakken, C. Adamo, J. Jaramillo, R. Gomperts, R. E. Stratmann, O. Yazyev, A. J. Austin, R. Cammi, C. Pomelli, J. W. Ochterski, R. L. Martin, K. Morokuma, V. G. Zakrzewski, G. A. Voth, P. Salvador, J. J. Dannenberg, S. Dapprich, A. D. Daniels, Ö. Farkas, J. B. Foresman, J. V. Ortiz, J. Cioslowski and D. J. Fox, Gaussian 09 Rev. C01, Gaussian, Inc, Wallingford CT, 2009 Search PubMed.
- P. J. Stephens and N. Harada, Chirality, 2010, 22, 229–233 CAS.
- J.-H. Lee, L.-G. Lin, P.-W. Xu, K. Saito, Q. Wei, A. G. Meadows, O. Y. Bongmba, G. Pradhan, H. Zheng, Y. Xu and Y.-X. Sun, Diabetes, 2016, 65, 2169–2178 CrossRef CAS PubMed.
- X.-J. Ma, L.-G. Lin, J. Yue, C.-S. Wu, C.-A. Guo, R.-T. Wang, K.-J. Yu, S. Devaraj, P. Murano, Z. Chen and Y.-X. Sun, Int. J. Mol. Sci., 2017, 18, 1302 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Lipid profile in livers, a listing of NMR, IR and HRESIMS spectra of compound 1, as well as the conformational analysis of compound 1. See DOI: 10.1039/c7ra09793j |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2017 |