Open Access Article
Open Access ArticleEnzymatic ring-opening polymerization (ROP) of lactides and lactone in ionic liquids and organic solvents: digging the controlling factors†
Hua Zhao *ab,
Gabrielle A. Nathanielb and
Princess C. Mereninib
*ab,
Gabrielle A. Nathanielb and
Princess C. Mereninib
aDepartment of Chemistry and Biochemistry, University of Northern Colorado, Greeley, CO 80639, USA. E-mail: hua.zhao@unco.edu; huazhao98@gmail.com
bDepartment of Chemistry and Forensic Science, Savannah State University, Savannah, GA 31404, USA
First published on 17th October 2017
Abstract
Polylactides (PLAs) and polylactones are sustainable and biodegradable polymers with important applications in packaging, horticultural materials, and biomedical fields. The enzymatic ring-opening polymerization (ROP) method represents a ‘green’ approach to the synthesis of these polyesters. However, there are lots of discrepancies in the literature on the lipase specificity towards a particular monomer (such as L-lactide), and many disagreements on the molecular weights produced by similar methods. To provide a better understanding of the key factors regulating the ROP reaction, we carried out a systematic investigation of the polymerization process under different temperatures catalyzed by different types and batches of lipases with various water contents, in different solvents including ionic liquids with known water contents, and by using different initiators. Our data clearly indicate that the key reaction parameters include the types of lipases and solvents, the amount of solvents, water contents in both enzymes and solvents, and the reaction temperature. N,N-Dimethylacetamide (DMA) and 1-butyl-3-methylimidazolium hexafluorophosphate ([BMIM][PF6]) at low concentrations were found suitable for the ROPs of lactides and ε-caprolactone leading to Mw of about 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (polydispersity indexes mostly below 1.8) and moderately high yields (up to 60%). Our study suggests that all of these reaction parameters need to be fully controlled during the enzymatic ROP reaction although some earlier studies often neglected some conditions (such as the exact water content in enzymes and solvents).
000 (polydispersity indexes mostly below 1.8) and moderately high yields (up to 60%). Our study suggests that all of these reaction parameters need to be fully controlled during the enzymatic ROP reaction although some earlier studies often neglected some conditions (such as the exact water content in enzymes and solvents).
Introduction
Polylactide (PLA), often referred to as poly(lactic acid), is a sustainable biopolymer because the monomer lactic acid or lactide can be produced from the microbial fermentation of agricultural by-products including carbohydrate-rich substances.1 PLA and its derivatives, as an important part of aliphatic polyesters, are thermoplastic, biodegradable, renewable and biocompatible polymers with mechanical properties similar to those of polystyrene or polyethylene terephthalate.2 PLA has a wide range of applications in packaging and horticultural materials;3 in addition, due to its high biocompatibility and the lack of toxicity, PLA has major uses in biomedical fields including controlled drug delivery carriers, tissue engineering scaffolds, surgical suture and bone fixation materials.1,4–6There are several common routes for preparing PLA but each of them has some drawbacks: (a) a direct polycondensation polymerization usually results in a low molecular weight PLA with poor mechanical properties; (b) azeotropic condensation polymerization can yield high molecular weight PLA (such as Mn up to 300![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000),7 but this method has several disadvantages including the need for high temperature, the continuous removal of byproducts (such as water) and long reaction times; (c) solid state polymerization (SSP) operates at a temperature above the glass transition temperature but below the melting temperature,8 and has the advantage of producing high molecular weight and fine control of side reactions but requires a much longer reaction time than in melt state or solutions; (d) on the other hand, ring-opening polymerization (ROP) of lactide (LA) (cyclic dimer of lactic acid) through coordination polymerization with metal derivatives such as tin(II) octoate [Sn(Oct)2] or tin(II) butoxide (PLA Mn up to ∼106),9 anionic polymerization (such as strong bases with alcohols), or cationic polymerization is industrially preferred to achieve high molecular weight PLA in bulk (in the melt/absence of solvent);1,10 however, the drawback from this option is the residues of metal catalysts in polyesters; this could be disadvantageous for medical and electronic applications although 10 ppm Sn(Oct)2 residue in PLA is generally considered to be safe.10
000),7 but this method has several disadvantages including the need for high temperature, the continuous removal of byproducts (such as water) and long reaction times; (c) solid state polymerization (SSP) operates at a temperature above the glass transition temperature but below the melting temperature,8 and has the advantage of producing high molecular weight and fine control of side reactions but requires a much longer reaction time than in melt state or solutions; (d) on the other hand, ring-opening polymerization (ROP) of lactide (LA) (cyclic dimer of lactic acid) through coordination polymerization with metal derivatives such as tin(II) octoate [Sn(Oct)2] or tin(II) butoxide (PLA Mn up to ∼106),9 anionic polymerization (such as strong bases with alcohols), or cationic polymerization is industrially preferred to achieve high molecular weight PLA in bulk (in the melt/absence of solvent);1,10 however, the drawback from this option is the residues of metal catalysts in polyesters; this could be disadvantageous for medical and electronic applications although 10 ppm Sn(Oct)2 residue in PLA is generally considered to be safe.10
Enzymatic ROP of lactides using lipases and esterases represents a ‘greener’ alternative to metal-based catalysts. Lipase-catalyzed polymerization of lactides has been carried out in bulk, in organic solvents (such as toluene), and recently in supercritical carbon dioxide (sc-CO2) and ionic liquids.1,6,10 Several lipases are commonly used in these reactions, such as Candida antarctica lipase B (CALB) (free form, or immobilized on acrylic resin known as Novozym 435), lipase PS from Burkholderia cepacia, Pseudomonas cepacia lipase PS, and porcine pancreatic lipase (PPL). Representative CALB-catalyzed reactions in each one of these solvents which led to high-molecular-weight PLAs are briefly discussed below for comparison purpose (see our recent review11 for details). In the absence of solvent, the Yoshizawa-Fujita group12 carried out the free CALB-catalyzed ROP of L-lactide at 130 °C, and obtained the polyester with Mw 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, polydispersity index (PDI) 1.13 and yield 54.1%. When the ROP reaction in toluene was catalyzed by free CALB at 100 °C for 24 h, Yoshizawa-Fujita et al.12 obtained PLA with Mn = 44
000, polydispersity index (PDI) 1.13 and yield 54.1%. When the ROP reaction in toluene was catalyzed by free CALB at 100 °C for 24 h, Yoshizawa-Fujita et al.12 obtained PLA with Mn = 44![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100, PDI = 1.15 and yield = 26.9%. Using the biphasic system of supercritical CO2 and melted L-lactide, Mw up to 12
100, PDI = 1.15 and yield = 26.9%. Using the biphasic system of supercritical CO2 and melted L-lactide, Mw up to 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900 of PLA was obtained when the ROP was catalyzed by Novozym 435 under a low initial water activity (aw < 0.16) at 65 °C.13 Several common ionic liquids have been examined in the enzymatic polymerization of lactide catalyzed by CALB. The Yoshizawa-Fujita group12 carried out CALB-catalyzed ring-opening polymerization of L-lactide in 1-butyl-3-methylimidazolium tetrafluoroborate ([BMIM][BF4]) at 110 °C, achieving Mn 54
900 of PLA was obtained when the ROP was catalyzed by Novozym 435 under a low initial water activity (aw < 0.16) at 65 °C.13 Several common ionic liquids have been examined in the enzymatic polymerization of lactide catalyzed by CALB. The Yoshizawa-Fujita group12 carried out CALB-catalyzed ring-opening polymerization of L-lactide in 1-butyl-3-methylimidazolium tetrafluoroborate ([BMIM][BF4]) at 110 °C, achieving Mn 54![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600, 1.25 PDI and 24.3% yield; in comparison, the solvent-free condition at 130 °C resulted in Mn 40
600, 1.25 PDI and 24.3% yield; in comparison, the solvent-free condition at 130 °C resulted in Mn 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, and 54.1% yield while toluene as the solvent at 120 °C led to Mn 42
000, and 54.1% yield while toluene as the solvent at 120 °C led to Mn 42![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600 and 17.3% yield. This group also noted the lipase activity in the polymerization reaction decreasing with different ionic liquids as: [BMIM][BF4] > [BMIM][Tf2N] > [BMIM][PF6] > [BMIM][dca] (Tf2N− = bis(trifluoromethylsulfonyl)imide, and dca− = dicyanamide). Several other studies also reported a lower degree polymerization of lactide in [BMIM][PF6], such as Mn 581 and 29.5 yield at 65 °C,14 and Mn 19
600 and 17.3% yield. This group also noted the lipase activity in the polymerization reaction decreasing with different ionic liquids as: [BMIM][BF4] > [BMIM][Tf2N] > [BMIM][PF6] > [BMIM][dca] (Tf2N− = bis(trifluoromethylsulfonyl)imide, and dca− = dicyanamide). Several other studies also reported a lower degree polymerization of lactide in [BMIM][PF6], such as Mn 581 and 29.5 yield at 65 °C,14 and Mn 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600 at 90 °C; however, a higher molecular weight Mn of 37
600 at 90 °C; however, a higher molecular weight Mn of 37![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800 and a higher yield of 63.2% were achieved in [HMIM][PF6] at 90 °C.15
800 and a higher yield of 63.2% were achieved in [HMIM][PF6] at 90 °C.15
On the other side, there have been inconsistent reports on “which enzyme is more active towards L-lactide?” and “what is the role of solvents?”. As summarized in Table S1 (ESI†), most reactions were conducted under so called “dry conditions”, but the exact water contents in solvents, enzymes and monomers were not determined. It is well known that water is the initiator of the ROP reaction, but an excess amount of water leads to the enzymatic hydrolysis of polyester.6,16,17 In addition, there have been mixed reports on the lipase specificity on L- and D-lactide. Matsumura et al.18 observed no activity of Novozym 435 toward the ROP of D,L-lactide, but a high activity for Pseudomonas cepacia lipase PS and a modest activity for Cundidu cylindruceu lipase and porcine pancreatic lipase (PPL). Under the optimum conditions, lipase PS gave weight-average molecular weights (Mw) up to 126![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 and 16% yield at 130 °C. This group19 further demonstrated that PPL exhibited a high activity for the copolymerization of lactide and trimethylene carbonate, resulting in Mw up to 21
000 and 16% yield at 130 °C. This group19 further demonstrated that PPL exhibited a high activity for the copolymerization of lactide and trimethylene carbonate, resulting in Mw up to 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 and 40% yield. Therefore, it is suggested20,21 that CALB has a better selectivity toward D-lactide than L-isomer, whilst lipase from Burkholderia cepacia (known as lipase PS) is more specific toward L-lactide. Duchiron et al.21 indicated that the addition of triethylamine could activate the lipases, resulting in 4900 Mn and 89% yield for Novozym 435-catalyzed ROP of D-lactide, and 1800 Mn and 80% yield for lipase PS-catalyzed ROP of L-lactide. On the contrary, a number of studies still reported the synthesis of relatively high molecular weights of polylactide catalyzed by Novozym 435 in organic solvents and ionic liquids (see Table S1†). For example, Omay et al.22 carried out the polymerization of D,L-lactide in dry toluene at 80 °C, and obtained Mn of 26
000 and 40% yield. Therefore, it is suggested20,21 that CALB has a better selectivity toward D-lactide than L-isomer, whilst lipase from Burkholderia cepacia (known as lipase PS) is more specific toward L-lactide. Duchiron et al.21 indicated that the addition of triethylamine could activate the lipases, resulting in 4900 Mn and 89% yield for Novozym 435-catalyzed ROP of D-lactide, and 1800 Mn and 80% yield for lipase PS-catalyzed ROP of L-lactide. On the contrary, a number of studies still reported the synthesis of relatively high molecular weights of polylactide catalyzed by Novozym 435 in organic solvents and ionic liquids (see Table S1†). For example, Omay et al.22 carried out the polymerization of D,L-lactide in dry toluene at 80 °C, and obtained Mn of 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 and 21
000 and 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 when using Novozym 435 and free CALB, respectively.
000 when using Novozym 435 and free CALB, respectively.
To clarify the inconsistent results of the enzymatic ROP of lactides, we aim to examine some key factors of the ROP reaction including different organic solvents and ionic liquids, water contents in the reaction system, different lipases and initiators. These evaluations will provide some guidance towards the future rational design of the enzymatic polymerization for polyester synthesis.
Experimental section
Materials
The items purchased from Sigma-Aldrich include L-(−)-lactide (catalog # 367044), DL-lactide (catalog # 303143), lipase from Pseudomonas fluorescens (Sigma 534730, Batch # MKCB1125V), Candida antarctica lipase B (CALB) immobilized on acrylic resin known as Novozym 435 (catalog # L4777), Candida antarctica lipase B (catalog # 62288), CALB immobilized on Immobead 150 (catalog # 54326), CALB-CLEA (Cross-Linked Enzyme Aggregate) (catalog # 16698), Amano lipase PS from Burkholderia cepacia (catalog # 534641), Amano lipase from Pseudomonas fluorescens (catalog # 534730), lipase from porcine pancreas (PPL) type II (catalog # L3126), Amano lipase A from Aspergillus niger (catalog # 534781), lipase from Pseudomonas cepacia immobilized in sol–gel-AK (catalog # 62279), lipase from Candida cylindracea immobilized in sol–gel-AK (catalog # 62278), and Resomer® R203H poly(D,L-lactide) (catalog # 719943, Lot # STBF1681V) with Mw 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–24
000–24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000. Lipase PS-C Amano I (catalog # ILPSAC0350403R) and lipase PS-D Amano I (catalog # ILPSAB0152305R) were kind gifts from Amano Enzyme USA (Elgin, IL). D-(+)-Lactide obtained from Sigma-Aldrich was a product of Ark Pharm (catalog # AK-57455). ε-Caprolactone was acquired from TCI America. Anhydrous dimethylformamide (DMF, 99.8%) was obtained from Alfa Asear (Ward Hill, MA). 1-Butyl-3-methylimidazolium hexafluorophosphate ([BMIM][PF6], high purity), 1-butyl-3-methylimidazolium bis(trifluoromethylsulfonyl)imide ([BMIM][Tf2N], synthesis grade), and 1-butyl-3-methylimidazolium tetrafluoroborate ([BMIM][BF4], high purity) obtained from VWR were products of Merck KGaA (EMD Millipore Corporation, Billerica, MA). Cholinium bis(trifluoromethylsulfonyl)imide ([Choline][Tf2N]), triethyl (2-(2-methoxyethoxy)ethoxy)ethylammonium bis(trifluoromethanesulfonyl)imide ([CH3(OCH2CH2)3-Et3N][Tf2N]), triethyl (2-(2-methoxyethoxy)ethoxy)ethylammonium acetate ([CH3(OCH2CH2)3-Et3N][OAc]), 1-ethyl-3-(2-(2-methoxyethoxy)ethoxy)ethyl)piperidinium bis(trifluoromethanesulfonyl)imide ([CH3(OCH2CH2)3-Et-Pip][Tf2N]), 1-ethyl-3-(2-(2-methoxyethoxy)ethoxy)ethyl)piperidinium acetate ([CH3(OCH2CH2)3-Et-Pip][OAc]) were prepared and characterized in our earlier studies.23,24
000. Lipase PS-C Amano I (catalog # ILPSAC0350403R) and lipase PS-D Amano I (catalog # ILPSAB0152305R) were kind gifts from Amano Enzyme USA (Elgin, IL). D-(+)-Lactide obtained from Sigma-Aldrich was a product of Ark Pharm (catalog # AK-57455). ε-Caprolactone was acquired from TCI America. Anhydrous dimethylformamide (DMF, 99.8%) was obtained from Alfa Asear (Ward Hill, MA). 1-Butyl-3-methylimidazolium hexafluorophosphate ([BMIM][PF6], high purity), 1-butyl-3-methylimidazolium bis(trifluoromethylsulfonyl)imide ([BMIM][Tf2N], synthesis grade), and 1-butyl-3-methylimidazolium tetrafluoroborate ([BMIM][BF4], high purity) obtained from VWR were products of Merck KGaA (EMD Millipore Corporation, Billerica, MA). Cholinium bis(trifluoromethylsulfonyl)imide ([Choline][Tf2N]), triethyl (2-(2-methoxyethoxy)ethoxy)ethylammonium bis(trifluoromethanesulfonyl)imide ([CH3(OCH2CH2)3-Et3N][Tf2N]), triethyl (2-(2-methoxyethoxy)ethoxy)ethylammonium acetate ([CH3(OCH2CH2)3-Et3N][OAc]), 1-ethyl-3-(2-(2-methoxyethoxy)ethoxy)ethyl)piperidinium bis(trifluoromethanesulfonyl)imide ([CH3(OCH2CH2)3-Et-Pip][Tf2N]), 1-ethyl-3-(2-(2-methoxyethoxy)ethoxy)ethyl)piperidinium acetate ([CH3(OCH2CH2)3-Et-Pip][OAc]) were prepared and characterized in our earlier studies.23,24
Methods
The overall water content of the reaction mixture was determined by mixing the reaction mixture (monomer, lipase and solvent) with 5.0 mL anhydrous methanol under gentle stirring at room temperature for 24 h. The water contents in methanol before and after the mixing were determined by the Karl Fischer titration. The overall water content in the reaction mixture was calculated from the water contents in methanol before and after the mixing.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 (Mw).22 The polyester structure was also confirmed by 1H and 13C NMR (JEOL 300 MHz) in CDCl3 and FT-IR (Shimadzu IR Prestige-21).12,22,29–32 The specific optical rotation of the polymers, [α], was determined in chloroform at a concentration of 10 mg mL−1 at room temperature using a Rudolph Autopol III polarimeter at the wavelength of 589 nm.
500 (Mw).22 The polyester structure was also confirmed by 1H and 13C NMR (JEOL 300 MHz) in CDCl3 and FT-IR (Shimadzu IR Prestige-21).12,22,29–32 The specific optical rotation of the polymers, [α], was determined in chloroform at a concentration of 10 mg mL−1 at room temperature using a Rudolph Autopol III polarimeter at the wavelength of 589 nm.Results and discussion
In our experiments, the water contents in all lipases, monomers and solvents were determined by the Karl Fisher titration with the detection limit of 1 ppm water (see Table S2 in ESI†). All solvents were obtained as the anhydrous grade or the high purity grade with the lowest possible water content. In this study, molecular weights (Mw and Mn) were determined by GPC analysis using polystyrene calibration; those of poly(ε-caprolactone) were also determined by 1H NMR analysis.Effect of water content in lipases on ROP
Water molecules associated with enzymes (including lipases) could be classified into two categories: water at the surface layer of enzyme molecules is called ‘essential water’ (or ‘free water’, or ‘hydration water’), and water buried inside enzyme molecules is known as the ‘bond water’ (or ‘structural water’).21,33,34 It was observed that under intensive drying over P2O5, lysozyme and subtilisin Carlsberg lost the layer of ‘essential water’, but still kept 3–4 and 14–16 structural water molecules per enzyme molecule, respectively.35 Since the Karl Fischer titration only determines ‘free water’ available in the solution, not any trapped or inaccessible water to the Karl Fischer reagents;33,36 therefore, it is most likely that only the ‘free water’ could be determined by the Karl Fischer titration. Table 1 reports the (‘free’) water contents in different batches of commercial Novozym 435, and in immobilized lipases calibrated with different thermodynamic water activities (aw = 0.11 via equilibrium with saturated LiCl solution, and aw = 0.33 via equilibrium with saturated MgCl2 solution).37–39 The ‘free water’ contents in commercial Novozym 435 range from 0.77–1.97 wt% among those five batches in Table 1 (trials 1, 4, 6, 9, and 13). The enzymatic ROP of L-lactide was catalyzed by different batches of Novozym 435 using a small amount (0.25 mL) of [BMIM][PF6] (containing 0.02 wt% water). In general, the molecular weight of PLA decreases (trials 1 > 4, 6 > 9, 13) with the increase in initial water content in the immobilized lipase. When the enzyme was exposed to saturated LiCl solution (aw = 0.11), the lipase's water content increased to 2.26–2.62 wt%, and the molecular weight of PLA slightly decreased (trial 2 vs. 1, 5 vs. 4, 7 vs. 6, and 11 vs. 9 in Table 1). The Gross group33 varied the water contents (e.g. 0.6, 1.15 and 1.95 wt%) of Novozym 435 and found a higher water content resulted in a lower molecular weight of PCL. The Sayer group40 indicated that drying Novozym 435 (the water content decreased from 1.74 wt% to 0.77 wt%) lead to a drop in polyester yield but an increase in molecular weight during the ROP of ω-pentadecalactone in supercritical CO2 using dichloromethane or chloroform as co-solvent. On the other hand, when the thermodynamic water activity further increased to aw = 0.33 (2.77–3.62 wt% water), the PLA's molecular weight maintained unchanged (such as trials 3 and 12 in Table 1 when comparing with those at aw = 0.11), or even increased (trial 8). Therefore, the free water content in the enzyme plays a critical role in the enzymatic polymerization reaction.| Trial | N435 batch # (purchase date) | Enzyme water (%) | Conversion (%) | Yield (%) | Mw | PDI |
|---|---|---|---|---|---|---|
| a Note: reactions conditions: 0.5 g lactide, 0.25 mL [BMIM][PF6] (0.02 wt% water), 100 mg Novozym 435 (Sigma L4777), gentle stirring at 130 °C for 7 days. Mw values were determined by GPC as calibrated by polystyrene standards. | ||||||
| 1 | 097K1155 (05/2009) | 0.77 | 93.5 | 20 | 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
1.71 |
| 2 | 2.62 (aw 0.11) | 85.0 | 22 | 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600 600 |
1.68 | |
| 3 | 3.62 (aw 0.33) | 93.4 | 44 | 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900 900 |
1.69 | |
| 4 | 067K3522 (09/2009) | 1.09 | 84.9 | 33 | 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 400 |
1.72 |
| 5 | 2.52 (aw 0.11) | 91.0 | 18 | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900 900 |
1.65 | |
| 6 | SLBP0766V (03/2016) | 1.09 | 63.0 | 12 | 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 500 |
1.41 |
| 7 | 2.26 (aw 0.11) | 76.5 | 19 | 9800 | 1.45 | |
| 8 | 3.49 (aw 0.33) | 90.8 | 50 | 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 100 |
1.73 | |
| 9 | SLBS9524 (03/2017) | 1.94 | 72.0 | 30 | 8600 | 1.52 |
| 10 | 1.94 (14 days) | 87.3 | 28 | 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700 700 |
1.50 | |
| 11 | 2.38 (aw 0.11) | 70.9 | 28 | 8300 | 1.44 | |
| 12 | 2.77 (aw 0.33) | 71.9 | 28 | 8100 | 1.47 | |
| 13 | Novozym (05/2017) | 1.97 | 56.4 | 32 | 9672 | 1.44 |
Effect of different solvents and water contents on ROP
The enzymatic ROP of L-lactide catalyzed by Novozym 435 was evaluated with the presence of different co-solvents (see Table 2, trials 1–3 for solvent-free/in bulk, 4–25 for organic solvents, and 26–44 for ionic liquids). At first, we noted that a higher temperature (e.g. 130 °C) led to higher molecular weights and conversions than lower temperatures (80–110 °C) either in bulk (trial 2 and 3 vs. 1), in triglyme (trial 8 vs. 7), or in [BMIM][PF6] (trial 36 vs. 34 and 35). Thus, we selected 130 °C as the reaction temperature for this polymerization process in most studies. The baseline data established by the reaction without any solvent (trial 3 in Table 2) were Mw 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900, PDI 1.75, conversion 72.2%, and isolated yield 14%. A variety of organic solvents were examined as media of the ROP reactions, including toluene, xylene, N,N-dimethylformamide (DMF), triglyme, tetraglyme, 1-methyl-2-pyrrolidone, and N,N-dimethylacetamide (DMA) because most of them have boiling points above 130 °C (except toluene 110 °C). Most organic solvents led to much lower molecular weights and yields than that in bulk (16
900, PDI 1.75, conversion 72.2%, and isolated yield 14%. A variety of organic solvents were examined as media of the ROP reactions, including toluene, xylene, N,N-dimethylformamide (DMF), triglyme, tetraglyme, 1-methyl-2-pyrrolidone, and N,N-dimethylacetamide (DMA) because most of them have boiling points above 130 °C (except toluene 110 °C). Most organic solvents led to much lower molecular weights and yields than that in bulk (16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900, 14% yield) except those in xylene (16
900, 14% yield) except those in xylene (16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300, 42%, but a higher PDI 2.03) and in DMA (trials 15–25 in Table 2). DMA seems to be an exceptionally benign solvent for this enzymatic polymerization reaction, affording the highest Mw 18
300, 42%, but a higher PDI 2.03) and in DMA (trials 15–25 in Table 2). DMA seems to be an exceptionally benign solvent for this enzymatic polymerization reaction, affording the highest Mw 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300 (with 20% yield and PDI 1.68, in trial 23) after 14 days of reaction. Novozym 435 recycled from the ROP reaction in DMA showed lower molecular weights of PLA being produced (trials 24 and 25), but the enzyme still retained certain activities after the vigorous reaction condition (unlike in bulk or in other organic solvents, the enzyme typically turned into char after the reaction). Most ionic liquids (26–44) failed to produce a higher molecular weight and/or yield comparing with the solvent-free condition except [BMIM][PF6], where the highest Mw observed was 17
300 (with 20% yield and PDI 1.68, in trial 23) after 14 days of reaction. Novozym 435 recycled from the ROP reaction in DMA showed lower molecular weights of PLA being produced (trials 24 and 25), but the enzyme still retained certain activities after the vigorous reaction condition (unlike in bulk or in other organic solvents, the enzyme typically turned into char after the reaction). Most ionic liquids (26–44) failed to produce a higher molecular weight and/or yield comparing with the solvent-free condition except [BMIM][PF6], where the highest Mw observed was 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (with 20% yield and PDI 1.71, in trial 36). In addition to the type of solvents, the amount of solvents is also crucial to the enzymatic ROP process. When comparing different amounts of solvents (trials 10/11/12 in 1-methyl-2-pyrrolidone, 16/20/21 in DMA, 36/40/41 in [BMIM][PF6]), a lower solvent content (such as 0.25 mL for 0.5 g L-lactide) seems beneficial to produce higher molecular weights (except trial 41). However, if no solvent is present, the oligo(lactide) easily solidifies which creates the mass transfer barrier for continuing the enzymatic reaction.
000 (with 20% yield and PDI 1.71, in trial 36). In addition to the type of solvents, the amount of solvents is also crucial to the enzymatic ROP process. When comparing different amounts of solvents (trials 10/11/12 in 1-methyl-2-pyrrolidone, 16/20/21 in DMA, 36/40/41 in [BMIM][PF6]), a lower solvent content (such as 0.25 mL for 0.5 g L-lactide) seems beneficial to produce higher molecular weights (except trial 41). However, if no solvent is present, the oligo(lactide) easily solidifies which creates the mass transfer barrier for continuing the enzymatic reaction.
| Trial | Lactide | Lipase | Solvent (water content) | T (°C) | Time (days) | Conversion (%) | Yield (%) | GPC Mw | PDI |
|---|---|---|---|---|---|---|---|---|---|
| a Note: general reaction conditions (or otherwise noted): 0.5 g lactide, 0.25 mL solvent, 100 mg immobilized lipase such as Novozym 435 (03/2017 batch as noted in Table 1, unless indicated otherwise) (or 50 mg free lipase), gentle stirring at 130 °C for 7 days. GPC Mw values were calibrated by polystyrene standards. | |||||||||
| 1 | L | N435 (5/09) | Bulk (no solvent) | 110 | 2 | 26.9 | — | 2700 | 1.49 |
| 2 | L | N435 | Bulk (no solvent) | 130 | 2 | 74.8 | — | 5900 | 1.79 |
| 3 | L | N435 | Bulk (no solvent) | 130 | 7 | 72.2 | 14 | 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900 900 |
1.75 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||||
| Different organic solvents | |||||||||
| 4 | L | N435 | Toluene (0.04 wt%) | 80 | 7 | 14.6 | — | 1600 | 1.33 |
| 5 | L | N435 | Xylene (0.03 wt%) | 130 | 7 | 90.3 | 42 | 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300 300 |
2.03 |
| 6 | L | N435 | DMF (0.05 wt%) | 130 | 7 | 94.4 | — | 4400 | 1.66 |
| 7 | L | N435 | Triglyme (0.03 wt%) | 80 | 7 | 21.0 | — | 1400 | 1.30 |
| 8 | L | N435 | Triglyme (0.03 wt%) | 130 | 7 | 95.5 | 2 | 8700 | 1.59 |
| 9 | L | N435 | Tetraglyme (0.07 wt%) | 130 | 7 | 87.0 | 1.8 | 5200 | 1.53 |
| 10 | L | N435 | 1-Methyl-2-pyrrolidone (0.25 mL) (0.03 wt%) | 130 | 7 | 89.1 | 8 | 7300 | 1.37 |
| 11 | L | N435 | 1-Methyl-2-pyrrolidone (0.5 mL) (0.03 wt%) | 130 | 7 | 69.1 | — | 2642 | 1.13 |
| 12 | L | N435 | 1-Methyl-2-pyrrolidone (1.0 mL) (0.03 wt%) | 130 | 7 | 62.5 | — | 2700 | 1.14 |
| 13 | L | N435 (2.77 wt% H2O, aw = 0.33) | 1-Methyl-2-pyrrolidone (0.25 mL) | 130 | 7 | 90.4 | — | 3600 | 1.26 |
| 14 | L | N435 (3.62 wt% H2O, aw = 0.33) | 1-Methyl-2-pyrrolidone (0.5 mL) | 130 | 7 | 86.5 | — | 4000 | 1.63 |
| 15 | L | N435 (5/09) | DMA (0.25 mL) (0.01 wt%) | 130 | 7 | 94.4 | 48 | 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800 800 |
1.44 |
| 16 | L | N435 (3/17) | DMA (0.25 mL) (0.01 wt%) | 130 | 7 | 90.3 | 32 | 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
1.37 |
| 17 | L | N435 (5/17) | DMA (0.25 mL) (0.01 wt%) | 130 | 7 | 94.6 | 28 | 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 400 |
1.37 |
| 18 | D | N435 (3/17) | DMA (0.25 mL) (0.01 wt%) | 130 | 7 | 95.8 | 26 | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 021 021 |
1.62 |
| 19 | DL | N435 (3/17) | DMA (0.25 mL) (0.01 wt%) | 130 | 7 | 80.0 | 8 | 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 811 811 |
2.05 |
| 20 | L | N435 | DMA (0.5 mL) (0.01 wt%) | 130 | 7 | 88.7 | 26 | 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 500 |
1.41 |
| 21 | L | N435 | DMA (1.0 mL) (0.01 wt%) | 130 | 7 | 90.9 | 34 | 9800 | 1.36 |
| 22 | L | N435 | DMA (0.25 mL) (0.01 wt%) | 130 | 3 | 55.4 | — | 4100 | 1.23 |
| 23 | L | N435 | DMA (0.25 mL) (0.01 wt%) | 130 | 14 | 94.9 | 20 | 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300 300 |
1.68 |
| 24 | L | N435 (recycled) | DMA (0.25 mL) (0.01 wt%) | 130 | 7 | 35.3 | 32 | 5800 | 1.32 |
| 25 | L | N435 (recycled) | DMA (0.5 mL) (0.01 wt%) | 130 | 7 | 44.5 | 14 | 6200 | 1.27 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||||
| Different ionic liquids | |||||||||
| 26 | L | N435 | [BMIM][BF4] (0.03 wt%) | 130 | 7 | 53.3 | — | 2700 | 1.37 |
| 27 | L | N435 | [BMIM][Tf2N] (0.02 wt%) | 130 | 7 | 83.7 | 10 | 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 100 |
1.28 |
| 28 | L | N435 | [Choline][Tf2N] (0.03 wt%) | 130 | 7 | 60.9 | — | 1400 | 1.19 |
| 29 | L | N435 | [CH3(OCH2CH2)3-Et3N][Tf2N] (0.03 wt%) | 130 | 7 | 69.0 | 8 | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 100 |
1.23 |
| 30 | L | N435 | [CH3(OCH2CH2)3-Et3N][OAc] (2.87 wt%) | 130 | 7 | 64.6 | 0.6 | 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 100 |
1.14 |
| 31 | L | N435 | [CH3(OCH2CH2)3-et-Pip][Tf2N] (0.06 wt%) | 130 | 7 | 82.3 | 10 | 9200 | 1.18 |
| 32 | L | N435 | [CH3(OCH2CH2)3-et-Pip][OAc] (2.43 wt%) | 130 | 7 | 62.1 | 4 | 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900 900 |
1.14 |
| 33 | L | No enzyme | [BMIM][PF6] (0.25 mL) (0.02 wt%) | 130 | 7 | 0 | 0 | — | — |
| 34 | L | N435 | [BMIM][PF6] (0.25 mL) (0.02 wt%) | 80 | 7 | 68.1 | 20 | 7100 | 1.31 |
| 35 | L | N435 | [BMIM][PF6] (0.25 mL) (0.02 wt%) | 110 | 7 | 68.8 | 27 | 8800 | 1.60 |
| 36 | L | N435 | [BMIM][PF6] (0.25 mL) (0.02 wt%), 0.26 wt% overall water | 130 | 7 | 93.5 | 20 | 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
1.71 |
| 37 | D | N435 | [BMIM][PF6] (0.25 mL) (0.02 wt%) | 130 | 7 | 92.1 | 44 | 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600 600 |
1.63 |
| 38 | DL | N435 | [BMIM][PF6] (0.25 mL) (0.02 wt%) | 130 | 7 | 78.3 | 12 | 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
1.54 |
| 39 | L | N435 | [BMIM][PF6] (0.25 mL) (0.02 wt%), purged (0.19 wt% overall water) | 130 | 7 | 89.2 | 30 | 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
1.45 |
| 40 | L | N435 | [BMIM][PF6] (0.5 mL) (0.02 wt%) | 130 | 7 | 84.1 | 25 | 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 400 |
1.80 |
| 41 | L | N435 | [BMIM][PF6] (1.0 mL) (0.02 wt%) | 130 | 7 | 44.4 | 16 | 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
1.46 |
| 42 | L | N435, aw = 0.11 | [BMIM][PF6] (0.25 mL) (0.02 wt%) | 130 | 7 | 85.0 | 22 | 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600 600 |
1.81 |
| 43 | L | N435, aw = 0.33 | [BMIM][PF6] (0.25 mL) (0.02 wt%) | 130 | 7 | 90.8 | 50 | 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 100 |
1.73 |
| 44 | L | N435 | [BMIM][PF6] (0.25 mL) (0.09 wt%) | 130 | 7 | 88.6 | 22 | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900 900 |
1.65 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||||
| Different enzyme preparations | |||||||||
| 45 | L | Free CALB (50 mg) | [BMIM][PF6] (0.25 mL) (0.02 wt%) | 130 | 7 | 93.5 | 34 | 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 500 |
1.80 |
| 46 | L | Free CALB (10 mg) | [BMIM][PF6] (0.25 mL) (0.02 wt%) | 130 | 7 | 39.2 | — | 2800 | 1.12 |
| 47 | L | CALB-CLEA (50 mg) | [BMIM][PF6] (0.25 mL) (0.02 wt%) | 130 | 7 | 92.4 | 22 | 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600 600 |
1.76 |
| 48 | L | CALB on Immobead 150 | [BMIM][PF6] (0.25 mL) (0.02 wt%) | 130 | 7 | 91.5 | 36 | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 200 |
2.15 |
| 49 | L | CALB on Immobead 150, aw = 0.11 | [BMIM][PF6] (0.25 mL) (0.02 wt%) | 130 | 7 | 73.6 | 24 | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800 800 |
1.38 |
| 50 | L | Amano P. fluorescens lipase | [BMIM][PF6] (0.25 mL) (0.02 wt%) | 130 | 7 | 74.3 | — | 3800 | 1.30 |
| 51 | L | Amano lipase PS | [BMIM][PF6] (0.25 mL) (0.02 wt%) | 130 | 7 | 58.3 | — | 3400 | 1.32 |
| 52 | L | Amano lipase PS | Tetraglyme (0.07 wt%) | 110 | 7 | 96.6 | 6.5 | 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 100 |
2.57 |
| 53 | L | PPL (Sigma) | [BMIM][PF6] (0.25 mL) (0.02 wt%) | 130 | 7 | 83.0 | 30 | 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 400 |
1.41 |
| 54 | L | lipase PS-C Amano I | [BMIM][PF6] (0.25 mL) (0.02 wt%) | 130 | 7 | 63.6 | — | 3000 | 1.12 |
| 55 | L | lipase PS-D Amano I | [BMIM][PF6] (0.25 mL) (0.02 wt%) | 130 | 7 | 15.3 | 0.2 | 1100 | 1.10 |
| 56 | L | lipase from Pseudomonas cepacia immobilized in sol–gel-AK | [BMIM][PF6] (0.25 mL) (0.02 wt%) | 130 | 7 | 69.5 | — | 1600 | 1.40 |
| 57 | L | lipase from Candida cylindracea immobilized in sol–gel-AK | [BMIM][PF6] (0.25 mL) (0.02 wt%) | 130 | 7 | 61.7 | — | 1800 | 1.23 |
| 58 | L | Amano lipase A from Aspergillus niger | [BMIM][PF6] (0.25 mL) (0.02 wt%) | 130 | 7 | 90.0 | 10 | 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
1.36 |
| 59 | L | Amano lipase A from Aspergillus niger | Tetraglyme (0.07 wt%) | 130 | 7 | 92.0 | 1600 | 1.80 | |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||||
| Different polymerization initiators | |||||||||
| 60 | L | N435, 10 μL ethylene glycol | DMA (0.25 mL) (0.01 wt%) | 130 | 7 | 95.5 | 18 | 7900 | 1.34 |
| 61 | L | N435, 5 μL ethylene glycol | DMA (0.25 mL) (0.01 wt%) | 130 | 7 | 97.5 | 4 | 3500 | 1.22 |
| 62 | L | N435, 10 μL triethylene glycol | DMA (0.25 mL) (0.01 wt%) | 130 | 7 | 94.4 | 12 | 8600 | 1.46 |
| 63 | L | N435, 10 μL glycerol | DMA (0.25 mL) (0.01 wt%) | 130 | 7 | 95.0 | 3700 | 1.24 | |
| 64 | L | N435, 10 μL 1-phenylethanol | DMA (0.25 mL) (0.01 wt%) | 130 | 7 | 94.6 | 30 | 6400 | 1.37 |
Earlier studies might provide some insights of why the enzymatic ROP reactions were not successful in most ionic liquids but in [BMIM[PF6]. The Monticelli group41 studied the melt-blending of PLA with an imidazolium IL (e.g. [BMIM]Cl, [BMIM]I, or [BMIM][PF6]), and found PLA could be miscible with up to 10% [BMIM][PF6] by mass; in addition, [BMIM][PF6] induced the lowest amount of decomposition of polymer matrix during the melt-blending process (at 180–230 °C), and even provided slightly plasticizing effect. The presence of ILs could lead to the hydrolytic degradation of PLA. Park and Xanthos42 evaluated the degradation of PLA (in terms of molecular weight reduction) in the presence of 5 wt% phosphonium-based ILs (decanoate and tetrafluoroborate), and found the decanoate-IL led to much more severer thermal degradation (160 °C) and hydrolytic degradation (60 °C in phosphate buffer) than the BF4−-based IL. Li et al.43 observed that acetate-based imidazoliums caused more thermal degradation (170 °C for 1 h) of PLA than a hydroxide-based IL ([BMIM][OH]) whilst the degradation in [BMIM]Cl, [BMIM][BF4] and [BMIM][HSO4] remained at a minimum level; they also found that a longer alkyl chain on the imidazolium cation led to a higher degree of degradation.
Another important factor is the water content in solvents as too much water often leads to unfavorable side reactions including polyester hydrolysis. As shown in earlier studies, with the increase in water content (above 0.2 wt% (ref. 44) or 0.5 wt% (ref. 45)), the enzymatic ROP rate in bulk increases, but the molecular weight of the polyester decreases.44,45 Thurecht et al.46 observed a lower molecular weight of polyester with a gradual increase in water content (from 0.004 to 2.004 wt%) in supercritical CO2 during the Novozym 435-catalyzed ROP of ε-caprolactone. In our study, when the water content in [BMIM][PF6] increased from 0.02 wt% (trial 36) to 0.09 wt% (trial 44), the molecular weight dropped from 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 to 10
000 to 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900. Although the water content in [BMIM][PF6] was only 0.02 wt%, the overall water content of the reaction system in trial 36 was determined to be 0.26 wt% (see Experimental section) because of the presence of water in the enzyme and monomer molecules. If the moisture was purged out of the reaction system after heating the reaction at 130 °C for 10 min (trial 39), the overall water content was reduced to 0.19 wt%; however, the molecular weight of PLA decreased to 13
900. Although the water content in [BMIM][PF6] was only 0.02 wt%, the overall water content of the reaction system in trial 36 was determined to be 0.26 wt% (see Experimental section) because of the presence of water in the enzyme and monomer molecules. If the moisture was purged out of the reaction system after heating the reaction at 130 °C for 10 min (trial 39), the overall water content was reduced to 0.19 wt%; however, the molecular weight of PLA decreased to 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 instead of increasing as predicted. Therefore, minimizing the water content in ROP is critical for achieving a high molecular weight of polyester, however, if the water content becomes too low, the enzymatic polymerization may not be efficient since water is also acting as the initiator.17,47
000 instead of increasing as predicted. Therefore, minimizing the water content in ROP is critical for achieving a high molecular weight of polyester, however, if the water content becomes too low, the enzymatic polymerization may not be efficient since water is also acting as the initiator.17,47
In terms of the lipase specificity towards L-, D-, and DL-lactide, we compared the respective reactions in DMA (trials 16, 18, and 19) and in [BMIM][PF6] (trials 36–38) and found that overall L-lactide resulted in the highest molecular weight and yield. Many literature examples concurred that Novozym 435 has the specificity towards L-lactide.5,48 However, some groups argued20,21 that CALB exhibits a better selectivity toward D-lactide than L-isomer, whilst lipase from Burkholderia cepacia (i.e. lipase PS) is more specific towards L-lactide. Matsumura et al.18 observed no activity of Novozym 435 towards the ROP of D,L-lactide, but a high activity for Pseudomonas cepacia lipase PS and a modest activity for Cundidu cylindruceu lipase and porcine pancreatic lipase (PPL).
Effect of enzyme preparations and different initiators on ROP
It is known that Novozym-435 contains about 10 wt% Candida antarctica lipase B (CALB) immobilized on a macroporous acrylic resin.49,50 We used the equivalent amount of free CALB (10 mg vs. 100 mg Novozym 435) and observed a low molecular weight of 2800 (trial 46 in Table 2). However, since free CALB takes much less volume in the reaction mixture than its immobilized form, we were able to add a high amount (50 mg) of free CALB in the ROP reaction. As a result, a high molecular weight (17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500) and a relatively high yield (34%) were achieved. Other forms of CALB also led to high molecular weights, such as Cross-Linked Enzyme Aggregate (CALB-CLEA, trial 47) and CALB on Immobead 150 (trials 48 and 49). Most other lipases (trials 50–59) showed relatively low activities leading to low molecular weights, except lipase PS (trial 52), PPL (trial 53), and lipase A from Aspergillus niger (trial 58).
500) and a relatively high yield (34%) were achieved. Other forms of CALB also led to high molecular weights, such as Cross-Linked Enzyme Aggregate (CALB-CLEA, trial 47) and CALB on Immobead 150 (trials 48 and 49). Most other lipases (trials 50–59) showed relatively low activities leading to low molecular weights, except lipase PS (trial 52), PPL (trial 53), and lipase A from Aspergillus niger (trial 58).
We also deployed different initiators (trials 60–64 in Table 2) for the enzymatic polymerization in DMA, such as ethylene glycol, triethylene glycol, glycerol and phenylethanol. But these initiators could not produce the high molecular weights as using water as the initiator (see trials 15–23 in Table 2).
Enzymatic ROP of ε-caprolactone
Based on the above understanding of reaction conditions for PLA synthesis, we further evaluated the enzymatic ROP of ε-caprolactone to produce poly(ε-caprolactone) (PCL). A solvent-free reaction at 70 °C for 2 days (trial 1 in Table 3) gave a relatively high Mw 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700 and a high yield of 56% (PDI = 1.45). The similar reactions in DMA (0.25 mL, trials 2 and 3 in Table 3) suggested a higher Mw up to 23
700 and a high yield of 56% (PDI = 1.45). The similar reactions in DMA (0.25 mL, trials 2 and 3 in Table 3) suggested a higher Mw up to 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, but even higher DMA contents (trials 4 and 5) caused lower molecular weights, which is in line with earlier study on PLA synthesis. A longer reaction time (7 days, trials 6 and 7) and a higher temperature (130 °C, trials 8 and 9) led to lower molecular weights, possibly due to the polyester degradation under these conditions. Surprisingly, free CALB (trial 10 in Table 3) failed to synthesize a high molecular weight of PCL, which is in contrast to the PLA reaction (trial 45 in Table 2). The use of [BMIM][PF6] produced a comparable molecular weight (Mw 20
000, but even higher DMA contents (trials 4 and 5) caused lower molecular weights, which is in line with earlier study on PLA synthesis. A longer reaction time (7 days, trials 6 and 7) and a higher temperature (130 °C, trials 8 and 9) led to lower molecular weights, possibly due to the polyester degradation under these conditions. Surprisingly, free CALB (trial 10 in Table 3) failed to synthesize a high molecular weight of PCL, which is in contrast to the PLA reaction (trial 45 in Table 2). The use of [BMIM][PF6] produced a comparable molecular weight (Mw 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700) as in bulk, but a lower yield of 46%.
700) as in bulk, but a lower yield of 46%.
| Trial | Solvent (water content) | T (°C) | Time (days) | Conversion (%) | Yield (%) | GPC Mw | PDI | GPC Mn | 1H NMR Mn |
|---|---|---|---|---|---|---|---|---|---|
| a Note: general reaction conditions (or otherwise noted): 0.5 g ε-caprolactone, 0.25 mL solvent, 100 mg Novozym 435 (or otherwise noted), gentle stirring at 70 °C. GPC Mw values were calibrated by polystyrene standards. | |||||||||
| 1 | No solvent | 70 | 2 | 95.7 | 56 | 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700 700 |
1.45 | 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300 300 |
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 400 |
| 2 | DMA (0.25 mL) (0.01 wt%) | 70 | 1 | 96.5 | 34 | 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
1.43 | 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 200 |
| 3 | DMA (0.25 mL) (0.01 wt%) | 70 | 2 | 97.0 | 60 | 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
1.56 | 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 100 |
11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600 600 |
| 4 | DMA (0.50 mL) (0.01 wt%) | 70 | 2 | 97.0 | 44 | 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 100 |
2.16 | 8366 | 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 200 |
| 5 | DMA (1.0 mL) (0.01 wt%) | 70 | 2 | 98.1 | 48 | 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 100 |
1.51 | 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 287 287 |
9400 |
| 6 | DMA (0.25 mL) (0.01 wt%) | 70 | 7 | 96.8 | 50 | 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700 700 |
2.24 | 8400 | 9900 |
| 7 | DMA (0.50 mL) (0.01 wt%) | 70 | 7 | 94.3 | 38 | 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900 900 |
1.51 | 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800 800 |
12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 100 |
| 8 | DMA (0.25 mL) (0.01 wt%) | 130 | 7 | 97.6 | 48 | 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300 300 |
1.52 | 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 400 |
12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 200 |
| 9 | DMA (0.25 mL) (0.01 wt%) | 130 | 2 | 95.3 | 48 | 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600 600 |
1.92 | 7600 | 8900 |
| 10 | DMA (0.25 mL) (0.01 wt%), 50 mg free CALB | 70 | 2 | 43.4 | 2 | 2968 | 1.31 | 2258 | 2200 |
| 11 | [BMIM][PF6] (0.25 mL) (0.02 wt%) | 70 | 2 | 88.4 | 46 | 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700 700 |
1.86 | 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 100 |
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 500 |
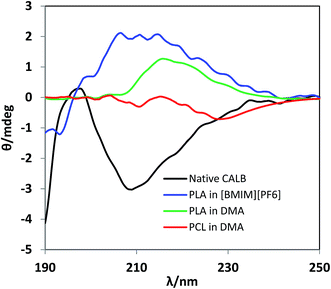
To understand the severity of the reaction conditions on lipase structures, circular dichroism (CD) spectra of CALB were recorded in the far-UV region (190–240 nm) for native CALB, and CALB samples after PLA and PCL synthesis (Fig. 1). The CD spectrum of native CALB shows a characteristic minimum band at 208 nm and a positive band at about 197 nm, which corresponding to about 34% α-helical structure.51,52 However, following the reactions of PLA and PCL syntheses, all CALB lost their characteristic bands indicating the disrupted secondary structures of lipase after the severe reaction conditions.
Confirmation of polyester structures and optical rotation
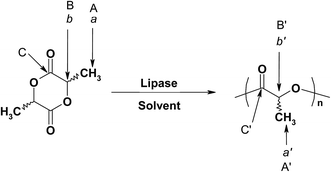
The structures of poly(L-lactide) (PLLA) and poly(D-lactide) (PDLA) were confirmed by 1H and 13C NMR spectra, and FT-IR spectra, as well as by the optical rotation of these polyesters. Fig. S1(a)–(d)† clearly confirmed the identities of PLLA and PDLA samples. As illustrated by Scheme 1, L-lactide shows two 1H NMR peaks at 1.67 ppm (a) and 5.04 ppm (b) while PLA shows two characteristic peaks at 1.57 ppm (a′) and 5.17 ppm (b′).12 In addition, major 13C NMR signals for PLA include 169.7 ppm (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 69.1 ppm (C–H), and 16.7 ppm (CH3), and for L-lactide monomer, peaks are 169.6 ppm (C
O), 69.1 ppm (C–H), and 16.7 ppm (CH3), and for L-lactide monomer, peaks are 169.6 ppm (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 69.0 ppm (C–H) and 16.7 ppm (CH3).22 As shown in Fig. S2,† the IR spectra of the PLLA and PDLA suggest peaks at 3000 and 2950 (C–H stretching vibration), 1760 (C–O stretching vibration), 1460 and 1380 (C–H bending vibration), and 1190 and 1090 cm−1 (C–O stretching vibration), respectively.12,53 The specific rotation [α]D (25 °C, CHCl3, 0.861 g/100 mL) of PLLA was reported to be −156°.54 Our PLLA (trial 1 in Table 1) and PDLA (trial 37 in Table 2) were determined to be −105° and +102° respectively, suggesting both polyesters are primarily isotactic. As shown in Fig. S1(e) and (f),† 1H NMR peaks for PCL are 4.06 ppm (t, 2H, –CH2O–], 2.30 ppm (t, 2H, –CH2O2–), 1.64 ppm (m, 4H, –(CH2)2–), 1.39 ppm (qunit, 2H, –CH2–]); 13C NMR peaks are 173.7 ppm, 64.3 ppm, 34.2 ppm, 28.4 ppm, 25.6 ppm, and 24.7 ppm. FT-IR bands (see Fig. S2†) are 3420 (νOH), 2947 (νCH), 1721 (νC
O), 69.0 ppm (C–H) and 16.7 ppm (CH3).22 As shown in Fig. S2,† the IR spectra of the PLLA and PDLA suggest peaks at 3000 and 2950 (C–H stretching vibration), 1760 (C–O stretching vibration), 1460 and 1380 (C–H bending vibration), and 1190 and 1090 cm−1 (C–O stretching vibration), respectively.12,53 The specific rotation [α]D (25 °C, CHCl3, 0.861 g/100 mL) of PLLA was reported to be −156°.54 Our PLLA (trial 1 in Table 1) and PDLA (trial 37 in Table 2) were determined to be −105° and +102° respectively, suggesting both polyesters are primarily isotactic. As shown in Fig. S1(e) and (f),† 1H NMR peaks for PCL are 4.06 ppm (t, 2H, –CH2O–], 2.30 ppm (t, 2H, –CH2O2–), 1.64 ppm (m, 4H, –(CH2)2–), 1.39 ppm (qunit, 2H, –CH2–]); 13C NMR peaks are 173.7 ppm, 64.3 ppm, 34.2 ppm, 28.4 ppm, 25.6 ppm, and 24.7 ppm. FT-IR bands (see Fig. S2†) are 3420 (νOH), 2947 (νCH), 1721 (νC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1171 cm−1 (δO–C
O), 1171 cm−1 (δO–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O). These characterizations are in line with literature data.55,56
O). These characterizations are in line with literature data.55,56
Evaluating the reliability of Mn from GPC
The Duda group9,57 and Soum group58 pointed out that the gel permeation chromatography (GPC) or size exclusion chromatography (SEC) using polystyrene standards tend to overestimated the value of Mn of polylactide; therefore, the experimental values need to be adjusted by a coefficient of 0.58,57,58 or 0.68 (ref. 9) (at least up to Mn ≤ 2 × 104, for the set of TSKgel G 2000 HXL and 6400 HXL columns with CH2Cl2 as an eluent). The correcting coefficients for polylactones are as the following: 0.56 ± 0.05 for poly(ε-caprolactone), 0.57 ± 0.05 for poly(δ-valerolactone), 0.54 ± 0.05 for poly(β-butyrolactone), and 0.54 ± 0.05 for poly(δ-caprolactone).58To verify the reliability of molecular weight obtained from GPC analysis, we analyzed a commercial poly(D,L-lactide) known as Resomer® R203H with reported Mw 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–24
000–24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, and determined its Mw as 27
000, and determined its Mw as 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, which is not too far from the reported value. In Table 3, we compared the Mn values from GPC analysis with those from the 1H NMR method, and found these values are much closer than that using the above-mentioned correcting coefficients. Therefore, the GPC method seems reliable for determining the molecular weights of PLA and PCL.
000, which is not too far from the reported value. In Table 3, we compared the Mn values from GPC analysis with those from the 1H NMR method, and found these values are much closer than that using the above-mentioned correcting coefficients. Therefore, the GPC method seems reliable for determining the molecular weights of PLA and PCL.
Conclusion
PLAs with weight-averaged molecular weight (Mw) values below 1 × 104, between 1 × 104 and 1 × 105, or above 1 × 105 are considered low molecular weight (LMW), medium molecular weight (MMW), and high molecular weight (HMW) respectively.59 Our PLAs have the highest molecular weights (Mw) in the range of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–20
000–20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, which places them as MMW polyesters. This type of polyesters could be used as the soft block of thermoplastic elastomers, or carriers for controlled drug delivery and release.
000, which places them as MMW polyesters. This type of polyesters could be used as the soft block of thermoplastic elastomers, or carriers for controlled drug delivery and release.
Our study suggests that the key factors controlling the ROP reactions include the types of lipases and solvents, the solvent concentration, water contents in enzymes, substrates and solvents, and the reaction temperature. DMA and [BMIM][PF6] at low concentrations are suitable for the ROPs of lactides and ε-caprolactone leading to Mw around 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 and moderately high yields. Molecular weights reported in literatures for similar enzymatic ROP methods (see Table S1† and reviews17,48,60) are not always in good agreement even in terms of magnitudes; this could be due to different reaction batches containing different amounts of water in enzymes and solvents, and using different methods for determining the molecular weight. In addition, the precipitation method could cause some discrepancies of molecular weights since precipitation of polyesters from reaction mixture could lead to an increase in molecular weight and a decrease in PDI.61
000 and moderately high yields. Molecular weights reported in literatures for similar enzymatic ROP methods (see Table S1† and reviews17,48,60) are not always in good agreement even in terms of magnitudes; this could be due to different reaction batches containing different amounts of water in enzymes and solvents, and using different methods for determining the molecular weight. In addition, the precipitation method could cause some discrepancies of molecular weights since precipitation of polyesters from reaction mixture could lead to an increase in molecular weight and a decrease in PDI.61
Conflicts of interest
There are no conflicts to declare.Acknowledgements
HZ acknowledges the supports by the Henry Dreyfus Teacher-Scholar Award (2012–2018), and the ACS Petroleum Research Fund (PRF# 54875-UR9).References
- K. M. Nampoothiri, N. R. Nair and R. P. John, Bioresour. Technol., 2010, 101, 8493–8501 CrossRef PubMed.
- R. E. Drumright, P. R. Gruber and D. E. Henton, Adv. Mater., 2000, 12, 1841–1846 CrossRef CAS.
- R. Auras, B. Harte and S. Selke, Macromol. Biosci., 2004, 4, 835–864 CrossRef CAS PubMed.
- P. Zinck, Rev. Environ. Sci. Bio/Technol., 2009, 8, 231–234 CrossRef CAS.
- A.-C. Albertsson and I. K. Varma, Biomacromolecules, 2003, 4, 1466–1486 CrossRef CAS PubMed.
- U. Piotrowska and M. Sobczak, Molecules, 2015, 20, 1–23 CrossRef PubMed.
- M. Ajioka, K. Enomoto, K. Suzuke and A. Yamaguchi, Bull. Chem. Soc. Jpn., 1995, 68, 2125–2131 CrossRef CAS.
- S. I. Moon, I. Taniguchi, M. Miyamoto, Y. Kimura and C. W. Lee, High Perform. Polym., 2001, 13, 189–196 CrossRef.
- A. Kowalski, J. Libiszowski, A. Duda and S. Penczek, Macromolecules, 2000, 33, 1964–1971 CrossRef CAS.
- J. Pretula, S. Slomkowski and S. Penczek, Adv. Drug Delivery Rev., 2016, 107, 3–16 CrossRef CAS PubMed.
- H. Zhao, J. Chem. Technol. Biotechnol., 2018, 93 DOI:10.1002/jctb.5444.
- M. Yoshizawa-Fujita, C. Saito, Y. Takeoka and M. Rikukawa, Polym. Adv. Technol., 2008, 19, 1396–1400 CrossRef CAS.
- R. García-Arrazola, D. A. López-Guerrero, M. Gimeno and E. Bárzana, J. Supercrit. Fluids, 2009, 51, 197–201 CrossRef.
- M. Mena, S. Chanfreau, M. Gimeno and E. Bárzana, Bioprocess Biosyst. Eng., 2010, 33, 1095–1101 CrossRef CAS PubMed.
- S. Chanfreau, M. Mena, J. R. Porras-Domínguez, M. Ramírez-Gilly, M. Gimeno, P. Roquero, A. Tecante and E. Bárzana, Bioprocess Biosyst. Eng., 2010, 33, 629–638 CrossRef CAS PubMed.
- A. Idris and A. Bukhari, Biotechnol. Adv., 2012, 30, 550–563 CrossRef CAS PubMed.
- A.-C. Albertsson and R. K. Srivastava, Adv. Drug Delivery Rev., 2008, 60, 1077–1093 CrossRef CAS PubMed.
- S. Matsumura, K. Mabuchi and K. Toshima, Macromol. Rapid Commun., 1997, 18, 477–482 CrossRef CAS.
- S. Matsumura, K. Tsukada and K. Toshima, Int. J. Biol. Macromol., 1999, 25, 161–167 CrossRef CAS PubMed.
- M. Takwa, M. W. Larsen, K. Hult and M. Martinelle, Chem. Commun., 2011, 47, 7392–7394 RSC.
- S. W. Duchiron, E. Pollet, S. Givry and L. Avérous, RSC Adv., 2015, 5, 84627–84635 RSC.
- D. Omay and Y. Guvenilir, Biocatal. Biotransform., 2013, 31, 132–140 CrossRef CAS.
- H. Zhao, G. A. Baker, Z. Song, O. Olubajo, T. Crittle and D. Peters, Green Chem., 2008, 10, 696–705 RSC.
- S. Tang, G. A. Baker, S. Ravula, J. E. Jones and H. Zhao, Green Chem., 2012, 14, 2922–2932 RSC.
- U. Piotrowska, M. Sobczak, E. Oledzka and C. Combes, J. Appl. Polym. Sci., 2016, 133, 43728 CrossRef.
- K. Sha, L. Qin, D. Li, X. Liu and J. Wang, Polym. Bull., 2005, 54, 1–9 CrossRef CAS.
- E. Ozsagiroglu, B. Iyisan and Y. Avcibasi-Guvenilir, Afr. J. Biotechnol., 2012, 11, 12688–12696 CAS.
- M. Mena, A. López-Luna, K. Shirai, A. Tecante, M. Gimeno and E. Bárzana, Bioprocess Biosyst. Eng., 2013, 36, 383–387 CrossRef CAS PubMed.
- K. A. Barrera-Rivera, A. Flores-Carreón and A. Martínez-Richa, J. Appl. Polym. Sci., 2008, 109, 708–719 CrossRef CAS.
- F.-X. Dong, L. Zhang, X.-Z. Tong, H.-B. Chen, X.-L. Wang and Y.-Z. Wang, J. Mol. Catal. B: Enzym., 2012, 77, 46–52 CrossRef CAS.
- G. A. R. Nobes, R. J. Kazlauskas and R. H. Marchessault, Macromolecules, 1996, 29, 4829–4833 CrossRef CAS.
- M. Sobczak, J. Appl. Polym. Sci., 2012, 125, 3602–3609 CrossRef CAS.
- Y. Mei, A. Kumar and R. Gross, Macromolecules, 2003, 36, 5530–5536 CrossRef CAS.
- C. S. Lee, M. T. Ru, M. Haake, J. S. Dordick, J. A. Reimer and D. S. Clark, Biotechnol. Bioeng., 1998, 57, 686–693 CrossRef CAS PubMed.
- M. Dolman, P. J. Halling, B. D. Moore and S. Waldron, Biopolymers, 1997, 41, 313–321 CrossRef CAS.
- P. M. Johnson, S. Kundu and K. L. Beers, Biomacromolecules, 2011, 12, 3337–3343 CrossRef CAS PubMed.
- M. Eckstein, M. Sesing, U. Kragl and P. Adlercreutz, Biotechnol. Lett., 2002, 24, 867–872 CrossRef CAS.
- G. V. Chowdary and S. G. Prapulla, Process Biochem., 2002, 38, 393–397 CrossRef CAS.
- P. J. Halling, Biotechnol. Tech., 1992, 6, 271–276 CrossRef CAS.
- A. E. Polloni, J. G. Veneral, E. A. Rebelatto, D. de Oliveira, J. V. Oliveira, P. H. H. Araújo and C. Sayer, J. Supercrit. Fluids, 2017, 119, 221–228 CrossRef CAS.
- L. Gardella, D. Furfaro, M. Galimberti and O. Monticelli, Green Chem., 2015, 17, 4082–4088 RSC.
- K. I. Park and M. Xanthos, Polym. Degrad. Stab., 2009, 94, 834–844 CrossRef CAS.
- X. Li, Q. Zhou, K. Yang and Y. Wang, Chem. Pap., 2014, 68, 1375–1380 CAS.
- K. S. Bisht, L. A. Henderson, R. A. Gross, D. L. Kaplan and G. Swift, Macromolecules, 1997, 30, 2705–2711 CrossRef CAS.
- K. S. Bisht, Y. Y. Svirkin, L. A. Henderson, R. A. Gross, D. L. Kaplan and G. Swift, Macromolecules, 1997, 30, 7735–7742 CrossRef CAS.
- K. J. Thurecht, A. Heise, M. deGeus, S. Villarroya, J. Zhou, M. F. Wyatt and S. M. Howdle, Macromolecules, 2006, 39, 7967–7972 CrossRef CAS.
- S. Namekawa, S. Suda, H. Uyama and S. Kobayashi, Int. J. Biol. Macromol., 1999, 25, 145–151 CrossRef CAS PubMed.
- S. Kobayashi, Macromol. Rapid Commun., 2009, 30, 237–266 CrossRef CAS PubMed.
- F. C. Loeker, C. J. Duxbury, R. Kumar, W. Gao, R. A. Gross and S. M. Howdle, Macromolecules, 2004, 37, 2450–2453 CrossRef CAS.
- F. Deng and R. A. Gross, Int. J. Biol. Macromol., 1999, 25, 153–159 CrossRef CAS PubMed.
- T. De Diego, P. Lozano, S. Gmouh, M. Vaultier and J. L. Iborra, Biomacromolecules, 2005, 6, 1457–1464 CrossRef CAS PubMed.
- J. Uppenberg, M. T. Hansen, S. Patkar and T. A. Jones, Structure, 1994, 2, 293–308 CrossRef CAS PubMed.
- S. Matsumura, K. Mabuchi and K. Toshima, Macromol. Symp., 1998, 130, 285–304 CrossRef CAS.
- F. Chabot and M. Vert, Polymer, 1983, 24, 53–59 CrossRef CAS.
- K. A. Barrera-Rivera, Á. Marcos-Fernández, R. Vera-Graziano and A. Martínez-Richa, J. Polym. Sci., Part A: Polym. Chem., 2009, 47, 5792–5805 CrossRef CAS.
- J. Cayuela, V. Bounor-Legaré, P. Cassagnau and A. Michel, Macromolecules, 2006, 39, 1338–1346 CrossRef CAS.
- A. Kowalski, A. Duda and S. Penczek, Macromolecules, 1998, 31, 2114–2122 CrossRef CAS.
- M. Save, M. Schappacher and A. Soum, Macromol. Chem. Phys., 2002, 203, 889–899 CrossRef CAS.
- H. Tsuji, Adv. Drug Delivery Rev., 2016, 107, 97–135 CrossRef CAS PubMed.
- C. Jérôme and P. Lecomte, Adv. Drug Delivery Rev., 2008, 60, 1056–1076 CrossRef PubMed.
- R. Marcilla, M. de Geus, D. Mecerreyes, C. J. Duxbury, C. E. Koning and A. Heise, Eur. Polym. J., 2006, 42, 1215–1221 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra09038b |
| This journal is © The Royal Society of Chemistry 2017 |