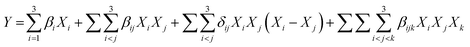Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Optimization of a minimal synergistic enzyme system for hydrolysis of raw cassava pulp†
Benjarat Bunterngsooka,
Thanaporn Laothanachareona,
Suda Natrchalayutha,
Sirithorn Lertphanichb,
Tatsuya Fujiic,
Hiroyuki Inouec,
Chalermkiart Youngthonga,
Duriya Chantasingha,
Lily Eurwilaichitra and
Verawat Champreda *a
*a
aEnzyme Technology Laboratory, Microbial Biotechnology and Biochemicals Research Unit, National Center for Genetic Engineering and Biotechnology, 113 Thailand Science Park, Phahonyothin Road, Khlong Luang, Pathumthani 12120, Thailand. E-mail: verawat@biotec.or.th; Fax: +66 2564 6707; Tel: +66 2564 6700 ext. 3446
bCassava and Starch Technology Research Laboratory, National Center for Genetic Engineering and Biotechnology, Bangkok 10900, Thailand
cResearch Institute for Sustainable Chemistry, National Institute of Advanced Industrial Science and Technology, 3-11-32 Kagamiyama, Higashi-Hiroshima, Hiroshima 739-0046, Japan
First published on 16th October 2017
Abstract
Cassava pulp is an underused agricultural by-product comprising residual starch granules entrapped in cell wall polysaccharides, making it unique from other lignocellulosic wastes in terms of enzymatic processing. In this study, a synergistic system comprising a minimal set of enzymes tailor-made for efficient bioprocessing of raw cassava pulp was developed. The fiber-degrading enzyme mixture comprises an endoglucanase (Cel12), β-glucosidase (BGL), endo-β-1,4-xylanase (XYN) and endo-polygalacturonase (EPG) from Aspergillus aculeatus and a cellobiohydrolase I (Cel7A) from Talaromyces cellulolyticus was initially optimized using the experimental design approach. A glucose recovery yield of 91.3% based on the total starch and cellulose content was obtained from saccharification of cassava pulp using the combination of 5.0 mg g−1 of fiber degrading enzyme mixture comprising Cel12, Cel7A, BGL, XYN and EPG in the ratio of 16.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25.5
25.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15.0
15.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 18.0
18.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25.0 together with 0.5 mg g−1 of raw starch degrading enzyme Stargen™ 002 with the degree of synergy of 1.43. Efficient hydrolysis was achieved without an energy-intensive pretreatment step, showing industrial applicability on saccharification and modification of cassava pulp and for further incorporation of these enzymes into ethanologens for consolidated bioprocessing.
25.0 together with 0.5 mg g−1 of raw starch degrading enzyme Stargen™ 002 with the degree of synergy of 1.43. Efficient hydrolysis was achieved without an energy-intensive pretreatment step, showing industrial applicability on saccharification and modification of cassava pulp and for further incorporation of these enzymes into ethanologens for consolidated bioprocessing.
1. Introduction
Conversion of agricultural wastes to value-added products such as biofuels, biochemicals, and biomaterials can help alleviate the dependence on non-renewable resources for making these products, while at the same time solves environmental problems related to agricultural waste disposal.1 Cassava (Manihot esculanta) is one of the most economically important crops in tropical and subtropical regions, ranking as the world's 5th most staple crop with an annual production of 288 million tons in 2016.2 Processing of cassava in the starch industry generates cassava pulp as the major by-product with approximately 10% of the weight of the starting material. The pulp contains approximately 60% residual starch by weight, while the rest comprises cell wall polysaccharides. The high residual starch content makes it a unique starting material for biorefineries compared with other second-generation cellulosic substrates, such as sugarcane bagasse, rice straw, and corn wastes. With its combined starch–cellulosic based nature, cassava pulp can be considered as a generation 1.5 feedstock for biorefinery. It is currently used for low-value products such as animal feed, or discarded as waste.3,4 However, its potential for conversion to a range of commodity fermentation products including various biofuels and chemicals has recently emerged.5–7Bioprocessing of cassava pulp, in general, is an energy-intensive process due to the requirement for pre-gelatinization of raw starch granules entrapped in the fibrous network of cell wall polysaccharides and liquefaction of released starch by a thermostable α-amylase at high temperature, followed by maltodextrin saccharification by a glucoamylase.8 Efficient non-thermal enzymatic hydrolysis of dried cassava pulp with no prior high temperature pre-gelatinization step using various combinations of crude commercial cellulolytic, hemicellulolytic, pectinolytic, and amylolytic enzymes compared with a crude complex enzyme with multi-cell wall and starch degrading activities from Aspergillus aculeatus has been reported.9 However, there has been no systematic dissection of the specific composite enzyme activities required for direct hydrolysis of cassava pulp in deeper details into their specificity levels, which is necessary to develop a more effective enzyme for processing of cassava pulp in bio-industry and for genetic modification of ethanologens for consolidated bioprocessing of this unique substrate as demonstrated for other cellulosic wastes.10,11
Decomposition of lignocellulosic materials requires a repertoire of core cellulolytic and accessory hemicellulolytic enzymes in different glycosyl hydrolase families, which function in synergistic and cooperative manners in order to hydrolyze the recalcitrant cell wall structure. Designing an effective enzyme mixture comprising a minimal set of hydrolytic enzymes is a promising strategy to develop highly efficient enzyme systems with specificity towards the target cellulosic substrates. Several tailor-made enzyme mixtures from various microbial origins have been reported for hydrolysis of pretreated wheat straw, rice straw and corn stover.12,13 Most works have tested different combinations of crude or commercial enzymes however, with very few reports on synergistic mixtures of recombinant or isolated hydrolases from different families with defined specificities, for examples, on hydrolysis of wheat straw and rice straw.13–15 Further enhancements of the released sugar yield with additional non-hydrolytic enzymes or auxiliary components (e.g. bacterial expansins and lytic polysaccharide monooxygenases) have also been reported.16,17 Owing to the unique starch–cellulosic characteristic of cassava pulp and its high content of pectic substance,5 optimization of an active enzyme system with defined specificities for its decomposition with no prior high-temperature pretreatment step is a great challenge. In this study, an efficient synergistic enzyme system comprising a minimal set of glycosyl hydrolases has been created by a systematic mixture design approach using selected recombinant enzymes from different fungal origins. The work gives insights into the development of a highly-effective enzyme system for direct saccharification of cassava pulp as well as modification of this unique feedstock in bio-industry.
2. Experimental
2.1 Substrate
Cassava pulp were kindly supplied by Chorchaiwat Industry Co. Ltd. (Bangkok, Thailand). Starch content was determined using the polarimetric method, according to the standard method of the Commission of the European Communities (1999)18 and Thai Industrial Standard for Tapioca Flour/Starch.19 Neutral and acid detergent fibers (NDF and ADF) and lignin were analyzed using the standard AOAC method (1998) (Method 992.16 and Method 973.18). The native pulp was de-starched by α-amylase (Speczyme®Alpha, DuPont, Rochester, NY) at 100 °C, pH 6.0 for 2 h. Thereafter, the sample was then washed with water thoroughly and the pulp was dried in an oven at 45 °C for overnight. The chemical compositions of the cassava pulp fiber were determined according to the standard NREL analysis method for lignocellulosic components.20 The native and de-starched cassava pulps were physically processed by milling, and were sieved through a 3 mm screen. The biomass was stored at room temperature before experimental study.2.2 Strains, enzymes, and plasmids
Expression vector pPICZαA (Invitrogen, Carlsbad, CA) was used for expression of the recombinant enzymes. Escherichia coli DH5a was used as the host strain for DNA cloning. Pichia pastoris (Invitrogen, Carlsbad, CA) was used for recombinant protein expression. The recombinant GH7 cellobiohydrolase I (Cel7A) from Talaromyces cellulolyticus was produced using the homologous expression system and purified as described previously.21 Recombinant plasmid pPICZα-Xyn encoding a GH10 endo-β-1,4-xylanase from A. aculeatus BCC17849 was constructed and reported in Laothanachareon et al., 2015,13 and the recombinant xylanase (XYN) was prepared as described below. The commercial raw starch degrading enzyme STARGEN™ 002, containing α-amylase from Aspergillus kawachii and gluco-amylase from Trichoderma reseei was purchased from Dupont (Rochester, NY). A. aculeatus BCC17849 was obtained from the BIOTEC Culture Collection, Thailand (http://www.biotec.or.th/bcc) and maintained on potato dextrose agar (PDA). Polysaccharides used as substrates in enzymatic activity analysis were obtained from Sigma-Aldrich.2.3 Recombinant plasmid construction and enzyme expression
The mature genes encoding a GH12 endoglucanase (Cel12) and an endopolygalacturonase (EPG) were amplified from cDNA of A. aculeatus BCC17849 prepared according to the method described in Laothanachareon et al., 2015.13 Cel12 gene was amplified using Cel12/F (5′GAATTCCAACAGACCCAACTCTGCGATCAA3′) and Cel12/R (5′TCTAGACTACTGTACGCTGGC AGACCAGTC3′) primers; while EPG gene was amplified using EFP/F (5′GCATGAATTCGCTCCCACCGACATCGAGAAGCG3′) and EPG/R (5′TACATCTAGATTAGCAAGAGGCGC CGGAAGG3′) primers, EcoRI and XbaI sites were underlined, respectively. The mature gene encoding a GH1 β-glucosidase (BGL) from A. aculeatus (GenBank accession no. P48825) was synthesized with codon optimization for expression in P. pastoris (ESI data Fig. S1†) (Genescript, Piscataway NJ). The gene fragments were digested with EcoRI and XbaI, and cloned into pPICZα expression vector digested with the same restriction enzymes. The recombinant plasmids were transformed into E. coli DH5α using the heat shock method22 and selected on LB agar containing 25 μg mL−1 of Zeocin. The inserted genes were verified by colony PCR and subsequently by DNA sequencing (Macrogen, Seoul, South Korea). The recombinant plasmids pPICZα-Cel12, pPICZα-EPG and pPICZα-BGL were linearized by PmeI and transformed into P. pastoris KM71 by electroporation at 1.5 kV cm−1, 200 Ω and 25 μF. The yeast transformants were selected on YPD agar containing 100 μg mL−1 of Zeocin and then Zeocin-resistant clones were screened for gene-integrated clones by colony PCR screening using AOX1 forward and AOX1 reverse primers.The recombinant strains containing plasmids pPICZα-XYN, pPICZα-Cel12, pPICZα-EPG and pPICZα-BGL were cultured in 200 mL BMGY [2% peptone, 1% yeast extract, 100 mM potassium phosphate pH 6.0, 1.34% yeast nitrogenous base (w/o amino acid), 0.4 μg mL−1 biotin, and 1% glycerol] until the OD600 of the culture reached 8–10, and then the cells were pelleted by centrifugation. The cell pellets were resuspended in 20 mL BMMY [BMGY containing 3% methanol instead of glycerol] for induction of the target gene. The cells were then cultivated under the same conditions for 3 d, and then the supernatant was collected by centrifugation at 5000 × g for 5 min at 4 °C. The enzyme was concentrated and desalted using a Macrosep column molecular weight cut-off 10 kDa (Pall, Port Washington, NY, USA). The enzyme was exchanged to 100 mM sodium acetate buffer (pH 5.0) prior to subsequent experimental studies. Concentration of the purified enzyme was determined using the Bradford assay kit (Biorad, Hercules, CA, USA).
2.4 Enzyme activity assay
Polysaccharide degrading activities were analyzed based on the amount of liberated reducing sugars using the 3,5-dinitrosalisylic acid (DNS) method.23 A one milliliter reaction mixture contained the appropriate dilution of enzyme in 100 mM sodium acetate buffer, pH 5.0 and the appropriate substrate: 1% (w/v) carboxymethyl cellulose for endo-glucanase activity; beechwood xylan for xylanase activity; microcrystalline cellulose Avicel PH101 for cellobiohydrolase activity; polygalacturonic acid for endopolygalacturonase activity and cassava starch for raw starch degrading activity. The reaction was incubated at 50 °C for 10 min for endo-glucanase, xylanase endopolygalacturonase and raw starch degrading activities, and 60 min for cellobiohydrolase activity. The amount of reducing sugars was determined from the absorbance measurement at 540 nm and interpolated from a standard curve of the corresponding sugar. The β-glucosidase activity was analyzed based on the amount of p-nitrophenol liberated from p-nitrophenyl-β-D-glucopyranoside in the same buffer at 50 °C. One unit of enzyme activity was defined as the amount of enzyme that liberated 1 μmole of product per min. For optimal temperature determination, the enzyme activity was determined in the temperature range of 30 to 70 °C in 100 mM sodium acetate (pH 5.0). For optimal pH determination, the activities were measured at 50 °C in the pH range of 4.0 to 9.0 using 100 mM sodium acetate (pH 4.0 to 5.5), 100 mM sodium phosphate (pH 6.0 to 8.0) and 100 mM Tris–HCl (pH 8 to 9.0) under the standard assay conditions. The kinetic parameters (Vmax and Km) were determined using by fitting the initial velocity data to Michaelis–Menten equation using Kaleida Graph data analysis software (Synergy Software, Reading, PA).2.5 Experimental mixture design
The interactions among the four recombinant fiber degrading enzymes on hydrolysis of de-starched cassava pulp were studied using an experimental mixture design approach.24 A [3,3]-augmented simplex lattice design implemented in the Minitab 17.0 software (Minitab Inc., State College, PA) was used to define an optimal enzyme mixture based on the glucose yield. The design contained 15 experimental points, which were examined in triplicate with four components and a lattice degree of two. In the mixture design, the sum of all components was fixed as 5 mg g−1 biomass on a dried weight basis. Three independent variables in the mixture design consisted of Cel12 (X1), Cel7A (X2), BGL (X3) and XYN (X4) for hydrolysis of 1% (w/v) of de-starched cassava pulp in 100 mM sodium acetate buffer (pH 5.0). The reaction was incubated at 50 °C with shaking at 200 rpm for 72 h. The reducing sugar released was measured using the DNS method (Miller, 1959). The sugar profile in the liquid fractions of the hydrolysis reaction was determined on a UltiMate® 3000 high performance liquid chromatography (Dionex, Sunnyvale, CA) equipped with a refraction index (RI) detector (Shodex, New York, NY) using an Aminex HPX-87H column (Bio-Rad, Hercules, CA) operating at 65 °C with 5 mM H2SO4 as the mobile phase at a flow rate of 0.5 mL min−1. The reducing sugar yield (Y1) was applied as dependent variables for the analysis and simulation of the respondent model. After the regression analysis, the full cubic model was used to predict the optimized ratio of the mixture components. The canonical form of the full cubic model is described by the following equation:whereas Y is the predicted response, βi is a linear coefficient, βij is a quadratic coefficient and βijk is a cubic coefficient. δij is a parameter of the model. βiXi represents the linear blending portion and the parameter βij represents either synergistic or antagonistic blending.
The degree of synergism was calculated according to the following equation:25
2.6 Scanning electron microscopy
The microstructure of the native and enzyme-treated cassava pulp was analyzed using a SU5000 scanning electron microscope (SEM) (Hitachi, Tokyo, Japan). The samples were dried and coated with gold. An electron beam energy of 10 kV was used for analysis.3. Results and discussion
3.1 Composition of raw and de-starched cassava pulp
The chemical composition of raw and de-starched cassava pulp used in this study is shown in Table 1. Raw cassava pulp principally comprised starch (51.01% of dry matter), with low contents of cellulose (14.75% of dry matter) and hemicellulose (3.31% of dry matter). It also contained 4.50% ash and sand. De-starched cassava pulp was mainly composed of fibrous material containing cellulose and hemicellulose (35.95 and 22.41% of dry matter, respectively) and 17.78% lignin with a minor starch content (0.61% of dry matter). The starch granules in raw cassava pulp are typically attached and trapped in the fibrous cell wall structure (ESI data Fig. S2†). In contrast, the de-starched pulp demonstrates a fibrous structure containing mainly of lignocellulosic substances.26 The compositions of the native cassava pulp in this study are within the range previously reported where they varied according to the processing conditions and crop varieties.5,27 The content of lignin in de-starched pulp was relatively low, suggesting in its higher hydrolysable nature compared to most lignocellulosic materials.3.2 Characterization of recombinant glycosyl hydrolases
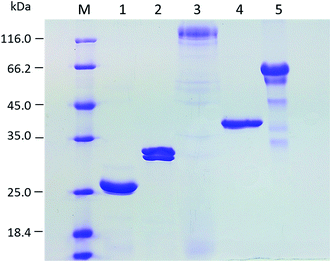
The set of enzymes used in this study has been selected from our in-house enzyme collection in a pre-screening experiment for their activities on hydrolysis of cassava pulp (data not shown). The sources of the enzymes with their specific activities are summarized in ESI data Table S1.† Four of the composite enzymes (Cel12, BGL, XYN, and EPG) in the optimized mixture for degradation of cassava pulp fiber are of A. aculeatus origin. This fungus has been previously reported as a source of raw starch degrading enzyme systems and a lignocellulose–degrading enzyme complex according to proteomic analysis.28 Its applications on direct low-temperature saccharification of cassava pulp9 and viscosity reduction of cassava root mash29,30 have been demonstrated. Heterologous expression of a debranching α-arabinofuranosidase and a pectin esterase from this fungus and their synergy with the commercial cellulase Accelerase® 1500 has been recently reported, showing potential of its vast array of plant biomass degrading hydrolytic and non-hydrolytic enzymes for hydrolysis of lignocelluloses.13 T. cellulolyticus is a potent cellulose producer possessing a highly efficient cellulase system. The strong activity of its cellobiohydrolase on synergistic hydrolysis of crystalline cellulose has been demonstrated.31Induction of heterologous gene expression in recombinant yeast strains led to production of the target enzymes in the culture supernatants (Fig. 1). The culture supernatant of yeast expressing Cel12 showed a single protein band with the molecular weight of 25 kDa, with more than 90% homogeneity. Two protein species of Mw between 30–35 kDa were observed in the culture supernatant of yeast expressing XYN. The culture supernatant of yeast expressing BGL showed a major band at 116 kDa with a series of minor bands with lower molecular weights, suggesting hyper-glycosylation of the target protein. EPG was expressed as a single band with the expected Mw of 35 kDa. The purified cellobiohydrolase Cel7A from T. cellulolyticus showed >90% purity with an apparent Mw of 60 kDa.
The enzymes used in this study showed the optimal working conditions in the range of 55 to 60 °C and pH 5.0 to 6.0 (ESI data Fig. S3†). Their kinetic parameters are provided in ESI data Table S1.† The conditions for the hydrolysis biomass reactions in this study was set at 50 °C, pH 5.0 which was slightly under their optimal temperature for maintaining enzyme stability during the hydrolysis process. Under biomass saccharification conditions in this work (50 °C at pH 5.0, see ESI data Table S2†), cellobiohydrolase Cel7A showed a specific activity of 0.075 U mg−1 against Avicel with a minor xylanase activity (0.05 U mg−1) against beechwood xylan. Cel12 showed CMCase activity with the specific activity of 9.31 U mg−1 with the side activity of 2.75 U mg−1 on beechwood xylan. XYN exhibited a specific activity of 389.80 U mg−1 against beechwood xylan with a trace CMCase activity (0.15 U mg−1 protein). BGL showed β-glucosidase activity of 0.33 U mg−1. EPG showed a specific activity of 169![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 089 U mg−1 based on hydrolysis of polygalacturonic acid. All enzymes retained >75% relative activity under the operational conditions in this study.
089 U mg−1 based on hydrolysis of polygalacturonic acid. All enzymes retained >75% relative activity under the operational conditions in this study.
3.3 Synergistic action of cellulase and hemicellulase on saccharification of de-starched cassava pulp
De-starched cassava pulp represents the fibrous part of the native pulp and consists mainly of lignocellulosic components. In this step, an optimal mixture of the key cellulolytic and hemicellulolytic enzymes for effective hydrolysis of this substrate was sought and synergistic interactions among these enzymes were systematically studied using the four component mixture design approach. The quaternary enzyme mixture comprised the core cellulases (Cel12, Cel7A, and BGL), and an endo-β-1,4-xylanase (XYN) selected from different ascomycete origins. The amount of each component was indicated as the amount in the mixture based on the component weight ratio. According to this method, the measured response, i.e. reducing sugar and glucose yields, was assumed to depend on the proportions of the composite enzymes present in the mixture. All 15 experimental points were located inside the triangular graph, which means that the sum of all enzyme components for every experimental point was always 100%, which was equal to a total protein loading of 5 mg g−1 biomass. The sugar yields obtained from the mixture design experiment are shown in Table 2.| Run no. | Enzyme loading (mg g−1 biomass) | Sugar released (mg g−1 biomass) | ||||
|---|---|---|---|---|---|---|
| Cel12 | Cel7A | BGL | XYN | Glucose | Reducing sugar | |
| 1 | 5.000 | 0.000 | 0.000 | 0.000 | 4.08 | 12.10 |
| 2 | 2.500 | 2.500 | 0.000 | 0.000 | 12.29 | 87.93 |
| 3 | 2.500 | 0.000 | 2.500 | 0.000 | 7.85 | 12.01 |
| 4 | 2.500 | 0.000 | 0.000 | 2.500 | 4.46 | 14.40 |
| 5 | 0.000 | 5.000 | 0.000 | 0.000 | 1.91 | 10.79 |
| 6 | 0.000 | 2.500 | 2.500 | 0.000 | 25.57 | 21.46 |
| 7 | 0.000 | 2.500 | 0.000 | 2.500 | 1.66 | 24.80 |
| 8 | 0.000 | 0.000 | 5.000 | 0.000 | 3.41 | 4.16 |
| 9 | 0.000 | 0.000 | 2.500 | 2.500 | 2.25 | 6.87 |
| 10 | 0.000 | 0.000 | 0.000 | 5.000 | 1.26 | 5.84 |
| 11 | 1.250 | 1.250 | 1.250 | 1.250 | 91.44 | 124.27 |
| 12 | 3.125 | 0.625 | 0.625 | 0.625 | 77.17 | 103.35 |
| 13 | 0.625 | 3.125 | 0.625 | 0.625 | 102.03 | 138.64 |
| 14 | 0.625 | 0.625 | 3.125 | 0.625 | 76.65 | 96.90 |
| 15 | 0.625 | 0.625 | 0.625 | 3.125 | 77.45 | 110.43 |
Only a small amount of glucose (1.26–4.08 mg g−1 biomass) was obtained from the individual enzymes (run no. 1, 5, 8 and 10). The binary combinations of Cel12 with Cel7A (run no. 2) and Cel7A and BGL (run no. 6) led to increased glucose yields (12.28 and 25.56 mg g−1 biomass, respectively). On the other hand, the supplementation of Cel7A and XYN (no. 7) exhibited only minor synergistic effect on releasing of glucose. The highest glucose yield of 102.02 mg g−1 biomass was achieved using the combination of four component enzymes in the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio of Cel12
1 ratio of Cel12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Cel7A
Cel7A![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) BGL
BGL![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) XYN (no. 13). This yield was slightly higher than that from reaction no. 11 comprising a 1
XYN (no. 13). This yield was slightly higher than that from reaction no. 11 comprising a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio of Cel12
1 ratio of Cel12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Cel7A
Cel7A![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) BGL
BGL![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) XYN (91.43 mg g−1 biomass). The highest reducing sugar yield of 138.64 mg g−1 was also achieved under the same conditions. The difference between the released glucose and reducing sugar could reflect the presence of cello-oligomers due to the absence of the downstream processing β-glucosidase activity, or the effects of XYN on releasing hemicellulose derived oligo products.
XYN (91.43 mg g−1 biomass). The highest reducing sugar yield of 138.64 mg g−1 was also achieved under the same conditions. The difference between the released glucose and reducing sugar could reflect the presence of cello-oligomers due to the absence of the downstream processing β-glucosidase activity, or the effects of XYN on releasing hemicellulose derived oligo products.
The response data for the glucose yield were then analyzed using a multiple regression analysis based on the full cubic model (R2 = 89.80%, p-value < 0.05). The results of the analysis of variance (ANOVA) analysis are shown in Table 3. For a single factor, positive correlations of all components on sugar yield were observed with the coefficients of 3.23–6.98. Interestingly, among the binary combinations, only Cel7A × BGL showed a significantly positive interaction with the coefficient of 94.03 at p-value < 0.05. The highest degree of synergism was found on the ternary combinations comprising Cel12/Cel7A/XYN with the maximal coefficient of 2803.32 followed by Cel7A/BGL/XYN, and Cel12/Cel7A/BGL, respectively. The results thus indicated strong synergistic interaction among the four enzymes on hydrolysis of the cassava pulp fiber.
| Factor | Coefficient | SE | T | p-Value |
|---|---|---|---|---|
a S = 14.3316 PRESS = 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 246.3, R2 = 89.80% R2(pred) = 81.98% R2(adj) = 85.52%. 246.3, R2 = 89.80% R2(pred) = 81.98% R2(adj) = 85.52%. |
||||
| Cel12 | 6.9 | 8.27 | <0.05 | <0.05 |
| Cel7A | 4.6 | 8.27 | <0.05 | <0.05 |
| BGL | 5.4 | 8.27 | <0.05 | <0.05 |
| Xyn | 3.2 | 8.27 | <0.05 | <0.05 |
| Cel12 × Cel7A | 45.8 | 40.51 | 1.13 | 0.267 |
| Cel12 × BGL | 25.4 | 40.51 | 0.63 | 0.536 |
| Cel12 × Xyn | 16.0 | 40.51 | 0.39 | 0.696 |
| Cel7A × BGL | 94.0 | 40.51 | 2.32 | 0.027 |
| Cel7A × Xyn | 6.8 | 40.51 | 0.17 | 0.868 |
| BGL × Xyn | 7.6 | 40.51 | 0.19 | 0.853 |
| Cel12 × Cel7A × BGL | 1790.2 | 1009.12 | 1.77 | 0.086 |
| Cel12 × Cel7A × Xyn | 2803.3 | 1009.12 | 2.78 | 0.009 |
| Cel12 × BGL × Xyn | 381.8 | 1009.12 | 0.38 | 0.708 |
| Cel7A × BGL × Xyn | 2474.3 | 1009.12 | 2.45 | 0.020 |
The fitted equation for the glucose yield based on the component amount after removal of the insignificant terms (p > 0.05) is shown as follows:
| Glucose yield (mg g−1 biomass) = 1.21 × Cel12 + 0.78 × Cel7A + 1.08 × BGL + 0.65 × XYN + 3.98 × Cel7A × BGL + 14.49 × Cel12 × Cel7A × BGL + 19.65 × Cel7A × BGL × XYN |
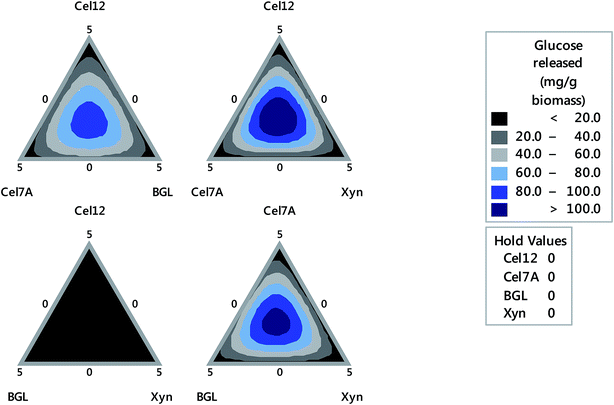
According to the ternary contour plot (Fig. 2), the area showing the greatest relative glucose yield was in the middle between the combination of Cel12, Cel7A and XYN and the combination of Cel7A, BGL and XYN axes that contributed to a glucose yield of more than 100 mg g−1 biomass. The optimized core cellulolytic/hemicellulolytic enzyme ratio was defined as the point where the maximal glucose yield was obtained, which was predicted to be 1.1, 1.7, 1.0 and 1.2 mg g−1 biomass for Cel12, Cel7A, BGL and XYN, respectively, with the expected reducing sugar yield of 112.83 mg g−1 biomass (R2 = 100%, p-value < 0.05). The experimental sugar yield of 101.72 ± 14.08 mg g−1 was obtained, which was 9.84% lower than that of the predicted value. A relatively low sugar yield for cellulose in the fiber suggested the requirement for additional enzyme activities besides the core cellulase and xylanase previously reported for most lignocellulosic substrates.
3.4 Effect of endo-polygalacturonase (EPG) on saccharification of destarched cassava pulp
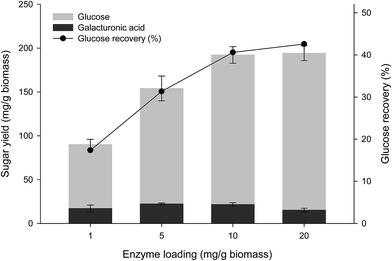
Cassava pulp is reported to contain approximately a substantial content of pectin on a dried weight basis.5 Therefore, the cooperative effects of EPG (which is the main enzyme for attacking the backbone of pectic substances) to the optimized (hemi)cellulolytic enzyme mixture were investigated. Enzymatic saccharification by the core (hemi) cellulases released 101.72 mg g−1-biomass glucose using the enzyme dosage of 5 mg g−1 (Table 4). Supplementation of EPG led to increased glucose and reducing sugar yields. The highest glucose yield of 134.79 mg g−1 was obtained from the combination of 75% core (hemi) cellulases and 25% EPG after 72 h hydrolysis. This was equivalent to the Cel12/Cel7A/BGL/XYN/EPG composition of 16.5/25.5/15.0/18.0/25.0. Regression analysis indicated a statistically significant synergistic interaction between the core cellulose mixture and EPG with the coefficient of 462.39 (p-value < 0.05) Table 5. The optimized enzyme mixture was defined as the cassava fiber degrading enzyme mixture (FD). The effect of FD dosage on substrate hydrolysis was then evaluated by varying the total enzyme dosage on hydrolysis of the de-starched cassava pulp. Glucose yield increased (72.97–179.12 mg g−1 biomass) with enzyme dosage from 1 to 20 mg g−1 biomass. Galacturonic acid was detected as a minor product component. The highest glucose yield was equivalent to a recovery yield of 42.5% based on the total glucan content at 20 mg g−1 biomass enzyme dosage (Fig. 3). A similar trend was observed for the released reducing sugar, with the highest yield of 204.33 mg g−1 at the enzyme dosage of 20 mg g−1. The low xylose and galacturonic acid yields observed were due to the presence of only the endo-acting enzymes in the mixture with no downstream β-xylosidase or galactosidase activities. The relatively low hydrolysis efficiency by the FD enzyme compared with that obtained using other cellulosic substrates (e.g. steam exploded pretreated bagasse where 51% glucose recovery was obtained from 7.5 mg g−1 enzyme dosage) would reflect physical resistance of the native fiber in cassava pulp as no prior physical or chemical pretreatment step was performed, resulting in the intact lignocellulose structure of the fiber fraction.32| Run no. | Enzyme (mg g−1 biomass) | Sugar released (mg g−1 biomass) | |||
|---|---|---|---|---|---|
| Core cellulase Opt 4° mixa | EPG | Glucose | Xylose | Galacturonic acid | |
| a Optimized Cel12/Cel7A/BGL/XYN mixture. | |||||
| 1 | 5.00 | 0.00 | 101.72 | 0.00 | 33.36 |
| 2 | 3.75 | 1.25 | 134.79 | 8.89 | 30.18 |
| 3 | 2.50 | 2.50 | 116.91 | 7.51 | 26.14 |
| 4 | 1.25 | 3.75 | 117.98 | 10.61 | 24.71 |
| 5 | 0.00 | 5.00 | 0.00 | 14.76 | 0.00 |
| Factor | Coefficient | SE | T | p-Value |
|---|---|---|---|---|
| a S = 17.9590 PRESS = 7944.75, R2 = 88.42% R2(pred) = 84.15% R2(adj) = 87.13%. | ||||
| Core cellulase (Opt 4° mix) | 65.68 | 9.573 | <0.05 | <0.05 |
| EPG | 48.68 | 9.573 | <0.05 | <0.05 |
| Core cellulase × EPG | 462.39 | 39.706 | 11.65 | 0.000 |
3.5 Saccharification of raw cassava pulp
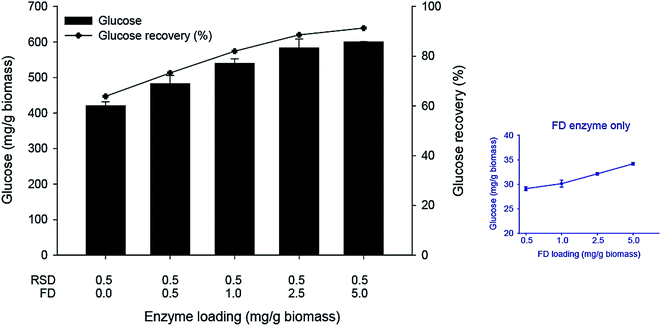
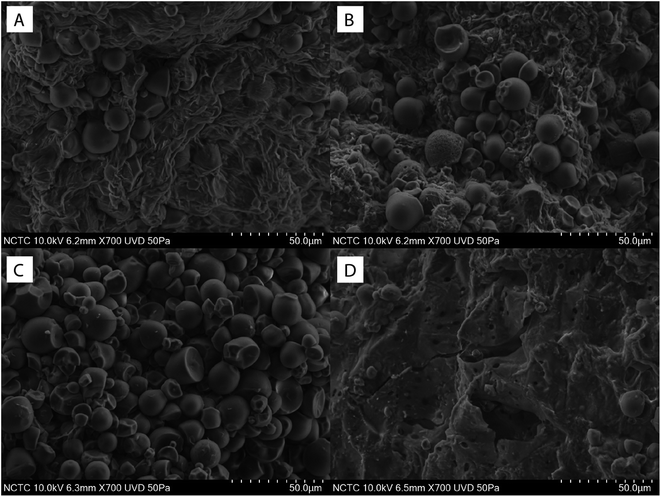
The minimal enzyme mixture for saccharification of native cassava pulp was then investigated using the optimized 5-component core fiber degrading enzymes (FD) attacking the fibrous non-starch polysaccharide part and a commercial raw starch degrading (RSD) enzyme Stargen™ 0002. The commercial enzyme showed a specific activity of 2.77 mg g−1 against raw cassava pulp with side activities on hydrolysis of lignocellulosic substrates comprising 0.44, 2.40, and 0.10 mg g−1 for CMCase, xylanase, and β-glucosidase activities, respectively under the operational conditions. The hydrolysis reactions were performed at 50 °C, which is below the onset gelatinization temperature of cassava starch (T0 = 58.5 °C).33,34 According to Fig. 4, hydrolysis of native pulp with the raw starch degrading enzyme (RSD) (0.5 mg g−1 biomass) led to a glucose yield of 420 mg g−1 biomass equivalent to 63.8% glucose recovery. In contrast, the use of FD only released a much smaller amount of glucose (29.12–34.19 mg g−1 biomass) at enzyme dosage ranging from 0.5–5.0 mg g−1 biomass. The low sugar released from raw pulp by FD suggested by steric hindrance of starch granules in the pulp structure which limited access of the FD enzymes to the fibrous part. The relatively high sugar yield from hydrolysis of raw pulp with the RSD enzyme suggested that majority of starch in the substrate was accessible to the enzymes under the experimental conditions used in which the milling step exposes the starch granules. Furthermore, the presence of low cell wall polysaccharide degrading enzyme activities in Stargen™ could also contribute to greater substrate accessibility.Supplementation of FD (0.5–5.0 mg g−1 biomass) with RSD (0.5 mg g−1 biomass) led to further increases in glucose yield (481.80–600.53 mg g−1 biomass), with the highest yield equivalent to 91.3% of the theoretical yield of the available glucose from starch and cellulose, suggesting almost complete saccharification of the substrate. The glucose yield achieved by the FD + RSD enzyme mixture was higher than that obtained using the FD (34.19 mg g−1) and RSD (420.03 mg g−1) individually at the enzyme dosage of 5.0 and 0.5 mg g−1, respectively. The calculated degree of synergism was 1.43 using the enzyme mixture. This suggested the cooperative action of multi-non starch polysaccharide hydrolyzing activities for degradation of the fibrous cell wall structure together with the amylolytic RSD activity acting on the accessible starch granules. The microstructure of enzymatically hydrolyzed cassava pulp was analyzed by SEM. The native cassava pulp showed an intact structure containing starch granules entrapped in the network of the fibrous cell wall structure (Fig. 5A). Hydrolysis by the 0.5 mg g−1 RSD enzyme led to partial degradation of the starch granules (Fig. 5B). The fiber part was effectively degraded by the core FD enzyme, resulting in the release of intact starch granules (Fig. 5C). Supplementation of RSD enzyme (0.5 mg g−1) with FD enzyme (5.0 mg g−1) resulted in decomposition of the fibrous cell wall network and hydrolysis of the released starch granules, resulting in the residual fibrous fraction (Fig. 5D). The results thus indicate that efficient saccharification of cassava pulp with no pre-gelatinization is possible by the combined actions of multiple non-starch polysaccharide hydrolyzing enzymes and RSD amylolytic enzymes.
Synergism among the composite cellulases i.e. endo-glucanase and a cellobiohydrolase (exo-glucanase) on hydrolysis of lignocellulosic structure is based on substrate binding specificity catalytic mechanism of the enzymes.31 The addition of a β-glucosidase can alleviate the inhibitory effects of cellobiose, the main product from the upstream enzymes.35 Their strong synergism with XYN, an endo-β1,4-xylanase which attacks xylan, the main hemicellulose in most agricultural wastes, is relevant to hydrolysis of the hemicellulose network covering the cellulose microfibers. Removal of xylan thus helps to increase the accessibility of cellulase for digestion of cellulose microfibers.36 The synergy of enzymes acting on different substrates is reflected in the increased glucose yield; however, the level of released pentoses observed in our study was limited since xylan is incompletely digested by XYN to short oligomers (2–5 mers).37 Complete hydrolysis of the accumulated xylose oligomers can be achieved by addition of an external β-xylosidase.36 Sawisit and coworkers (2015) reported that a relatively low glucose (<20% glucose recovery) was obtained from cassava pulp hydrolysis using only endocellulase or xylanase activity.38 The relatively low released sugar by the cellulase/xylanase mixture thus suggested the requirement for an additional downstream enzyme.
Cassava pulp contains a relatively high content of pectin (approximately 7%)5 compared with most other cellulosic agricultural wastes such as those from sugarcane and miscanthus.39 According to the result in this study, supplementation of EPG to the cellulase/xylanase mixture led to a further increase in the liberated glucose (134.8 mg g−1) while the enzyme dosage was kept constant. This was equivalent to a 35.0% increase compared with that of the cellulase/xylanase mixture (101.72 mg g−1 biomass). Pectic substance in cassava pulp is a heterogeneous structure comprising galacturonic acid backbone linked with sugar side chains, which acts as cement between the plant cells.40 Synergism between cellulolytic and pectinolytic enzymes on disruption of the fibrous cell wall structure in cassava pulp has been previously reported.41 Treatment with both types of enzyme led to enhanced release of the entrapped starch granules and higher yield of the starch extracted from cassava pulp. Rattanachomsri et al., 2009 also reported the cooperative effect of crude commercial Aspergillus pectinase with T. reesei cellulase on direct saccharification of cassava pulp, which led to a 57% increase in glucose yield compared with that obtained using cellulase alone.9
Compared to previously reported cooperative enzyme systems comprising crude and/or isolated single activity enzymes, the optimized enzyme system developed in this study could release >90% glucose from the substrate using an enzyme dosage of only 5.5 mg g−1, whereas >20 mg g−1 loading is reported as necessary to achieve >80% yield for most cellulosic wastes e.g. corn stover pretreated by ammonia fiber expansion12 and steam pretreated corn stover42 using synergistic enzyme systems comprising combinations of different isolated glycosyl hydrolases. It should be noted that the lower enzyme loading required for the system may be due to the more easily hydrolyzable nature of the cassava pulp with its high starch content compared with other cellulosic agricultural wastes.43 The synergism between the FD and RSD enzyme systems can be explained by a mechanism in which destruction of the plant cell wall leads to increased accessibility of the amylolytic enzyme to the exposed starch granule.41,44 Raw starch degrading enzymes have been identified from several fungal origins e.g. Penicilium and Rhizopus.45,46 The combination of non-starch polysaccharide degrading activities (crude cellulase from T. reesei and β-glucosidase from A. niger) and amylolytic activities (α-amylase from Bacillus licheniformis and glucoamylase from A. niger) has been reported to release 460 mg of reducing sugars per g biomass from hydrolysis of untreated cassava pulp, equivalent to 24.32% and 64.29% higher, respectively than that obtained using the non-starch polysaccharide degrading enzymes and amylolytic enzymes alone.47 Supplementation of crude cellulase at 2.85 U g−1 of dry pulp was also shown to increase glucose yield from cassava pulp fourfold compared with that obtained using amylase alone.27
With its high efficiency on hydrolysis of both the fibrous and starch parts in cassava pulp, the optimized enzyme system reported here can potentially find applications in various aspects of cassava processing. These involve low-temperature non-thermal saccharification of the pulp with no prior high-temperature pre-gelatinization and liquefaction steps. This process was reported to allow substantial energy saving compared with the conventional process.45,48,49 The high performance optimized FD enzyme mixture can also give increased starch yield from processed cassava pulp.41 Application of the fiber degrading enzymes would be also promising for viscosity reduction of cassava substrates in very high gravity fermentation processes for increasing ethanol titer in the final fermentation products.29 Transferring of the gene set encoding for the optimized enzyme system into ethanologic microbes for equipping capability for consolidated bioprocessing of cassava pulp is promising.
4. Conclusion
For the first time, the design and optimization of a synergistic multi-enzyme system comprising a minimal set of recombinant non-starch polysaccharide degrading and raw starch degrading enzymes have been reported. The system efficiently saccharify cassava pulp without the need for an energy-intensive pretreatment step. Application of the system on different biotechnological processes relevant to cassava pulp processing is warranted.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This work was financially supported by National Science and Technology Development Agency (Grant No. P-15-50502) and the BIOTEC postdoctoral program. We thank Philip Shaw for suggestions and editing of the manuscript.References
- J. Saini, R. Saini and L. Tewari, 3 Biotech, 2014, 1–17, DOI:10.1007/s13205-014-0246-5.
- Food Outlook: Biannual Report on Global Food Markets, Food and Agriculture Organization of the United Nations [FAO], October 2016 Search PubMed.
- P. Kurdi and C. Hansawasdi, LWT--Food Sci. Technol., 2015, 63, 1288–1293 CrossRef CAS.
- P. Paengkoum and S. Paengkoum, J. Anim. Physiol. Anim. Nutr., 2010, 94, e59–e65 CrossRef CAS PubMed.
- Djuma'ali, N. Soewarno, Sumarno, D. Primarini and W. Sumaryono, Makara Journal of Technology, 2011, 15, 183–192 Search PubMed.
- N. Sarkar, S. K. Ghosh, S. Bannerjee and K. Aikat, Renewable Energy, 2012, 37, 19–27 CrossRef CAS.
- M. Zhang, L. Xie, Z. Yin, S. K. Khanal and Q. Zhou, Bioresour. Technol., 2016, 215, 50–62 CrossRef CAS PubMed.
- W. Apiwatanapiwat, Y. Murata, A. Kosugi, R. Yamada, A. Kondo, T. Arai, P. Rugthaworn and Y. Mori, Appl. Microbiol. Biotechnol., 2011, 90, 377–384 CrossRef CAS PubMed.
- U. Rattanachomsri, S. Tanapongpipat, L. Eurwilaichitr and V. Champreda, J. Biosci. Bioeng., 2009, 107, 488–493 CrossRef CAS PubMed.
- J.-J. Chang, F.-J. Ho, C.-Y. Ho, Y.-C. Wu, Y.-H. Hou, C.-C. Huang, M.-C. Shih and W.-H. Li, Biotechnol. Biofuels, 2013, 6, 19 CrossRef CAS PubMed.
- L.-H. Fan, Z.-J. Zhang, S. Mei, Y.-Y. Lu, M. Li, Z.-Y. Wang, J.-G. Yang, S.-T. Yang and T.-W. Tan, Biotechnol. Biofuels, 2016, 9, 137 CrossRef PubMed.
- D. Gao, N. Uppugundla, S. P. Chundawat, X. Yu, S. Hermanson, K. Gowda, P. Brumm, D. Mead, V. Balan and B. E. Dale, Biotechnol. Biofuels, 2011, 4, 1–11 CrossRef PubMed.
- T. Laothanachareon, B. Bunterngsook, S. Suwannarangsee, L. Eurwilaichitr and V. Champreda, Bioresour. Technol., 2015, 198, 682–690 CrossRef CAS PubMed.
- A. Karnaouri, L. Matsakas, E. Topakas, U. Rova and P. Christakopoulos, Front. Microbiol., 2016, 7, 177 Search PubMed.
- I. J. Kim, J. Y. Jung, H. J. Lee, H. S. Park, Y. H. Jung, K. Park and K. H. Kim, Bioprocess Biosyst. Eng., 2015, 38, 929–937 CrossRef CAS PubMed.
- L. P. Huang, B. Jin, P. Lant and J. Zhou, Biochem. Eng. J., 2005, 23, 265–276 CrossRef CAS.
- S. Jung, Y. Song, H. M. Kim and H. J. Bae, Enzyme Microb. Technol., 2015, 77, 38–45 CrossRef CAS PubMed.
- The Commission of the European Communities, Commission Directive 1999/79/EC, 1999.
- Thai Industrial Standard Institute, Standard for tapioca flour/starch, UDC664.272, Ministry of Industry, Bangkok, 1978 Search PubMed.
- A. Sluiter, B. Hames, R. Ruiz, C. Scarlata, J. Sluiter and D. Templeton, Determination of Structural Carbohydrates and Lignin in Biomass, Report NREL/TP51042618, National Renewable Energy Laboratory, 2008 Search PubMed.
- H. Inoue, T. Fujii, M. Yoshimi, L. E. Taylor, S. R. Decker, S. Kishishita, M. Nakabayashi and K. Ishikawa, J. Ind. Microbiol. Biotechnol., 2013, 40, 823–830 CrossRef CAS PubMed.
- J. Sambrook and D. W. Russell, Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press, 3rd edn, New York, 2001 Search PubMed.
- G. L. Miller, Anal. Chem., 1959, 31, 426–428 CrossRef CAS.
- J. Cornell, Experiments with Mixtures: Designs, Models, and the Analysis of Mixture Data, 3rd edn, Wiley, USA, 2002 Search PubMed.
- I. J. Kim, K. H. Nam, E. J. Yun, S. Kim, H. J. Youn, H. J. Lee, I.-G. Choi and K. H. Kim, Appl. Microbiol. Biotechnol., 2015, 99, 8537–8547 CrossRef CAS PubMed.
- S. Menggred, P. Chatakanonda, K. Piyachomkwan and K. Sriroth, Preparation of microcrystalline cellulose from destarched cassava pulp, Miracle Grand Convention Hotel, Bangkok, Thailand, 2013 Search PubMed.
- N. Thongchul, S. Navankasattusas and S.-T. Yang, Bioprocess Biosyst. Eng., 2010, 33, 407–416 CrossRef CAS PubMed.
- S. Suwannarangsee, J. Arnthong, L. Eurwilaichitr and V. Champreda, J. Microbiol. Biotechnol., 2014, 24, 1427–1437 CrossRef CAS PubMed.
- A. Poonsrisawat, S. Wanlapatit, A. Paemanee, L. Eurwilaichitr, K. Piyachomkwan and V. Champreda, Process Biochem., 2014, 49, 1950–1957 CrossRef CAS.
- A. Poonsrisawat, S. Wanlapatit, R. Wansuksri, K. Piyachomkwan, A. Paemanee, C. Gamonpilas, L. Eurwilaichitr and V. Champreda, Process Biochem., 2016, 51, 2104–2111 CrossRef CAS.
- H. Inoue, S. Decker, L. Taylor, S. Yano and S. Sawayama, Biotechnol. Biofuels, 2014, 7, 151–163 CrossRef PubMed.
- C. M. P. Braga, P. d. S. Delabona, D. J. d. S. Lima, D. A. A. Paixão, J. G. d. C. Pradella and C. S. Farinas, Bioresour. Technol., 2014, 170, 316–324 CrossRef CAS PubMed.
- S. V. Gomand, L. Lamberts, L. J. Derde, H. Goesaert, G. E. Vandeputte, B. Goderis, R. G. F. Visser and J. A. Delcour, Food Hydrocolloids, 2010, 24, 307–317 CrossRef CAS.
- T. d. S. Rocha, A. P. d. A. Carneiro and C. M. L. Franco, Food Sci. Technol., 2010, 30, 544–551 CrossRef.
- R. R. Singhania, A. K. Patel, R. K. Sukumaran, C. Larroche and A. Pandey, Bioresour. Technol., 2013, 127, 500–507 CrossRef CAS PubMed.
- Q. Qing and C. E. Wyman, Biotechnol. Biofuels, 2011, 4, 18 CrossRef CAS PubMed.
- K. K. Podkaminer, A. M. Guss, H. L. Trajano, D. A. Hogsett and L. R. Lynd, Appl. Environ. Microbiol., 2012, 78, 8441–8447 CrossRef CAS PubMed.
- A. Sawisit, S. S. Jantama, S. Kanchanatawee and K. Jantama, Bioprocess Biosyst. Eng., 2015, 38, 175–187 CrossRef CAS PubMed.
- M. B. G. Latarullo, E. Q. P. Tavares, G. P. Maldonado, D. C. C. Leite and M. S. Buckeridge, Front. Plant Sci., 2016, 7, 1401 Search PubMed.
- C. Xiao and C. T. Anderson, Front. Plant Sci., 2013, 4, 67 Search PubMed.
- K. Sriroth, R. Chollakup, S. Chotineeranat, K. Piyachomkwan and C. G. Oates, Bioresour. Technol., 2000, 71, 63–69 CrossRef CAS.
- J. Hu, V. Arantes and J. Saddler, Biotechnol. Biofuels, 2011, 4, 36 CrossRef CAS PubMed.
- P. Li, H. t. Chen and M. j. Zhu, Ethanol fermentation from cassava pulp by a novel sequential co-culture, International Conference on Materials for Renewable Energy & Environment, 20–22 May 2011, pp. 413–417 Search PubMed.
- R. M. Collares, L. V. S. Miklasevicius, M. M. Bassaco, N. P. G. Salau, M. A. Mazutti, D. A. Bisognin and L. M. Terra, J. Zhejiang Univ., Sci., B, 2012, 13, 579–586 CrossRef CAS PubMed.
- B. Jin, L. P. Huang and P. Lant, Biotechnol. Lett., 2003, 25, 1983–1987 CrossRef CAS PubMed.
- H.-J. Lin, L. Xian, Q.-J. Zhang, X.-M. Luo, Q.-S. Xu, Q. Yang, C.-J. Duan, J.-L. Liu, J.-L. Tang and J.-X. Feng, J. Ind. Microbiol. Biotechnol., 2011, 38, 733–742 CrossRef CAS PubMed.
- M. Zhu, P. Li, X. Gong and J. Wang, Biosci., Biotechnol., Biochem., 2012, 76, 671–678 CrossRef CAS PubMed.
- B. Y. Jeon, S. J. Kim, D. H. Kim, B. K. Na, D. H. Park, H. T. Tran, R. Zhang and D. H. Ahn, Biotechnol. Bioprocess Eng., 2007, 12, 566 CrossRef CAS.
- Q.-S. Xu, Y.-S. Yan and J.-X. Feng, Biotechnol. Biofuels, 2016, 9, 216 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra08472b |
| This journal is © The Royal Society of Chemistry 2017 |