Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Glycerol–water mediated centrifuge controlled green synthesis of oleic acid capped PbS quantum dots for live cell imaging†‡
M. Vijaya Bharathiab,
Kaustab Ghosh*a and
Priyankar Paira*b
aSchool of Electronics Engineering(SENSE), VIT University, Chennai Campus, Tamilnadu, India. E-mail: kaustab@vit.ac.in
bDepartment of Chemistry, School of Advanced Sciences, VIT University, Vellore-632014, Tamilnadu, India. E-mail: priyankar.paira@vit.ac.in; Fax: +91-416-2243092; Tel: +91-416-2243091
First published on 21st August 2017
Abstract
Glycerol–water mediated convenient synthesis of PbS quantum dots (QDs) is introduced utilizing distinctive precipitation strategies. It is exceptionality fascinating to observe that the aforementioned factor assumes a major role in the capping process, stability as well as purity and crystalline nature of the QD. Strong red fluorescence from QDs in the HeLa cell makes these materials suitable for deep tissue imaging.
Introduction
Quantum dots (QDs) have received incredible attention from researchers as functional materials due to their quantum confinement effect, shape and size dependent optical properties and stable, narrow fluorescence peaks. Colloidal semiconductor PbS QDs, having a vast exciton Bohr radius of 18 nm,1 offer tunable luminescence over visible and NIR regions (400–2500 nm) by controlling the dot size.2–4 In the limit of strong confinement, the third order nonlinear optical response of PbS QDs is relied upon to be thirty times that of GaAs and one thousand times that of CdSe materials, which is exceptionally attractive for photonic and optical switching device applications.5 Different routines have been suggested to synthesize PbS quantum dots.6–13 PbS QDs capped with b-lactoglobulin were synthesized in aqueous medium using microwaves and used for examining 293T cells.14 Core–shell PbS–CdS QDs were synthesised using cation exchange method for a duration of 1–48 hours. Likewise, core PbS QDs were synthesized using GSH and used for examining the lymph system, cerebral blood vessels, and breast tumor.15 The RNase-A assisted PbS QDs in the attractive NIR-II window were also synthesized in aqueous medium using microwaves.16 Oleic acid capped PbS QDs with a size <2 nm were synthesized previously.17 One-step synthetic strategy for the preparation of recombinant protein (EGFP-protein G)-coated PbS QDs for the imaging of breast tumors at the cellular and whole-body level was already developed.18It should be noted that amongst these routines is the Cademartiri synthesis route19 that employs lead chloride with elemental sulphur as the precursor in oleylamine as the solvent. Herein, trioctylphosphine (TOP) can be added to oleylamine solvent as a capping agent to accomplish great control of QD size tenability and reproducibility. Notwithstanding, the utilization of TOP and trioctylphosphine oxide (TOPO) improves the toxicity of the nanoparticles that are unacceptable for in vivo applications and are risky, expensive and unsatisfactory for mass production of PbS QDs.20 Comparable issues emerged when these PbS QDs were procured in aqueous solution utilizing a mixture of thioglycerol and dithioglycerol as stabilizing agents or utilizing dihydrolipoic acid as a stabilizer at room temperature.21 PbS QDs were also prepared in aqueous medium utilizing capping material such as 1-thioglycerol/dithioglycerol,22 dihydrolipoic acid,23 L-cysteine,24 apoferritin25 and luciferase.26 Among these coated QDs, those coated with 1-thioglycerol/dithioglycerol have tuneable emission in a second NIR window. However, PbS QDs capped with 1-thioglycerol and dithioglycerol are non-biocompatible and cytotoxic. An alternate paramount perspective is the virtue of the colloidal QDs as the impurities create a deep energy level in the band gap and disable device performance. The carrier mobility of these materials, which relies upon the scattering mechanism, is a function of crystal flawlessness and purity.27,28
Results and discussion
From the abovementioned perspective, in this study, we report the synthesis of pure PbS colloidal QDs utilizing sol–gel method with less poisonous and green precursors, for instance, sodium sulfide, which is inodorous and less dangerous than organic sulphur.29 We utilized different precipitation techniques for eliminating the unreacted precursor and solvents for obtaining purified PbS QDs for different applications. The synthesis was carried out using a hot injection method; first, the sulfur precursor was prepared by the reaction of sodium sulfide and oleic acid in glycerol/water under reflux conditions at 180 °C in nitrogen atmosphere. Likewise, the lead precursor was also prepared from lead acetate trihydrate and oleic acid following the similar protocol. Then, the lead precursor was quickly injected to the sulfur precursor under reflux conditions at constant temperature, turning the solution darkish brown, indicating the formation of PbS QDs. The reaction mechanism is given in eqn (1) and (2).| Pb(OAc)2·5H2O + oleic acid (OA) → Pb(OA)2 | (1) |
| Pb(OA)2 + S2− → PbS + 2(OA) | (2) |
The reaction continued for an appropriate time for the growth of PbS QDs. After that, the reaction mixture was cooled to room temperature, centrifuged and dried for further use. Samples A and B were obtained by following a different centrifugation technique (see ESI†). PbS nanoparticles (sample A) were acquired by precipitation in ethanol, followed by centrifugation for 15 minutes at the speed of 4000 rpm. This precipitation was repeated five times for complete evacuation of unreacted precursor and solvents. Likewise, the purification of sample B was done by precipitation with ethanol, followed by centrifugation for 10 minutes at a speed of 7000 rpm. This precipitation was repeated 7 times to completely remove the impurities. With increased centrifugation speed and washing times, the insoluble aggregates, unreacted precursors and free ligands could be removed.30,31 Density Gradient Centrifugation (DGC) techniques are widely used for separation in colloid science and in cellular and molecular biology. Objects that are heavier than the solvent settle spontaneously due to gravity, which takes a very long time. The centrifugal force can be increased with speed. For nanoparticles, gravitational energy is commensurate with thermal energy, and due to this, the particles will not settle at all. To overcome this, centrifugal forces help particles to move away from the axis of rotation and separate these particles by size and shape.32 The purity of sample B over sample A is clearly supported by UV-vis spectra and EDX results (Fig. S1,† 1 and 2). A green solvent mixture (glycerol–water) mediated convenient synthesis of oleic acid capped pure crystalline PbS QD was obtained with high yield by controlling the speed of centrifugation only. The simple procedure, use of green solvent, ease of isolation, high yield and photostability of PbS QD make this method green and environmentally benign.
Fig. S1 demonstrates the absorption spectra of sample A with an absorption peak centered at 250 nm, portraying strong absorption in the ultraviolet region and good quantum confinement effect in contrast with the bulk PbS absorption region of 3200 nm. We attribute the source of this absorption band to 1pe–1ph transitions in these PbS dots.33 No other visible absorption peaks can be seen, which can imply that the transitions occur from other quantized states of the QD. Sample B demonstrates a few absorption peaks, i.e. the onset at 500 nm, and an alternate strong absorption peak at 259 nm, which can be credited to 1se–1sh and 1pe–1ph transitions individually. More numbers of excitonic peaks in the absorption spectra of sample B dots can be attributed to the good crystalline quality of the nanoparticles. Broad absorption spectrum in the visible region indicates the fluorescence efficiency of sample B in the near-infrared region (NIR). Once we excited sample B at 550 nm, we observed a significant emission peak at 855 nm with a high Stokes shift of 300 nm (Fig. S2†). Fluorescence peak in the NIR window is useful for deep tissue imaging because scattering of light is reduced in NIR compared to that in the visible region, which helps in obtaining good results from bio-imaging techniques for live cancer cell detection in the human body. All these exciton transitions additionally demonstrate a great quantum confinement impact, in contrast with bulk PbS.
To affirm the binding of the capping agents to the surface of PbS QDs, we performed Fourier transform infrared (FTIR) spectroscopy analysis in the range of 400–4000 cm−1 as delineated in Fig. S3.† The C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bond stretching peak at 1743 cm−1 in the IR spectra of sample B can be ascribed to the great capping of the oleate ligand to the surface of the QDs, which vanished in the IR spectra of sample A. Presence of oleic acid capping is additionally evidenced from the IR bands of 2853 and 2922 cm−1 of sample B, which is because of the symmetric and asymmetric C–H stretching modes of the CH2 groups of immaculate oleic acid. These bands are not seen in the spectra of sample A, demonstrating poor oleate ligand capping on this QD surface. Sample A demonstrated a strong and broad absorption peak at 3447 cm−1, which can be attributed to the hydrogen bonded hydroxyl group on the surface of the QDs, which very nearly vanished in the spectra of sample B. We assume that sample A, with lower centrifugal speed and precipitation carried out five times, could not effectively evacuate the solvents, which prompted a broad absorption peak at 3447 cm−1 attributed to an exchangeable proton from the alcohol and carboxylic acid group. This prompted poor oleate capping on the QD surface. Nonetheless, fast centrifugation at 7000 rpm and seven precipitations of sample B prompted the complete evacuation of organic impurities, which prompted strong bonding of the capping ligand on the QD, hence providing better stability and restraining particle growth.
O bond stretching peak at 1743 cm−1 in the IR spectra of sample B can be ascribed to the great capping of the oleate ligand to the surface of the QDs, which vanished in the IR spectra of sample A. Presence of oleic acid capping is additionally evidenced from the IR bands of 2853 and 2922 cm−1 of sample B, which is because of the symmetric and asymmetric C–H stretching modes of the CH2 groups of immaculate oleic acid. These bands are not seen in the spectra of sample A, demonstrating poor oleate ligand capping on this QD surface. Sample A demonstrated a strong and broad absorption peak at 3447 cm−1, which can be attributed to the hydrogen bonded hydroxyl group on the surface of the QDs, which very nearly vanished in the spectra of sample B. We assume that sample A, with lower centrifugal speed and precipitation carried out five times, could not effectively evacuate the solvents, which prompted a broad absorption peak at 3447 cm−1 attributed to an exchangeable proton from the alcohol and carboxylic acid group. This prompted poor oleate capping on the QD surface. Nonetheless, fast centrifugation at 7000 rpm and seven precipitations of sample B prompted the complete evacuation of organic impurities, which prompted strong bonding of the capping ligand on the QD, hence providing better stability and restraining particle growth.
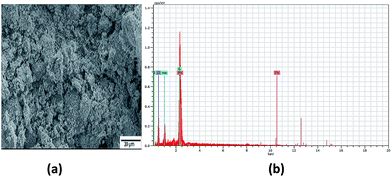
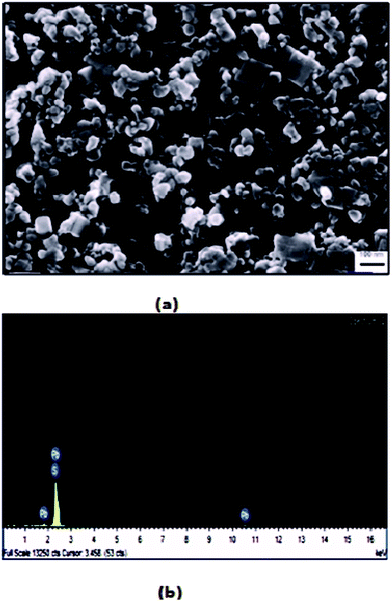
The SEM images of sample A are displayed in Fig. 1(a), which portrays complete cluster formation of the QDs because of poor oleate ligand capping, as affirmed through the FTIR spectra. This brought on quicker detachment of the capping ligand, which initiated ‘Ostwald ripening’34 where the QD molecules became larger to diminish the surface energy. This prompted the agglomeration of QDs in the form of a cluster, as seen in the image, and no dots were particularly noticeable here. The EDX spectra of sample A dots in Fig. 1(b) exhibit the existence of sulfur, sodium and oxygen. The SEM image of sample B in Fig. 2(a) shows highly dense QDs of different shapes and sizes. Herein, despite the fact that the dots tend to agglomerate, they can be notably visible. Nonetheless, in sample B, no impurity elements are seen in the EDX spectra, as indicated in Fig. 2(b), in contrast to sample A.
The TEM images of sample A in Fig. 3(a) demonstrated spherical dots of different sizes. Patches can likewise be seen in the background owing to the impurities present in the sample. Conversely, the TEM image of sample B in Fig. 3(b) exhibits well-defined and distinct spherical dots of different sizes with no background patches. Presence of clear lattice planes can likewise be seen in the image, which confirms the well-crystallized structure of the QDs. The HRTEM image of sample B is likewise demonstrated in Fig. 3(c), which demonstrates distinct dot formation exhibiting lattice planes with a diameter of 15 nm.
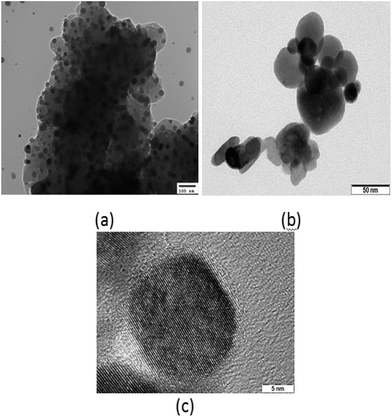 | ||
| Fig. 3 (a) TEM image of sample A dots; (b) TEM image of sample B dots; (c) HRTEM image of sample B dots. | ||
Fig. S4 exhibits the X-ray diffraction pattern of PbS QDs. Sample B shows sharp diffraction peaks that exactly match the standard values and are, thus, indicative of its highly crystalline structure and purity. XRD spectra of sample A also demonstrate its crystalline nature, but the diffraction peak is not so sharp compared to that of sample B, as revealed by the diffraction patterns with the peaks at 25°, 30°, 42°, 51°, 53°, 62°, 68°, 71° and 79° corresponding to the (111), (200), (311), (222), (400), (331), (420), (422) and (511) planes, respectively, of the Pbs QDs, as confirmed as per the JCPDS file no. 77 0244. The average nano-crystalline size of the QDs can be obtained using Debye–Scherrer equation:
D = αλ/β![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cos cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θ θ
| (3) |
Undoubtedly, cellular imaging is one of the imperative practices in biology and medicine, and it is an important technique of cellular analysis, particularly analysis of biological procession in cells. Nevertheless, two key factors could influence on the usage of QDs in cellular imaging, that is, cytotoxicity and fluorescence stability. Fluorescence stability was measured by time dependent fluorescence spectroscopy. As shown in Fig. S5,† with the extension of storage time, the fluorescence intensity of PbS QDs (sample B) did not decrease but instead increased. Therefore, oleic acid capped Pbs QDs (sample B) are suitable for cellular imaging.
The cytotoxicity of as-prepared PbS QDs, which was used to evaluate their biocompatibility, was studied using cervical cancer HeLa cells and human embryonic kidney HEK 293 cells as the model cell lines for the standard MTT method. Percentage (%) of cell viability was measured from the absorption of different concentration of QD (0–800 μg mL−1) treated cells at 570 nm. In Fig. S6,† it was observed that oleic acid capped QDs are cytotoxic in both the cell lines at extremely higher concentrations only. Log IC50 values of sample B in HeLa and HEK293 were observed to be 1.78 ± 0.01 μg mL−1 and 1.93 ± 0.006 μg mL−1, respectively.
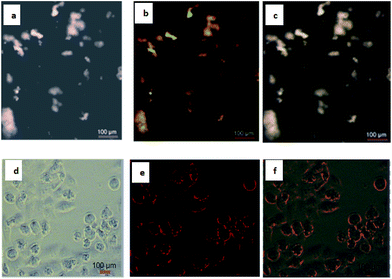
To utilize the QDs in cellular imaging, we studied the cellular uptake of PbS QDs (sample B) by cancerous HeLa cells. Cells were incubated with 3 × 10−5 M of QDs in cell culture DMEM for 4 hours and monitored using Olympus fluorescence microscopy. After 4 hours of incubation with QDs, cells showed bright green and red fluorescence under two different filters of the microscope. The aggregation of nanoparticles in the cytoplasm and nucleus is clearly visible (Fig. 4).
Conclusions
In summary, the present study reports a convenient green synthetic approach for the preparation of PbS QDs with less toxic sodium sulfide and oleic acid as the capping agent rather than TOP and TOPO chemicals, which can open its plausibility for applications in biological systems. We obtained good crystalline PbS QDs as we expanded the centrifugation speed to 7000 rpm, repeated the precipitation 7 times and dried the product for six hours. This technique could effectively eliminate the hydrogen bonded hydroxyl group and encouraged great capping of the oleate ligand on the surface of the QD for hindering particle agglomeration and growth. The XRD and EDX spectra revealed the formation of profoundly immaculate and crystalline QDs, which can be suitably utilized for optoelectronic applications. Fluorescence imaging study with the HeLa cell clearly indicates that these QDs can be utilized for deep tissue imaging.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors thank VIT University for providing us with research seed funding and laboratory facilities. We acknowledge DST, New Delhi, India for DST-FIST project. We also acknowledge DST-SERB, India for young scientist grant. We acknowledge Honeywell Technology Solutions Lab, Bangalore, for providing the facilities to carry out the SEM with EDX analysis and Indian Institute of Science, Bengaluru, for providing the facilities to carry out the TEM analysis.Notes and references
- J. L. Machol, F. W. Wise and R. C. Patel, Phys. Rev. B, 1993, 48, 2819 CrossRef CAS.
- F. W. Wise, Acc. Chem. Res., 2000, 33, 773 CrossRef CAS PubMed.
- L. Cademartiri, E. Montanari, G. Calestani, A. Migliori, A. Guagliardi and G. A. Ozin, J. Am. Chem. Soc., 2006, 128, 10337 CrossRef CAS PubMed.
- M. Kowshik, W. Vogel, J. Urban, S. K. Kulkarni and K. M. Paknikar, Adv. Mater., 2002, 14, 815 CrossRef CAS.
- M. A. Hines and G. D. Scholes, US Pat., 7, 118, 627 B2, 2006.
- S.-M. Lee, Y.-W. Jun, S.-N. Cho and J. Cheon, J. Am. Chem. Soc., 2002, 124, 11244 CrossRef CAS PubMed.
- K. Singh, A. A. McLachlan and D. G. Marangoni, Colloids Surf., A, 2009, 82, 345 Search PubMed.
- H. Cao, Q. Gong, X. Qian, H. Wang, J. Zai and Z. Zhu, Cryst. Growth Des., 2007, 7, 425 CAS.
- Y. K. A. Lau, D. J. Chernak, M. J. Bierman and S. Jin, J. Mater. Chem., 2009, 19, 934 RSC.
- T. Thongtem, S. Kaowphong and S. Thongtem, Ceram. Int., 2008, 34, 1691 CrossRef CAS.
- Z. Quan, C. Li, X. Zhang, J. Yang, P. Yang, C. Zhang and J. Lin, Cryst. Growth Des., 2008, 8, 2384 CAS.
- S. Xiong, B. Xi, D. Xu, C. Wang, X. Feng, H. Zhou and Y. Qian, J. Phys. Chem. C, 2007, 111, 16761 CAS.
- F. Zuo, S. Yan, B. Zhang, Y. Zhao and Y. Xie, J. Phys. Chem. C, 2008, 112, 2831 CAS.
- J. Chen, Y. Kong, W. Wang, H. Fang, Y. Wo, D. Zhou, Z. Wu, Y. Li and S. Chen, Chem. Commun., 2016, 52, 4025 RSC.
- T. Jin and Y. Imamura, ECS J. Solid State Sci. Technol., 2016, 5, 3138 CrossRef.
- Y. Kong, J. Chen, H. Fang, G. Heath, Y. Wo, W. Wang, Y. Li, Y. Guo, S. D. Evans, S. Chen and D. Zhou, Chem. Mater., 2016, 28, 3041 CrossRef CAS PubMed.
- A. Shrestha, N. A. Spooner, S. Z. Qiao and S. Dai, Phys. Chem. Chem. Phys., 2016, 18, 14055 RSC.
- A. Sasaki, Y. Tsukasaki, A. Komatsuzaki, T. Sakata, H. Yasuda and T. Jin, Nanoscale, 2015, 7, 5115 RSC.
- L. Cademartiri, G. von Freymann, A. C. Arsenault, J. Bertolotti, D. A. Wiersma, V. Kitaev and G. A. Ozin, Small, 2005, 1, 1184 CrossRef CAS PubMed.
- Q. Wang, S. Li, P. Liu and X. Min, Mater. Chem. Phys., 2012, 137, 580 CrossRef CAS.
- Y. Jiao, X. Gao, J. Lu, Y. Chen, J. Zhou and X. Li, Mater. Lett., 2012, 72, 116 CrossRef CAS.
- X. Zhao, I. Gorelikov, S. Musikhin, S. Cauchi, V. Sukhovatkin, E. H. Sargent and E. Kumacheva, Langmuir, 2005, 21, 1086 CrossRef CAS PubMed.
- D. Deng, W. Zhang, X. Chen, F. Liu, J. Zhang, Y. Gu and J. Hong, Eur. J. Inorg. Chem., 2009, 23, 3440 CrossRef.
- Y. Yu, K. Zhang and S. Sun, Appl. Surf. Sci., 2012, 258, 7181 CrossRef CAS.
- B. Hennequin, L. Turyanska, T. Ben, A. M. Beltrán, S. I. Molina, M. Li, S. Mann, A. Patanè and N. R. Thomas, Adv. Mater., 2008, 20, 3592 CrossRef CAS.
- N. Ma, A. F. Marshall and J. Rao, J. Am. Chem. Soc., 2010, 132, 6884 CrossRef CAS PubMed.
- L. Kucher, J. Drapala and J. Lunacek, J. Cryst. Growth, 2006, 94, 161 Search PubMed.
- K. Ghosh, V. N. Mani and S. Dhar, J. Cryst. Growth, 2009, 31, 1521 CrossRef.
- Y. Dong, Z. Jian-Wen, Z. Jing-Bo and L. Yuan, Acta Phys.-Chim. Sin., 2011, 27, 1239 Search PubMed.
- X. He, L. Gao and N. Ma, Sci. Rep., 2013, 3, 2825 CrossRef PubMed.
- B. Shakeri and R. W. Meulenberg, Langmuir, 2015, 31, 13433 CrossRef CAS PubMed.
- B. Kowalczyk, I. Lagzi and B. A. Grzybowski, Curr. Opin. Colloid Interface Sci., 2011, 16, 135 CrossRef CAS.
- M. Flores-Acosta, M. Sotelo-Lerma, H. Arizpe-Chávez, F. F. Castillón-Barraza and R. Ramírez-Bon, Solid State Commun., 2003, 128, 407 CrossRef CAS.
- H. Zhao, M. Chaker and D. Ma, J. Phys. Chem. C, 2009, 113, 6497 CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra08443a |
| ‡ Led acetate, oleic acid, sodium sulphide, glycerol, PBS buffer, MTT reagent, DMSO, trypsin was purchased from Sigma Aldrich. DMEM media was purchased from Himedia. Cancer HeLa cell line and normal kidney cell line (HEK 293) was obtained from NCCS, Pune. |
| This journal is © The Royal Society of Chemistry 2017 |