Open Access Article
Open Access ArticleSurface decoration of selenium nanoparticles with curcumin induced HepG2 cell apoptosis through ROS mediated p53 and AKT signaling pathways†
Min Guo‡
,
Yinghua Li‡ ,
Zhengfang Lin,
Mingqi Zhao,
Misi Xiao,
Changbing Wang,
Tiantian Xu,
Yu Xia and
Bing Zhu
,
Zhengfang Lin,
Mingqi Zhao,
Misi Xiao,
Changbing Wang,
Tiantian Xu,
Yu Xia and
Bing Zhu *
*
Center Laboratory, Guangzhou Women and Children's Medical Center, Guangzhou Medical University, Guangzhou, 510120, China. E-mail: zhubing2016@hotmail.com
First published on 13th November 2017
Abstract
Hepatocellular carcinoma (HCC) is one of the most lethal cancers, and the morbidity and mortality are increasing continuously. Curcumin plays an important role in anticancer activity, while its clinical application is limited by its poor aqueous solubility. To develop an aqueous formulation and improve the anticancer activity of curcumin, curcumin-surface decorated selenium nanoparticles (Se@Cur) were designed in the present study. Transmission electron microscopy (TEM) images and MTT assays indicated Se@Cur dramatically suppressed the proliferation of HepG2 cells and showed low toxicity to normal cells. Compared with SeNPs and curcumin, Se@Cur significantly inhibited the migration ability of HepG2 cells. Moreover, induction of apoptosis in HepG2 cells by Se@Cur was proved by accumulation of the sub-G1 cell population, nuclear condensation and activation of caspase-3. Furthermore, Se@Cur promoted intracellular ROS overproduction and induced apoptosis via activating p53 and AKT signal pathways. Finally, in a xenograft nude mice model, Se@Cur suppressed the growth of tumors. Altogether, the findings in the present study demonstrated the application of Se@Cur as a safe and hopeful strategy for chemotherapeutics of HCC.
1 Introduction
Hepatocellular carcinoma (HCC) is considered as one of the most common malignancies and ranks third in cancer-associated deaths around the world.1–4 About 250![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 new cases of HCC increase every year, with nearly 6
000 new cases of HCC increase every year, with nearly 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 deaths occurring.5 The conventional therapies at the early stages of HCC include surgical restriction, interventional therapy, radiotherapy and radiofrequency ablation.6,7 The survival rate improves significantly after surgical restriction at the early phase of HCC.8,9 Unfortunately, the majority of patients with HCC were advanced stage at the time of diagnosis without proper screening tests and effective surveillance strategies.10 Besides, due to its insensitivity to chemotherapy and high metastatic potential, the survival of patients with HCC is very poor. Moreover, chemotherapy drugs possess potential drawbacks, such as short half-life, poor aqueous solubility and significant toxicity. Thus, it is urgent to develop novel therapeutic agents for the treatment of HCC.11
000 deaths occurring.5 The conventional therapies at the early stages of HCC include surgical restriction, interventional therapy, radiotherapy and radiofrequency ablation.6,7 The survival rate improves significantly after surgical restriction at the early phase of HCC.8,9 Unfortunately, the majority of patients with HCC were advanced stage at the time of diagnosis without proper screening tests and effective surveillance strategies.10 Besides, due to its insensitivity to chemotherapy and high metastatic potential, the survival of patients with HCC is very poor. Moreover, chemotherapy drugs possess potential drawbacks, such as short half-life, poor aqueous solubility and significant toxicity. Thus, it is urgent to develop novel therapeutic agents for the treatment of HCC.11
Curcumin, a hydrophobic polyphenol derived from turmeric Curcuma longa, has wide biological activities, such as anticancer, antioxidant, antibacterial, antifungal and antiviral.12–15 Curcumin plays a role in anticancer activity by regulating the expression of several cellular targets like AKT, MAPK, p53, AMPK and NF-κB, and thus controls cancer cell proliferation, growth, invasion and induces apoptosis.16,17 Furthermore, owing to no signs of toxicity in humans or animals treated with curcumin, it was proved to be a natural and safe agent.18 Unfortunately, the main problems for the clinical application of curcumin are compromised by its low oral bioavailability, rapid degradation and poor solubility in aqueous solution.19–21 Therefore, an aqueous formulation of curcumin is required for clinical application.
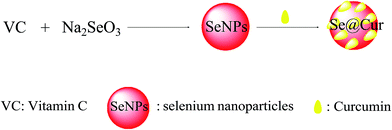
Nanotechnology has already been implied in different medical aspects, with a focus on the diagnosis of diseases and drug delivery systems for therapy of diseases.22–24 As potential drug delivery systems, nanomaterial has the characteristics of smart, surface functionalization and well soluble in aqueous solution, which provides a crucial way to develop aqueous formulations of hydrophobic drugs.25 Selenium is an important dietary trace element and can inhibit the growth of cancer cells.26–28 Numerous reports have shown that selenium is a potent cancer chemopreventive agent and is associated with risk of many cancers, such as stomach, bladder, prostate, ovarian, colon, lung, breast cancer, and so on.29–32 Due to the unique properties of selenium nanomaterial, it is applied in medicine, especially cancer gene and drug delivery.33 Thus, curcumin-loaded SeNPs nanoparticle (Se@Cur) was designed and synthesized as a potential cancer therapy against hepatocellular carcinoma.
The reactive oxygen species (ROS) play an important role in many physiological processes. The redox imbalance is associated with much pathology, such as cancer and other diseases.34 ROS are generated by the chemical reduction of O2, including superoxide (O2˙), superoxide anion (O2˙−), hydroxyl (˙OH) and hydrogen peroxide (H2O2). ROS plays a role of cell-signaling proteins, which are involved in growth, differentiation, and apoptosis of cell.35,36 Recent studies have indicated that ROS have impacts on many signaling pathways, such as MAPKs, NF-κB, Keap1-Nrf2-ARE, PI3K-AKT, etc.37 In this study, Se@Cur was designed as a new chemotherapeutic agent to achieve HepG2 cells apoptosis and its underlying anticancer mechanisms through ROS-mediated signaling pathway were also explored.
2 Experimental
2.1 Materials
HepG2 cells and LO2 cells were acquired from American Type Culture Collection (ATCC, Manassas, VA). Fetal bovine serum (FBS) and Dulbecco's modified Eagle's medium (DMEM) were obtained from Gibco. P53, AKT and cleaved caspase-3 antibody were provided by Cell Signaling Technology Inc. (Danvers, MA, USA). Na2SeO3, Vitamin C (VC), coumarin-6, thiazolyl blue tetrazolium bromide (MTT) were acquired from Sigma-Aldrich (EMD Millipore, Billerica, MA, USA). BCA protein assay kit, Reactive Oxygen Species Assay Kit and caspase-3 activity assay kit were purchased from Beyotime Institute of Biotechnology (Shanghai, People's Republic of China). Mice were obtained from the Guangdong Medical Laboratory Animal Center (Guangdong, China). All animal procedures were performed according to the guidelines of Guangdong Medical Laboratory Animal Center.2.2 Synthesis and characterization of Se@Cur
After SeNPs were synthetized as previous.38 1 mM curcumin was added into the SeNPs under magnetic stirring for half an hour. Then the Se@Cur complex was purified and re-dispersion in Milli-Q water. Finally, the production was dialyzed and measured by ICP-MS analysis. The particle morphology, elemental composition analysis, size distribution, zeta potential and stability in aqueous solutions of Se@Cur were analyzed by transmission electron microscopy (TEM), Energy Dispersive X-ray (EDX) and Malvern Zetasizer Nano ZS particle analyzer, respectively. FT-IR samples were recorded using the KBr-disk method on Equinox 55 IR spectrometer. XPS measurement was carried out on an ESCALab 250 spectrometer.2.3 Cell culture and MTT assay
The HepG2 and LO2 cells were maintained in DMEM supplemented with 10% FBS, 100 U ml−1 penicillin and 50 U ml−1 streptomycin at 37 °C in a humidified incubator (Thermo Scientific, USA) with 5% CO2. The cytotoxicity and viability were evaluated by MTT assay.39 HepG2 and LO2 cells were seeded in triplicate in 96 well flat bottom respectively. Thereafter, the cells were exposed to SeNPs, curcumin, Se@Cur at different concentrations for 48 h. After treatment, cells were incubated with MTT regent for 5 h. Then the culture medium was removed and 150 μl DMSO were added to each well. The conversion of MTT was read at 570 nm absorbance by microplate reader.2.4 Scratch assay
The anti-cell migration effect of Se@Cur on HepG2 cells was detected by a scratch assay. In brief, after the cell confluence reached 80%, cell monolayers were wounded with a sterile microtip and washed with PBS to discard detached cells. Then the cells were treated with Se, curcumin, Se@Cur and incubated for 24 h. After that, the wound closure was observed and photographed by an Olympus microscope at 0 and 24 h.2.5 Cell cycle assay
The cell cycle assay was performed as describe previously.40 After treatment with SeNPs, curcumin and Se@Cur for 48 h, the HepG2 cells were harvested by trypsinization. Then, the collected cells were resuspended in ice cold 70% ethanol and stored over night at −20 °C for fixation. Then the fixed cells were incubated with Propidium iodide at room temperature for 30 min away from light. The cells were measured by flow cytometry. Cell cycle phase quantification was analyzed by using ModFit LT 3.1 software.2.6 Measurement of reactive oxygen species (ROS) level
The level of intracellular ROS which is induced by Se@Cur is detected as describe previously.41,42 In brief, after treatment with Se@Cur in different time, the HepG2 cells were stained with DCFH-DA (10 μM) for 30 min in the dark. The ROS was detected by a fluorescence microscope. The same exposition time was acquired for all treatment groups to capture images. The fluorescence intensity was determined by a fluorescence plate reader with an excitation wavelength of 488 nm and emission wavelength of 525 nm respectively.2.7 Activation of caspase-3
Caspase-3 activity was measured according to the methods described in our previous paper.43,44 Cell lysates, detection buffer and specific caspase substrates Ac-DEVD-pNA (2 mM) were added into a 96-well plate for 2 h at 37 °C. The activation of caspase-3 was measured through the conversion of pNA, which was measured by 405 nm absorbance using a microplate reader.2.8 Western blotting analysis
The expression of proteins associated with different signaling pathways after treatment with Se@Cur in HepG2 cells were analyzed by Western blot as previously reported.45,46 In brief, HepG2 cells were lysed with RIPA solution after treatment with Se@Cur for 48 h, followed by concentration detection of the proteins with a BCA kit. After that, equivalent amount of proteins were loaded in SDS-PAGE, then transferred to polyvinylidene difluoride membranes, and finally visualized by X-ray film.2.9 HepG2 cell xenograft mice model
32 female nude mice were subcutaneously injected into the right armpit region with harvested HepG2 cells. When the volume of tumors reached about 75 mm3, the tumor-bearing mice were acquired. After that, the tumor-bearing mice were randomly allocated into a control group and 3 treatment groups. For study, tumor-bearing mice were treated with Se@Cur at dosages of 3 mg kg−1, 6 mg kg−1 and 9 mg kg−1 through intravenous injection every 2 days and continued for 16 days. As the control group, the mice were treated with equal volume of PBS (0.2 ml). 16 days later, the tumor-bearing mice were killed, of which the tumor weight and body weight were measured. The animal experiments were performed following an approved protocol of Animal Experimentation Ethics Committee.2.10 Statistical analysis
All results were all obtained from three independent tests and the data was analyzed using SPSS (ver. 19.0). Continuous variables were presented as mean ± SD. The differences among multiple groups were compared using one-way analysis of variance (ANOVA). The probability of P value of <0.05 (*) and P value of <0.01 (**) were deemed to be significant in all experiments.3 Results and discussion
3.1 Preparation and characterization of Se@Cur
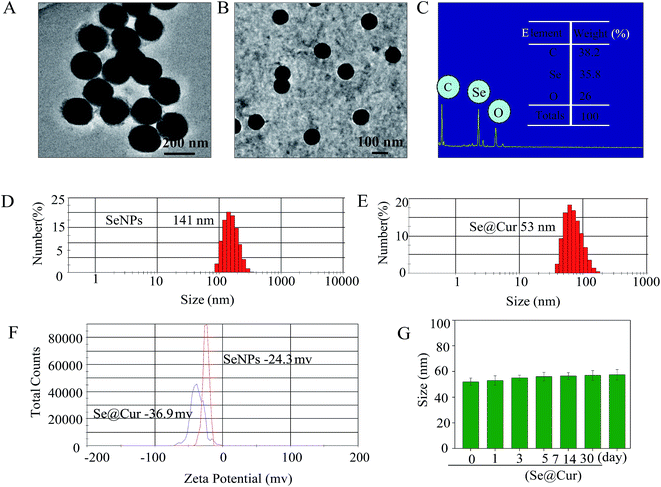
In present study, we constructed a chemical method to synthesize selenium nanoparticle coated with curcumin (Se@Cur) and the synthetic route was shown in Fig. 1. The morphology of SeNPs and Se@Cur was characterized by TEM firstly. As shown in Fig. 2A and B, SeNPs and Se@Cur all presented small and sphere particles. While, compared with SeNPs, Se@Cur was more uniform and dispersive. As shown in Fig. 2C, the EDX analysis of Se@Cur showed the presence of three main signals, including Se atoms signal from SeNPs (35.8%), O (38.2%) and C (26%) that from curcumin. Fig. 2D and E presented the size graphs of SeNPs and Se@Cur. SeNPs without curcumin decorated showed small sphere particles with a size of 141 nm, while Se@Cur apparently reduced the size to 53 nm (Fig. 2D and E). Se@Cur with the property of small size enhanced the cellular uptake and was more easily entered into cell membrane. Besides, the absolute value of zeta potentials generated by SeNPs was 24.3 mv, and increased to 36.9 mV when capped with curcumin (Fig. 2F). The results indicated that after surface modification with curcumin, Se@Cur showed higher stability than SeNPs. Furthermore, from Fig. 2G, the size of Se@Cur gradually increased and kept stable for at least 30 days. Taken together, the characterization of Se@Cur with small size and low zeta potential effectively promoted cellular uptake and nanostructures stability.3.2 The structure of Se@Cur
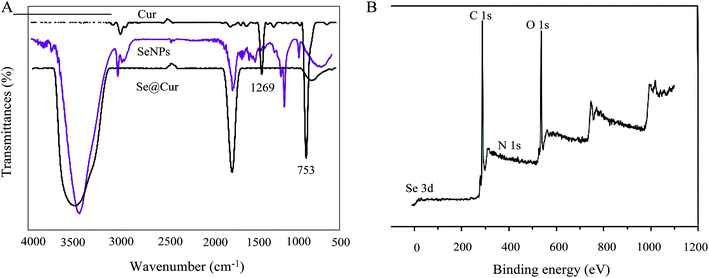
The FTIR spectrum of Se@Cur gave clear evidence that curcumin ligand formed part of the nanocomposite. Curcumin displays IR absorbance peaks at 1269 cm−1 and 753 cm−1 corresponded to –OH and –C–H. The absence of these peaks in Se@Cur indicated the formation of Se@Cur (Fig. 3A). The XPS spectra were also recorded to examine the interaction between SeNPs and curcumin. As shown in Fig. 3B, the O 1s peak in the spectrum of Se@Cur further confirmed that curcumin had been successfully conjugated to the SeNPs, the results support the formation of Se–O bond in Se@Cur.3.3 In vitro cytotoxicity and anti-proliferation activity of Se@Cur
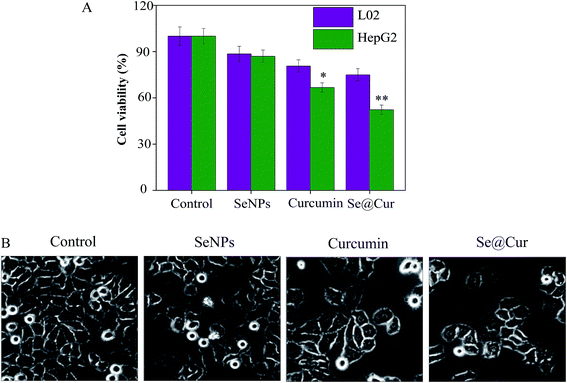
To measure the viability and anti-proliferation activity of HepG2 cells after treatment with SeNPs, curcumin and Se@Cur, the MTT assay was performed. As shown in Fig. 4A, the cell survival rate of LO2 cells was higher than HepG2 cells when exposed to SeNPs (88.45% vs. 86.94%), curcumin (80.65% vs. 66.72%) and Se@Cur (74.95% vs. 52.29%). Compared with SeNPs and curcumin, Se@Cur remarkably suppressed the viability of HepG2 cells and showed less cytotoxicity to normal cells. As shown in Fig. 4B, after treatment with SeNPs, curcumin and Se@Cur, the decrease of cell number, cell rounding, cell vacuoles and cytoplasm shrinkage were observed in different degree. The effects of SeNPs, curcumin and Se@Cur on the growth of HepG2 cells were also further confirmed. Synergy was evaluated by calculation of in vitro fractional inhibitory concentration-index values: minimum inhibitory concentration (MIC) of drug A combination present in Se@Cur of SeNPs (125 μM); MIC of drug B combination present in Se@Cur of Cur (20 μM); MIC of drug A alone corresponded to free SeNPs (1 mM); MIC of drug B alone corresponded to free Cur (62.5 μM). Fractional IC (FIC) was calculated as (MIC drug A combination/MIC drug A alone) + (MIC drug B combination/MIC drug B alone) = 125 μM/1 mM + 20 μM/62.5 μM = 0.445. FIC was 0.445, below 0.5, indicating synergy. In this study, the FIC index was basically interpreted as follows: FIC < 0.5, synergy; FIC 0.5–2, indifference; FIC > 2, antagonism. These results indicated Se@Cur suppressed the proliferation of HepG2 cells most effectively and induced more cancer cells death than non-cancer cell.3.4 Se@Cur inhibited the migration of HepG2 cells
As present in Fig. 5, the HepG2 cells were exposed to SeNPs, curcumin, Se@Cur and the cellular migration was analyzed by a scratch assay. In the control group, the migrating cell was almost completely occupied the wounded gap after 24 h. While, in the Se@Cur treated group, this gap was not occupied by migrating cells. The Migratory ratio of control, SeNPs, curcumin and Se@Cur in Fig. S1.† This result suggested that Se@Cur significantly inhibited the migration ability of HepG2 cells.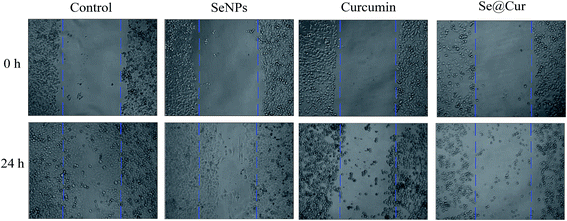 | ||
| Fig. 5 A wound-healing assay was conducted to analyze cell migration. The migration of HepG2 cells was dramatically suppressed after treatment with Se@Cur for 24 h. | ||
3.5 Se@Cur inhibited HepG2 cells proliferation by induction of apoptosis
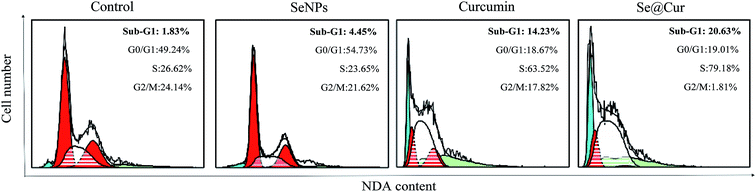
Most anticancer drugs can induce cancer cells apoptosis which is an important mechanism for anticancer effect. In order to prove whether apoptosis was involved in HepG2 cell death induced by SeNPs, curcumin, and Se@Cur, a cell cycle assay was analyzed by flow cytometry. As shown in Fig. 6, the results revealed that the sub-G1 apoptotic cell population was significantly increased in the DNA histogram in different treatments. For instance, the exposure of HepG2 cells to different treatments of control, SeNPs, curcumin, and Se@Cur significantly increased in apoptotic population from 1.83 to 20.63%. Compare to control group, Se@Cur treatment group had a significantly higher percentage of cells in the S phase indicating inhibition the proliferation of HepG2 cells. Translocation of phosphatidylserine induced by SeNPs, curcumin and Se@Cur in HepG2 cell was showed in Fig. S2.† Those results suggested that Se@Cur induced HepG2 cells apoptosis and S phase cell cycle arrest.3.6 Activation of caspase-3 by Se@Cur
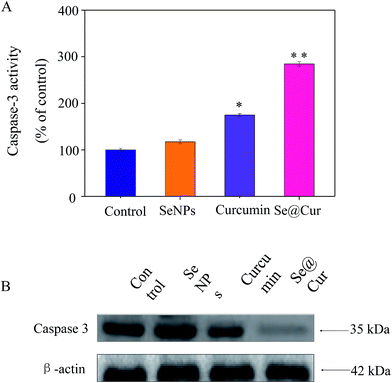
Caspases are some family of cysteine proteases and important components in response to apoptosis. Caspase-3 has been regarded as the main executioner caspases in the cell, which lead to cell disassembly and DNA fragmentation. As an effector caspase, activation of caspase-3 was engaged by activation of initiator caspases through extrinsic or intrinsic apoptosis pathways. Caspase-3 involved in proteolytic cleavage of some important proteins. Activation of caspase-3 was measured by western blotting and fluorometric test. Compared to control group, the activation of caspase-3 was significantly increased to 117%, 175%, 285% when the HepG2 cells were exposed to SeNPs, curcumin, Se@Cur, respectively (Fig. 7A). As shown in Fig. 7B, the level of caspase-3 expression was downregulated with treatment of Se@Cur. The results above suggested that nanosystem markedly increased the activation of caspase-3 to mediate apoptosis.3.7 Induction of ROS in HepG2 cells by Se@Cur
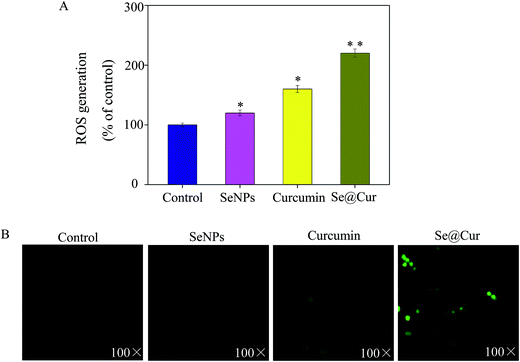
Caspase-3 was confirmed as an important mediator of cell apoptosis.It has been reported that ROS was a vital regulator of cell signaling pathways triggered by anticancer drugs. In order to investigate weather Se@Cur could trigger ROS-mediated apoptosis, the ROS level was detected by DCF fluorescence assay. After exposure to SeNPs, curcumin, and Se@Cur, the ROS generation of HepG2 cells markedly elevated to 120%, 160% and 220% of the control group, respectively (Fig. 8A). From photos in Fig. 8B, the average fluorescent intensity of HepG2 cells treated with Se@Cur was the strongest, followed by curcumin and SeNPs treatment group. TEM image of HepG2 cells treated with Se@Cur was showed in Fig. S3.† These results revealed that the involvement of ROS in the anticancer action of Se@Cur.
3.8 ROS-mediated signaling pathways by Se@Cur
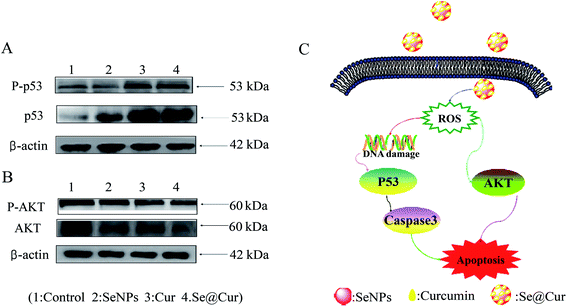
Furthermore, the molecular mechanisms of Se@Cur in inducing HepG2 cells apoptosis were investigated. Intracellular ROS overproduction could cause DNA damage response and lead to cell apoptosis via regulation of multiple signaling pathways, including JNK, AKT, ERK and P53, et al.47,48 The overproduction of ROS were significantly generated in cells exposed to Se@Cur as aforesaid, so the vital molecules expressed in ROS-mediated downstream signaling pathway were tested by Western blotting. As shown in Fig. 9A, compared with SeNPs and curcumin, the expression of p53 was in a down-regulated trend after treatment with Se@Cur. Similarly, for the AKT signaling pathway, Se@Cur markedly promoted the protein expression of AKT (Fig. 9B). In general, the present results indicated that Se@Cur induced HepG2 cells apoptosis by at least two ROS-mediated signaling pathways.3.9 In vivo anticancer activity of Se@Cur
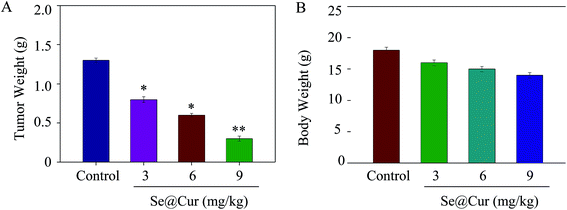
In vivo anticancer effect of Se@Cur is an important indicator for the application in the medical field.49 Therefore, the dose dependent experiment of tumor weight and body weight were performed to evaluate its in vivo anticancer activity. As shown in Fig. 10A, the HepG2 xenogragts nude mice were treated with Se@Cur at the dose of 3, 6, 9 mg kg−1 for a period of 16 days, and the tumor weight after last treatment were 0.81 ± 0.03 g, 0.60 ± 0.04 g and 0.31 ± 0.02 g, respectively. Similarly, after treatment with different concentration of Se@Cur, the body weights were 16 ± 0.46 g, 15 ± 0.41 g and 14 ± 0.42 g, respectively (Fig. 10B). Taken together, compare to control group, there was no statistically significant difference in body weight, but no distinct reduction was observed in the body weight. Images of the mice with different size of tumors were showed in Fig. S4.† The dates suggested that Se@Cur significantly inhibited the growth of tumor while exhibited low side effect. Taken together, these results indicated the effective in vivo anticancer capacity of Se@Cur.4 Conclusions
In conclusion, this paper demonstrated a chemical method to synthesize curcumin loaded selenium nanoparticle (Se@Cur). Se@Cur exhibited superior abilities to enhance the solubility of drug, inhibit cell proliferation and suppress cell migration. Furthermore, the mechanistic investigation revealed that Se@Cur activated caspase-3 mediated HepG2 cells apoptosis by ROS overproduction. Moreover, Se@Cur induced HepG2 cells apoptosis via P53 and AKT signaling pathways. The in vivo experiments showed injection of mice with Se@Cur effectively inhibited the tumor growth. Taken together, for achieving anticancer activity, the strategy to use SeNPs as a carrier of curcumin could be a highly efficient way. Se@Cur may be a candidate for next assessment as a chemotherapeutics of cancers, especially HCC.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Technology Planning Project of Guangzhou (2018-1002-SF-0673), the Technology Planning Project of Guangdong Province (No. 2014A020212697), the Technology Planning Project of Guangdong (No. 201607010120), The Science and Technology Planning Project of Guangdong Province (No. 2014A020212024),The Science and Technology Planning Project of Guangdong Province (No. 2015A020211002), the Medical science and technology project of Guangzhou Municipal Health Bureau (No. 0005559A11105033).Notes and references
- J. Lan, N. Wang, L. Huang, Y. Liu, X. Ma, H. Lou, C. Chen, Y. Feng and W. Pan, Eur. J. Med. Chem., 2017, 127, 554–566 CrossRef CAS PubMed
.
- B. Zhu, L. Sun, W. Luo, M. Li, D. H. Coy, L. Yu and W. Yu, Oncotarget, 2017, 8, 23289–23302 Search PubMed
.
- M. Li, W. Zhang, B. Wang, Y. Gao, Z. Song and Q. C. Zheng, Int. J. Nanomed., 2016, 11, 5645–5669 CrossRef PubMed
.
- A. G. Duffy, S. V. Ulahannan, O. Makorova-Rusher, O. Rahma, H. Wedemeyer, D. Pratt, J. L. Davis, M. S. Hughes, T. Heller, M. ElGindi, A. Uppala, F. Korangy, D. E. Kleiner, W. D. Figg, D. Venzon, S. M. Steinberg, A. M. Venkatesan, V. Krishnasamy, N. Abi-Jaoudeh, E. Levy, B. J. Wood and T. F. Greten, J. Hepatol., 2017, 66, 545–551 CrossRef CAS PubMed
.
- Z. H. Ye, L. Gao, D. Y. Wen, Y. He, Y. Y. Pang and G. Chen, OncoTargets Ther., 2017, 10, 1711–1723 CrossRef PubMed
.
- X. F. Zhang, X. Yang, H. L. Jia, W. W. Zhu, L. Lu, W. Shi, H. Zhang, J. H. Chen, Y. F. Tao, Z. X. Wang, J. Yang, L. X. Wang, M. Lu, Y. Zheng, J. Zhao, Q. Z. Dong and L. X. Qin, Cancer Biol. Med., 2016, 13, 459–468 CrossRef PubMed
.
- R. Lencioni, J. M. Llovet, G. Han, W. Y. Tak, J. Yang, A. Guglielmi, S. W. Paik, M. Reig, D. Y. Kim, G. Y. Chau, A. Luca, L. R. del Arbol, M. A. Leberre, W. Niu, K. Nicholson, G. Meinhardt and J. Bruix, J. Hepatol., 2016, 64, 1090–1098 CrossRef CAS PubMed
.
- R. Dutta and R. I. Mahato, Pharmacol. Ther., 2017, 173, 106–117 CrossRef CAS PubMed
.
- J. H. Yoon, J. I. Choi, Y. Y. Jeong, A. Schenk, L. Chen, H. Laue, S. Y. Kim and J. M. Lee, J. Hepatol., 2016, 65, 1155–1162 CrossRef CAS PubMed
.
- G. Roviello, L. Zanotti, M. R. Cappelletti, A. Gobbi, G. Borsella, C. Pacifico, A. G. Multari and D. Generali, Crit. Rev. Oncol. Hematol., 2016, 108, 62–68 CrossRef PubMed
.
- C. H. Wang, K. C. Wey, L. R. Mo, K. K. Chang, R. C. Lin and J. J. Kuo, Asian Pac. J. Cancer Prev., 2015, 16, 3595–3604 CrossRef PubMed
.
- R. Barbara, D. Belletti, F. Pederzoli, M. Masoni, J. Keller, A. Ballestrazzi, M. A. Vandelli, G. Tosi and A. M. Grabrucker, Int. J. Pharm., 2017, 526, 413–424 CrossRef CAS PubMed
.
- M. A. Abdel-Wahhab, A. S. Salman, M. I. Ibrahim, A. A. El-Kady, S. H. Abdel-Aziem, N. S. Hassan and A. I. Waly, Food Chem. Toxicol., 2016, 94, 159–171 CrossRef CAS PubMed
.
- W. R. Garcia-Nino and J. Pedraza-Chaverri, Food Chem. Toxicol., 2014, 69, 182–201 CrossRef CAS PubMed
.
- S. Kim, M. J. Stebe, J. L. Blin and A. Pasc, J. Mater. Chem. B, 2014, 2, 7910–7917 RSC
.
- W. Zhong, K. Qian, J. Xiong, K. Ma, A. Wang and Y. Zou, Biomed. Pharmacother., 2016, 83, 302–313 CrossRef CAS PubMed
.
- L. Wang, X. Chen, Z. Du, G. Li, M. Chen, X. Chen, G. Liang and T. Chen, J. Exp. Clin. Cancer Res., 2017, 36, 47 CrossRef PubMed
.
- M. Kumari, L. Ray, M. P. Purohit, S. Patnaik, A. B. Pant, Y. Shukla, P. Kumar and K. C. Gupta, Eur. J. Pharm. Biopharm., 2017, 117, 346–362 CrossRef CAS PubMed
.
- K. Margulis, S. Srinivasan, M. J. Ware, H. D. Summers, B. Godin and S. Magdassi, J. Mater. Chem. B, 2014, 2, 3745–3752 RSC
.
- K. V. Jardim, G. A. Joanitti, R. B. Azevedo and A. L. Parize, Mater. Sci. Eng., C, 2015, 56, 294–304 CrossRef CAS PubMed
.
- J. S. Baek and C. W. Cho, Eur. J. Pharm. Biopharm., 2017, 117, 132–140 CrossRef CAS PubMed
.
- M. M. Wen, N. S. El-Salamouni, W. M. El-Refaie, H. A. Hazzah, M. M. Ali, G. Tosi, R. M. Farid, M. J. Blanco-Prieto, N. Billa and A. S. Hanafy, J. Controlled Release, 2017, 245, 95–107 CrossRef CAS PubMed
.
- D. Sobot, S. Mura and P. Couvreur, J. Mater. Chem. B, 2016, 4, 5078–5100 RSC
.
- Y. F. Song, Y. Q. Xie, J. J. Yang, R. Q. Li, X. Jin and J. Yang, J. Mater. Chem. B, 2016, 4, 6588–6596 RSC
.
- L. B. Naves, C. Dhand, J. R. Venugopal, L. Rajamani, S. Ramakrishna and L. Almeida, Prog. Biomater., 2017, 6, 13–26 CrossRef PubMed
.
- Z. Cui, D. Liu, C. Liu and G. Liu, Medicine, 2017, 96, e5944 CrossRef CAS PubMed
.
- L. Wang, J. Wang, X. Liu, Q. Liu, G. Zhang and L. Liang, Biosci. Rep., 2016, 36, e000395 Search PubMed
.
- M. Vinceti, G. Dennert, C. M. Crespi, M. Zwahlen, M. Brinkman, M. P. Zeegers, M. Horneber, R. D'Amico and C. Del Giovane, Cochrane Database Syst. Rev., 2014, 3, CD005195 Search PubMed
.
- Y. Zhang, X. Li, Z. Huang, W. Zheng, C. Fan and T. Chen, Nanomedicine, 2013, 9, 74–84 CrossRef CAS PubMed
.
- P. D. Terry, B. Qin, F. Camacho, P. G. Moorman, A. J. Alberg, J. S. Barnholtz-Sloan, M. Bondy, M. L. Cote, E. Funkhouser, K. A. Guertin, E. S. Peters, A. G. Schwartz, J. M. Schildkraut and E. V. Bandera, J. Nutr., 2017, 147, 621–627 CrossRef CAS PubMed
.
- X. Cai, C. Wang, W. Yu, W. Fan, S. Wang, N. Shen, P. Wu, X. Li and F. Wang, Sci. Rep., 2016, 6, 19213 CrossRef CAS
.
- D. L. Hatfield, P. A. Tsuji, B. A. Carlson and V. N. Gladyshev, Trends Biochem. Sci., 2014, 39, 112–120 CrossRef CAS PubMed
.
- F. Maiyo and M. Singh, Nanomedicine, 2017, 12, 1075–1089 CrossRef CAS PubMed
.
- M. Zuberek, D. Wojciechowska, D. Krzyzanowski, S. Meczynska-Wielgosz, M. Kruszewski and A. Grzelak, J. Nanobiotechnol., 2015, 13, 72 CrossRef PubMed
.
- H. Zhang, A. M. Gomez, X. Wang, Y. Yan, M. Zheng and H. Cheng, Cardiovasc. Res., 2013, 98, 248–258 CrossRef CAS PubMed
.
- L. A. Sena and N. S. Chandel, Mol. Cell, 2012, 48, 158–167 CrossRef CAS PubMed
.
- J. Zhang, X. Wang, V. Vikash, Q. Ye, D. Wu, Y. Liu and W. Dong, Oxid. Med. Cell. Longevity, 2016, 2016, 4350965 Search PubMed
.
- Y. Li, Z. Lin, M. Zhao, T. Xu, C. Wang, H. Xia, H. Wang and B. Zhu, Int. J. Nanomed., 2016, 11, 3065–3076 CrossRef CAS PubMed
.
- Y. H. Li, Z. F. Lin, T. T. Xu, C. B. Wang, M. Q. Zhao, M. S. Xiao, H. Z. Wang, N. Deng and B. Zhu, RSC Adv., 2017, 7, 1453–1463 RSC
.
- Y. H. Li, Z. F. Lin, M. Q. Zhao, T. T. Xu, C. B. Wang, L. Hua, H. Z. Wang, H. M. Xia and B. Zhu, ACS Appl. Mater. Interfaces, 2016, 8, 24385–24393 CAS
.
- Y. H. Li, Z. F. Lin, M. Q. Zhao, M. Guo, T. T. Xu, C. B. Wang, H. M. Xia and B. Zhu, RSC Adv., 2016, 6, 89679–89686 RSC
.
- Y. Li, M. Guo, Z. Lin, M. Zhao, M. Xiao, C. Wang, T. Xu, T. Chen and B. Zhu, Int. J. Nanomed., 2016, 11, 6693–6702 CrossRef PubMed
.
- B. Zhu, Y. Li, Z. Lin, M. Zhao, T. Xu, C. Wang and N. Deng, Nanoscale Res. Lett., 2016, 11, 198 CrossRef PubMed
.
- Y. H. Li, X. L. Li, Y. S. Wong, T. F. Chen, H. B. Zhang, C. R. Liu and W. J. Zheng, Biomaterials, 2011, 32, 9068–9076 CrossRef CAS PubMed
.
- Z. F. Lin, Y. H. Li, M. Guo, T. T. Xu, C. B. Wang, M. Q. Zhao, H. Z. Wang, T. F. Chen and B. Zhu, RSC Adv., 2017, 7, 742–750 RSC
.
- Y. H. Li, X. L. Li, W. J. Zheng, C. D. Fan, Y. B. Zhang and T. F. Chen, J. Mater. Chem. B, 2013, 1, 6365–6372 RSC
.
- N. M. Khan, A. Haseeb, M. Y. Ansari, P. Devarapalli, S. Haynie and T. M. Haqqi, Free Radical Biol. Med., 2017, 106, 288–301 CrossRef CAS PubMed
.
- G. Wang, T. Zhang, W. Sun, H. Wang, F. Yin, Z. Wang, D. Zuo, M. Sun, Z. Zhou, B. Lin, J. Xu, Y. Hua, H. Li and Z. Cai, Free Radical Biol. Med., 2017, 106, 24–37 CrossRef CAS PubMed
.
- Y. Huang, L. He, W. Liu, C. Fan, W. Zheng, Y. S. Wong and T. Chen, Biomaterials, 2013, 34, 7106–7116 CrossRef CAS PubMed
.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra08796a |
| ‡ Min Guo and Yinghua Li contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2017 |