Open Access Article
Open Access ArticleBiological evaluation of dinuclear copper complex/dichloroacetic acid cocrystal against human breast cancer: design, synthesis, characterization, DFT studies and cytotoxicity assays†
Mohammad Usmana,
Farukh Arjmand a,
Rais Ahmad Khanb,
Ali Alsalmeb,
Musheer Ahmadc and
Sartaj Tabassum
a,
Rais Ahmad Khanb,
Ali Alsalmeb,
Musheer Ahmadc and
Sartaj Tabassum *a
*a
aDepartment of Chemistry, Aligarh Muslim University, Aligarh-202002, India. E-mail: tsartaj62@yahoo.com; Tel: +91 9358255791
bDepartment of Chemistry, College of Science, King Saud University, P.O. Box 2455, Riyadh 11451, Saudi Arabia
cDepartment of Applied Chemistry, Aligarh Muslim University, Aligarh-202002, India
First published on 12th October 2017
Abstract
Binuclear copper(II) cocrystal “[Cu2(valdien)2⋯2Cl2CHCOOH],” 1 was synthesized from H2valdien scaffold and anticancer drug pharmacophore “dichloroacetic acid” embedded with two Cu(II) connected via a hydrogen bonded network. [Cu2(valdien)2⋯2Cl2CHCOOH] 1 was thoroughly characterized by single-crystal XRD and by other spectroscopic techniques. The non-covalent interaction (NCI) index and Hirshfeld surface analysis were used to study the various kinds of interactions (O⋯H, N⋯H, H⋯Cl, Cu⋯H, C⋯O, N⋯O, C⋯Cl, and O⋯Cl, etc.) responsible for the stabilization of crystal lattice. [Cu2(valdien)2⋯2Cl2CHCOOH] 1 was validated as potential antitumor drug entity by studying its DNA binding profile, cleavage mechanism with pBR322 by gel electrophoretic assay and in vitro cytotoxicity on MCF-7 cancer cell lines. The mechanistic pathways were also deduced via dual staining AO/EB of cancer cells which confirmed the potential of the cocrystal [Cu2(valdien)2⋯2Cl2CHCOOH] 1 to act as effective anticancer agent towards breast cancers.
1. Introduction
Bio-inorganic and Pharmaceutical chemists are striving constantly to improve the physical and pharmacokinetic features of active pharmaceutical ingredients (APIs) viz., crystallinity, solubility, hygroscopicity, stability, particle size, flow, filterability, density etc.1–6 Current approaches involved in changing properties of APIs include the exploitation of hydrates, solvates, salts, and more recently, cocrystals. Cocrystals are single phase crystalline materials comprising of two or more different molecular and/or ionic compounds in a stoichiometric ratio, which are generally neither solvates nor simple salts.7 They are distinct from two conformers due to non-covalent interactions in the new material.Although the number of possible organic cocrystals is virtually infinite, the cocrystals of only metal complexes and containing metal complex-drug are rare.8–16 The combinatorial synthetic approach to obtain a single unit drug motif from conformers has invoked much interest in drug development strategy, because monotherapy (i.e., targeting a specific receptor) is not much efficacious in the treatment of chronic diseases, such as HIV/AIDS, cancer, diabetes, and cardiovascular disorders etc.17–24 In the present contribution, an example of unique cocrystallized bis(o-valdien copper) complex with dichloroacetic acid (DCA) is described. To the best of our knowledge, there is no cocrystal of a metal complex having drug moiety ‘dichloroacetic acid’ as one of the components.
Dichloroacetic acid (DCA) is directly involved in cell apoptosis and works synergistically with additional cancer therapies, viz., radio, gene, and viral therapy.25–28 Since it gets easily absorbed by the body and can permeate through blood–brain barrier; it poses risks of unexpected and severe neurological effects.29 Therefore, a cocrystals based prodrugs are designed for sustained release of free haloacetates in the circulatory system, which can be cleaved to release the haloacetates in a selective manner inside the cancer cell. The pro-drug molecule self-assembled in the presence of a dinuclear copper complex to generate stabilized cocrystals. It is worth mentioning here that many metalloenzymes and proteins contain in their active sites two copper ions that operate cooperatively.30 Thus, a great deal of consideration has been given to bimetallic copper compounds with two metal ions in close proximity because they provide an opportunity to study intramolecular binding, bimetallic catalysts, magnetic exchange interactions, multi-electron redox reactions and activity mimicking the possible activation of small substrate molecules by enzymes.31 In addition, the current studies showed that the binuclear Cu(II) compounds possess high anticancer activities.32 Therefore, we attempt to synthesized binuclear copper(II) complex as an anticancer chemotherapeutic.
The designing of the ligand motifs has an important role in tuning and regulating the drug candidate specificity towards the biological target molecules. Among the several organic ligands, our interest stems in Schiff bases derived from o-vanillin and their metal complexes are known to exhibit diverse biological applications viz., antitumor, anti-inflammatory, antiviral, antibacterial and cell imaging agents.33 The synthetic strategy to obtain bioactive cocrystal fundamentally depends on the use of polydentate bridging ligand as in the case of transition-metal therapeutics34,35 which can mediate synergistic interactions between the drug conformer dichloroacetic acid (DCA) via hydrogen bonding and its dinuclear copper complex. Copper demonstrates high affinity for nucleobases and has exhibited broad anticancer activity due to the selective permeability of cancer cell membranes to copper complexes.36,37 Besides this, copper is a physiologically important endogenous metal ion that plays a significant role in redox reactions and thus triggers the production of reactive oxygen species (ROS) which induce apoptosis in cancer cells.38
A novel dinuclear Cu(II) cocrystal “[Cu2(valdien)2⋯2Cl2CHCOOH]” as a potent antitumor drug motif has been achieved and thoroughly characterized via single crystal XRD and other spectroscopic methods. The chemotherapeutic potential was ascertained by binding with CT-DNA employing UV-visible, fluorescence, and circular dichroism techniques. DNA cleavage experiments were carried out to study the mechanistic insight into the binding phenomena at the target site. 1 was tested against MCF-7 breast cancer cells line to validate its cytotoxicity.
2. Results and discussion
2.1. Synthesis and characterization
The compartmental acyclic Schiff base H2valdien ligand was obtained from the condensation reaction of o-vanillin and diethylenetriamine. To obtain [Cu2(valdien)2], Cu(CH3COO)2·H2O was reacted with H2valdien in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ration in the presence of triethylamine in MeOH. The cocrystallization process involved combining [Cu2(valdien)2] with dichloroacetic acid (DCA) (1
1 ration in the presence of triethylamine in MeOH. The cocrystallization process involved combining [Cu2(valdien)2] with dichloroacetic acid (DCA) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) in methanol/acetonitrile (4
2) in methanol/acetonitrile (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) solution. The resulting solution was subjected to controlled evaporation to help crystallization. After 2–3 months, dark brown colored rectangular-shaped crystals were obtained (Scheme 1).
1) solution. The resulting solution was subjected to controlled evaporation to help crystallization. After 2–3 months, dark brown colored rectangular-shaped crystals were obtained (Scheme 1).
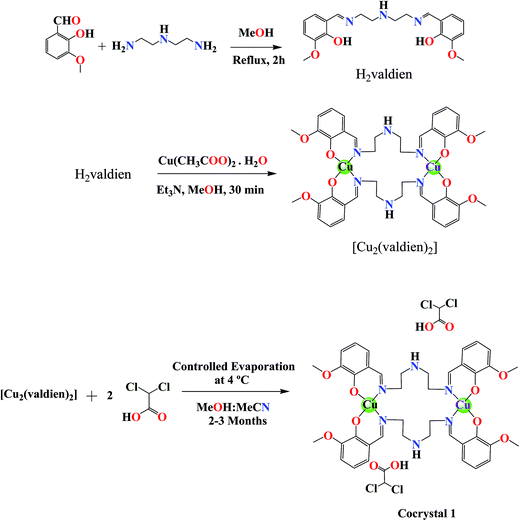 | ||
| Scheme 1 Schematic representation of the ligand and dinuclear copper(II) cocrystal “[Cu2(valdien)2⋯2Cl2CHCOOH]”. | ||
The IR spectrum of cocrystal [Cu2(valdien)2⋯2Cl2CHCOOH] 1 exhibited strong absorption bands between 1655 cm−1 and 1414 cm−1 attributed to carboxylic acid groups. Broad bands in the region 3510–3100 cm−1 indicate the presence of N–H group. The sharp peak at 1617 cm−1 was attributed to the imine bond.
The electronic spectrum (Fig. S1†) of cocrystal [Cu2(valdien)2⋯2Cl2CHCOOH] 1 exhibited an intense transition at 271 nm attributed to the intraligand π–π* transition and the absorption band at 404 nm characteristic of the LMCT was observed. The low energy d–d band appeared at 559 nm consistent with the distorted square planar environment around Cu(II).39
Emission spectra of cocrystal [Cu2(valdien)2⋯2Cl2CHCOOH] 1 (Fig. S2†) recorded at room temperature in MeOH, exhibited fluorescence at 458 nm when excited at 371 nm, while a weak band appears at 332 nm along with 458 nm upon excitation at 271 nm. The emission corresponds to intra-ligand charge transfer (ILCT), a ligand to ligand charge transfer (LLCT) or a combination of both.
To obtain the evidence for the stability of the dinuclear cocrystal [Cu2(valdien)2⋯2Cl2CHCOOH], 1 in solution, ESI-MS, and UV-vis spectra were measured. The ESI-MS shows an m/z of 1122.93 [M + H+] (Fig. S3†), corresponding to the stability of cocrystal [Cu2(valdien)2⋯2Cl2CHCOOH] 1 in solution phase.40 Also, the UV-vis spectra (Fig. S4†) of 1 in Tris–HCl buffer under physiological conditions (pH 7.3 and T 296 K) was measured over different time intervals (0, 1, 3, 6, 12, 24, 48 and 72 h) using UV-vis spectrophotometer. The small variation in measured spectra (ca. 10 nm blue shift with 0.2 hyperchromic shift), was observed which may be attributed to the solvation effect of [Cu2(valdien)2⋯2Cl2CHCOOH] under physiological conditions (Tris–HCl buffer, pH 7.3 & T 310 K). This spectral variation without appearing any new absorption band indicated the stability of the cocrystal in Tris–HCl buffer medium.
2.2. Crystal structure description of [Cu2(valdien)2⋯2Cl2CHCOOH] cocrystal (1)
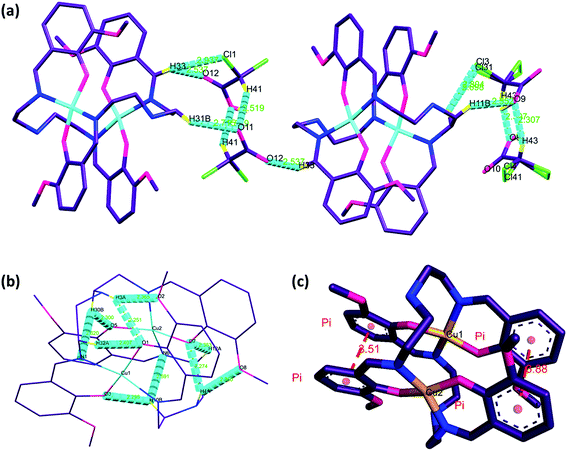
The dinuclear copper(II) cocrystal “[Cu2(valdien)2⋯2Cl2CHCOOH] 1 was structurally characterized by single-crystal X-ray crystallography (Fig. 1). Details of the crystallography data and refinement parameters are summarized in Table 1. Selected bond angles and distances are listed in Table S1 and S2.† Particulars of the hydrogen bonding parameters are shown in Table S3.† Single-crystal X-ray structural study revealed that 1 crystallized in the triclinic P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) space group possessing the lattice parameters, a = 12.790(5) Å, b = 14.351(5) Å, c = 15.034(5) Å, α = 109.079(5), β = 91.480(5), γ = 114.551(5), per unit cell. The asymmetric unit of 1 contains one [Cu2(valdien)2] and two DCA molecules, in which [Cu2(valdien)2] moiety link each other through the C33–H33⋯O12, C11–H11B⋯O9, C12–H12A⋯C13/Cl31 hydrogen bonds to form a one-dimensional chain (Fig. S5†), two adjacent chains are connected by dichloroacetic acid (DCA) molecules through C7–H7B⋯O10 hydrogen bonds to generate a 2D sheet (Fig. S5†). The 2D sheets are further held together by a network of C36–H36⋯Cl3/Cl31 hydrogen bonds to form the 3D framework of 1 (Fig. S5†).
space group possessing the lattice parameters, a = 12.790(5) Å, b = 14.351(5) Å, c = 15.034(5) Å, α = 109.079(5), β = 91.480(5), γ = 114.551(5), per unit cell. The asymmetric unit of 1 contains one [Cu2(valdien)2] and two DCA molecules, in which [Cu2(valdien)2] moiety link each other through the C33–H33⋯O12, C11–H11B⋯O9, C12–H12A⋯C13/Cl31 hydrogen bonds to form a one-dimensional chain (Fig. S5†), two adjacent chains are connected by dichloroacetic acid (DCA) molecules through C7–H7B⋯O10 hydrogen bonds to generate a 2D sheet (Fig. S5†). The 2D sheets are further held together by a network of C36–H36⋯Cl3/Cl31 hydrogen bonds to form the 3D framework of 1 (Fig. S5†).
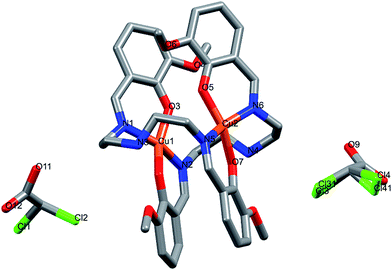 | ||
| Fig. 1 X-ray crystal structure of dinuclear cocrystal [Cu2(valdien)2⋯2Cl2CHCOOH] 1. Hydrogen atoms are omitted for clarity. | ||
| Parameters | 1 |
|---|---|
| a GOF is defined as {∑[w(F02 − Fc2)]/(n − p)}1/2 where n is the number of data and p is the number of parameters.b R = {∑∣∣F0∣ − ∣Fc∣∣/∑∣F0∣}, wR2 = {∑w(F02 − Fc2)2/∑w(F02)2}1/2. | |
| Formula | C88H100C19Cu4N12O24 |
| Fw (g mol−1) | 2283.00 |
| Crystal system | Triclinic |
| Space group | P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
| a (Å) | 12.790(5) |
| b (Å) | 14.351(5) |
| c (Å) | 15.034(5) |
| α (deg) | 109.079(5) |
| β (deg) | 91.480(5) |
| γ (deg) | 114.551(5) |
| U (Å3) | 2329.8(14) |
| Z | 1 |
| ρcalc (g cm−3) | 1.627 |
| μ (mm−1) | 1.241 |
| F(000) | 1173 |
| Crystal size (mm) | 0.28 × 0.21 × 0.15 |
| Temp (K) | 296(2) |
| Measured reflns | 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 813 813 |
| Unique reflns | 7323 |
| GOFa | 1.079 |
| Final Rb indices [I > 2σ(I)] | R1 = 0.0829, wR2 = 0.2178 |
| Rb indices (all data) | R1 = 0.0898, wR2 = 0.2229 |
| CCDC | 1402001 |
Furthermore, [Cu2(valdien)2] unit consists of two independent valdien2− ligands, adopting η1:η1:η1:η1:μ2 coordination mode (Fig. S6†) in bridging fashion to synchronize both the metal centers Cu1 and Cu2 through their phenoxide atoms O1, O3, O5, O6 and their iminic nitrogen atoms N1, N2, N5, N6. Each copper ion is, therefore, tetra-coordinated in a distorted square planar N2O2 environment which deviated significantly from planarity. The phenyl rings of the ligands are parallel two by two, giving rise to π⋯π stacking interactions with centroid⋯centroid distances of 3.515 and 3.884 Å (Fig. 2).
The hydrogen atoms on N3 & N4 are involved in intra-molecular hydrogen bonds with O1, O2 & O8, O7, respectively [N3–H3A⋯O1: 2.84 (11) Å, N3–H3A⋯O2: 2.97 (12) Å, N4–H4A⋯O7: 2.90 (11) Å, N4–H4A⋯O8: 3.01 (12) Å, C10–H10B⋯O3: 2.92 (11) Å, C10–H10B⋯N6: 3.23 (13) Å, C12–H12A⋯O7: 2.87 (11) Å, C30–H30B⋯O5: 2.94 (12) Å, C30–H30B⋯N1: 3.22 (13) Å and C32–H32⋯O1: 2.88 (11) Å] (see Fig. 2), which along with the stacking interactions, help stabilizing the conformation of the [Cu2(valdien)2] moiety.
2.3. Hirshfeld surface analyses
The Hirshfeld surfaces of the title cocrystal are illustrated in Fig. 3, showing that have been mapped over dnorm (−0.27 to 1.54 Å), shape index (−1.0 to 1.0 Å) and curvedness (−4.0 to 0.40 Å). The surfaces are shown in a similar orientation for 1, around which they are calculated. It is clear that the information present in the histogram (Fig. S7†) is summarized effectively in these spots, with large circular depressions (deep red) visible on the front and back views of the surfaces indicative of hydrogen bonding contacts. Other visible spots in the surfaces are because of H⋯H contacts.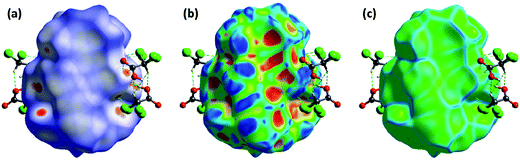 | ||
| Fig. 3 Hirshfeld surface mapped with dnorm (left), shape index (middle), and curvedness (right) for the title cocrystal [Cu2(valdien)2⋯2Cl2CHCOOH] 1. | ||
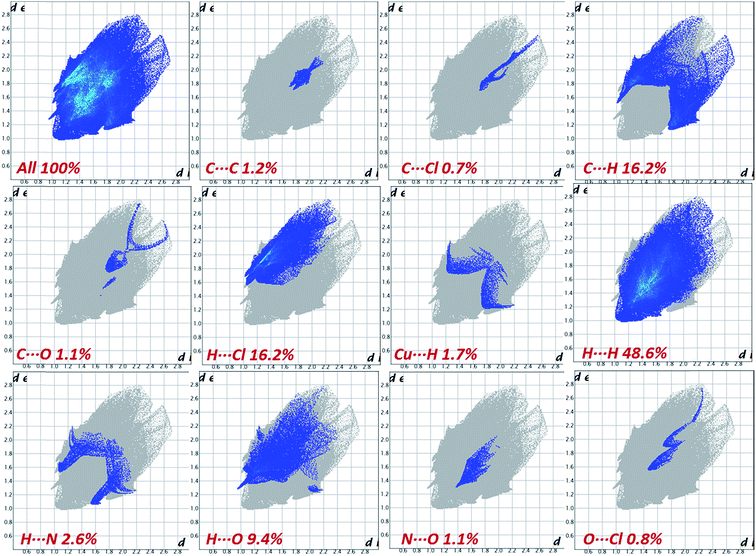
The 2D fingerprint plots and the relative contribution of the important intermolecular contacts of dichloroacetic acid (DCA) in the cocrystal are shown in Fig. 4. The most significant contributions come from H⋯H, Cl⋯H, C⋯H and H⋯O contacts, which correspond to the van der Waals interactions and hydrogen bonds, respectively. Since the dnorm values for these contacts are close (48.6%, 16.2%, 16.2% and 9.40% for H⋯H, H⋯Cl, C⋯H and H⋯O, respectively), the lattice is stabilized equally by both H-bonds and dispersion forces.
The synthon interactions, which have the closest interatomic distances of all non-covalent interactions, are seen on the fingerprint plot as two distinct spikes as in the case of C⋯H and N⋯H contacts. The H⋯H contacts, being less directed are presented on 2D fingerprint plots as bulk central areas. In contrast to H⋯O, C⋯H and H⋯Cl contacts, are attractive while the H⋯H contacts reside in the part of the Hirshfeld surface with positive dnorm, and thus should be considered repulsive.
The bright colored area in the upper right part of the fingerprint plot and large flat region across the molecule in Hirshfeld surface, which is most clearly visible on the curvedness surface corresponds to π–π stacking contacts. The red and blue triangles on the same region of the shape index surface is another characteristic of π–π stacking (C⋯C) interaction between the parallel aromatic ring of the copper-coordinated validien−2 ligand moiety which comprises 1.2% to the total Hirshfeld surfaces. Blue triangles represent convex regions because of the ring carbon atoms of the molecule inside the surface, while red triangles represent concave regions resulting from the carbon atoms of the π-stacked moiety above it.
The directional C⋯H/H⋯C close contacts that are associated with π⋯H interaction between metal complex and dichloroacetic acid (DCA) and metal complexes of adjacent layers are observed as broad spikes on each side in the fingerprint plot (Fig. 4). Interestingly, in the C–H⋯π interaction alkyl, C–H and O–H (DCA) are involved along with the C–H of aromatic rings, this could be due to the delocalization of electron density of nitrogen atoms to interacting alkyl carbon atoms of the valdien−2 ligand moiety. Additionally, some other non-traditional interactions (Cu⋯H, C⋯O, N⋯O, N⋯H, C⋯Cl, and O⋯Cl) to are also observed which are lesser contributing to Hirshfeld surface. Finally, this example underlines the utility of Hirshfeld surfaces, and in particular, fingerprint-plot analysis for “visual screening” and rapid detection of unusual crystal structures features through a whole structure view of intermolecular interactions.
2.4. Theoretical insight into intermolecular interactions via NCI approach
The non-covalent interaction (NCI) index based on the relationship between the electron density and reduced density gradient (RDG) has been introduced by Yang and co-workers.57,58 This technique is based on the analysis and the graphical interpretation of the electronic density (ρ) and its derivatives, namely the λ2 eigenvalues of its Hessian and its reduced gradient s(ρ):The non-covalent interaction (NCI) index was carried out based on the properties of the electronic density of the molecule, to visualize both attractive (hydrogen bonding, van der Waals) and repulsive (steric) interactions.41 An inter- or intra-molecular interaction causes a significant alteration in the reduced gradient of density (S) in between the interacting atoms resulting in density critical points and interacting fragments. In the 2D plots of S vs. ρ, the troughs represent critical points.42 To analyze the non-covalent interaction is attractive or repulsive, the sign of an eigenvalue λ2 of the electronic density Hessian matrix, ∇2ρ = λ1+ λ2 + λ3 (λ1 < λ2 < λ3) can be utilized.43 The reduced gradient of density S troughs are used to recognize non-covalent interactions, point at which the value of S is approaching zero, the strength of interaction is defined by quantity sign (λ2)ρ, i.e., higher the value, stronger will be interaction and −ve sign (attractive interaction) vs. +ve sign (repulsive interaction).
The NCI index calculation of 1 revealed various kinds of intra- and inter-molecular non-covalent interactions within the cocrystal which are responsible for the stabilization of crystal lattice. These interactions identified in the 2D plots as spikes with cutoff value (RDG = 2 and ρ = −0.25 to 0.25) and their corresponding electronic density map with color coding. The electronic density map of the title compound not only revealed the attractive and previously identified non-covalent contacts but also display valuable information about the destabilization factor as steric clashes which are repulsive in nature and responsible for the geometrical distortion.
These stabilization and destabilization contacts differ in the shape of isosurfaces whose color code reveals the nature of the interaction, where red is used for destabilizing, blue for stabilizing and green for delocalized weak interactions.
Fig. 5 displays the S vs. sign (λ2)ρ in the [Cu2(valdien)2⋯2Cl2CHCOOH] cocrystal exhibited four characteristic spikes with density −0.05, −0.03, −0.01, 0.006 which are corresponding to the strong intra-molecular, inter-molecular, H⋯O, H⋯N, CH⋯Cl hydrogen-bond and π⋯π stacking interactions in the cocrystal, respectively.
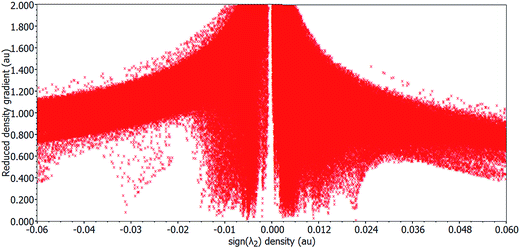 | ||
| Fig. 5 Plot of the reduced density gradient versus the electronic density multiplied by the sign of the second Hessian eigenvalue of cocrystal [Cu2(valdien)2⋯2Cl2CHCOOH] 1. | ||
Interestingly, we observed various sites where electronic density was distributed between the various fragments of the premise of the co-crystal revealing not only hydrogen bonding (H⋯O, H⋯N, H⋯Cl), CH⋯π and π⋯π stacking interaction responsible for the stabilization of geometrical conformation of the co-crystal, in fact various other non-traditional dispersion forces between the atom pairs (Cu⋯H, C⋯O, N⋯O, C⋯Cl, and O⋯Cl) were also responsible. These non-traditional dispersion forces are derived due the extensive delocalization of the electronic charge density within the cavity of the [Cu2(valdien)2] complex as green colored NCI surface is a signature for the delocalization of electronic density (Fig. 6). It is found that the result of NCI index calculation is in good agreement with the Hirshfeld surface analysis results.
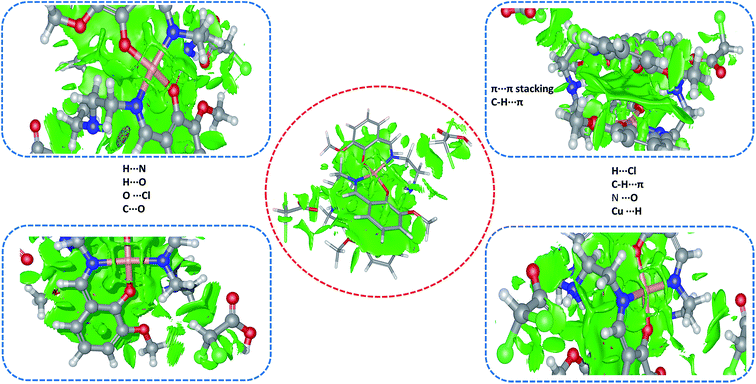 | ||
| Fig. 6 NCI electronic density distribution (isovalue: 0.8) in the [Cu2(valdien)2⋯2Cl2CHCOOH] 1 cocrystal. | ||
2.5. DNA binding profile and pBR322 nuclease activity
The binding study of 1 with CT-DNA was analyzed by spectroscopic optical methods via UV-vis, fluorescence, and circular dichroism. The band centered at 266 nm exhibited ‘hyperchromism’ with strong blue shift of 5–7 nm and the weak band at 399 nm exhibited ‘hypochromism’ of about 33% and 56%, respectively (Fig. S8a†) revealing the favorable electrostatic interaction of the cationic core to the polyanionic phosphate backbone of the DNA double helix, in addition to the active participation of ligand chromophores via partial intercalation.44,45 1 was quantitatively ascertained by intrinsic binding constant, Kb, value which was found to be 2.60 × 104 M−1 (Fig. S9†).From the emissive titration spectra, it is apparent that on the addition of increasing concentration of CT DNA (0.00–1.8 × 10−4 M) to fixed amount of 1 (Fig. S8c and S10†), displayed hyperchromism of about 73%, respectively at 458 nm with no shift in the wavelength was observed. The intrinsic binding constant K for 1 determined from Scatchard equation from the plot of r/CF versus r (= CB/[DNA]) were found to be 5.8 × 104 M−1. Furthermore, upon addition of increasing concentration of 1 to pretreated CT DNA with EB ([DNA]/[EB] = 1) solution, the emission band at 596 nm exhibited quenching of the emission intensity more than 70% of the initial fluorescence intensity when the molar ratio of the complex to DNA (r = [Complex]/[DNA]) range from 1.6 to 15.0 (Fig. S8d and S11†). The observed quenching of DNA-EB fluorescence indicated that the EB molecule was displaced by 1 from their DNA binding sites.46 The quenching efficiency (KSV) was evaluated, and quenching data is in agreement with the classical linear Stern–Volmer equation,46 which was found to be 5.30 × 104 M−1.
The CD spectrum of DNA exhibits characteristic B-type signature in the UV part of the spectra with a positive band at 276 nm (UV: λmax, 260 nm, CD [mdeg] 0.9602) because of base stacking and a negative band at 245 nm (CD [mdeg] −0.8893) due to the right-handed helicity of the B-DNA form (with a zero-crossover around 254 nm) which are quite sensitive to the mode of DNA interactions with small molecules.47 Upon successive addition of 1, the positive band decreased in intensity with a concomitant red shift to 280 nm while the negative band (245 nm) increased in intensity, (Fig. S8b†) which ascertains the potential of 1 to unwind the DNA by loosing its helicity.
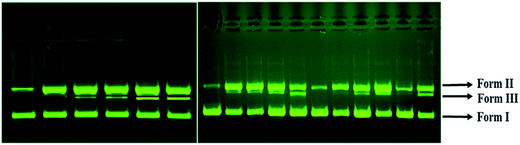
The ability of 1 to cleave DNA was examined by incubating different concentrations of 1 with supercoiled pBR322 DNA in the absence of reducing agents using standard physiological conditions (5 mM Tris–HCl/50 mM NaCl buffer, pH 7.2, incubation time 1 h). On increasing concentration of 1, Form I of pBR322 DNA gets converted into Form II (Lane 2–4), gradually, starting from 5 μM to 20 μM. But, when the concentration of 1 reached to 25 μM (Lane 6), Form II appeared with the formation of Form III, which migrates in between SC and NC form (Fig. 7a) revealing double strand DNA cleavage.48
To ascertain whether any adventitious agents present in the reaction mixture could account for the increased DNA degradation by “[Cu2(valdien)2⋯2Cl2CHCOOH],” 1, the DNA cleavage activity was studied in the presence of activators. The nuclease efficiency of the copper(II) complexes are known to depend on the activators used for initiating the DNA cleavage. Thus, the further activity of “[Cu2(valdien)2⋯2Cl2CHCOOH],” 1 has been studied with different activators viz. H2O2 as an oxidizing agent, ascorbate (Asc), 3-mercaptopropionic acid (MPA) and glutathione (GSH) as reducing agents. The significant enhancement in the cleavage activity was noticed and followed the order H2O2 > GSH > MPA > ASC (Fig. 7b).
To get insight into the pathway of nuclease activity, (Fig. 7b) 1 was treated with DMSO and EtOH (hydroxyl radical scavengers), SOD (superoxide scavenger) and NaN3 (singlet oxygen quencher). No apparent inhibition was noticed upon addition of DMSO and EtOH. However, in the presence of NaN3, significant inhibition was observed which revealed that ROS species viz., singlet oxygen; 1O2 was responsible for the DNA cleavage process via oxidative pathway.49
| Cu(II)-complex + DNA → Cu(II)-complex DNA |
| Cu(II)-complex + e− → Cu(I)-complex |
| Cu(I)-complex + O2 → Cu(II)-complex + O2− |
| O2− + 2H+ → O2 + H2O2 |
In Fig. 7b, the DNA cleavage by 1 was also carried out in the presence of DAPI (minor groove binder) and methyl green, MG (major groove binder). The cleavage pattern obtained showed no inhibition in case of MG whereas DAPI resulted in apparent inhibition, implicates the minor groove binding affinity of the complex 1.
2.6. Molecular docking studies
To account for the topology of target-specific binding between cocrystal [Cu2(valdien)2⋯2Cl2CHCOOH] 1 with DNA, blind molecular docking was carried out to mimic the interaction modes. From the resultant docked structures (Fig. 8), it is clear that 1 has slightly twisted the hydrophobic surface (interior) of DNA in such a way that planar part of appended pharmacophore ligand makes favorable stacking interactions between DNA base pairs and 1 lead to attractive charge electrostatic interaction with the sugar-phosphate backbone which defines the stability of groove, detail description given in Table 2. The binding energy of the docked structures cocrystal [Cu2(valdien)2⋯2Cl2CHCOOH] 1 was found to −9.2 kcal mol−1. Therefore, it can be concluded that it is in good agreement with the experimental studies.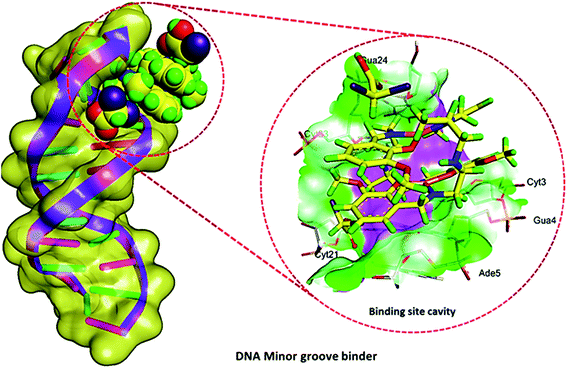 | ||
| Fig. 8 Molecular docked model of cocrystal [Cu2(valdien)2⋯2Cl2CHCOOH] 1 with DNA [dodecamer duplex of sequence d(CGCGAATTCGCG)2 (PDB ID: 1BNA)]. | ||
| Name | Distance (Å) | Category | Type | Binding affinity (kcal mol−1) |
|---|---|---|---|---|
| 1: H11-B: DC21: O2 | 1.92 | Conventional | ||
| A: DA5: H3-1: O12 | 2.19 | Conventional | ||
| 1: H26-B: DC23: O3′ | 2.36 | Hydrogen bond | C–hydrogen bond | −9.20 |
| 1: H32-A: DC3: O2 | 3.06 | C–hydrogen bond | ||
| 1: H32-A: DG4: O4′ | 2.65 | C–hydrogen bond | ||
| 1: H1-B: DC23: O2 | 2.52 | C–hydrogen bond |
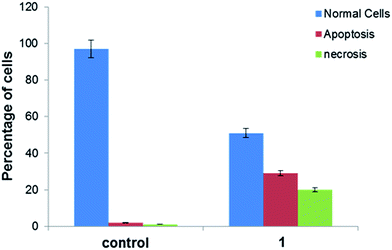
2.7. Cytotoxicity studies
| Compounds/synthetic drug | IC50 values (μM) (upon treatment of 24 h) |
|---|---|
| [Cu2(valdien)2⋯2Cl2CHCOOH] 1 | 15.0 ± 0.15 |
| Dichloroacetic acid (DCA) | >100 |
| Cisplatin | 20.2 ± 0.05 |
3. Conclusions
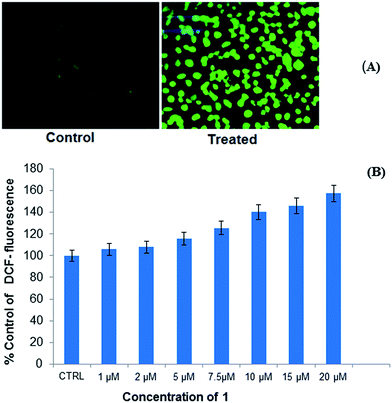
A novel dinuclear copper(II) cocrystal “[Cu2(valdien)2⋯2Cl2CHCOOH],” 1 was synthesized from H2valdien scaffold and dichloroacetic acid embedded with two Cu(II) connected via hydrogen bonded network. [Cu2(valdien)2⋯2Cl2CHCOOH] 1 was thoroughly characterized by single-crystal X-ray crystallography and by other spectroscopic techniques viz. FT-IR, UV-vis, ESI-mass and fluorescence spectroscopy. The non-covalent interaction (NCI) index and Hirshfeld surface analysis revealed the stabilization of cocrystal [Cu2(valdien)2⋯2Cl2CHCOOH] lattice from various kinds of non-covalent interactions (O⋯H, N⋯H, H⋯Cl, Cu⋯H, C⋯O, N⋯O, C⋯Cl, and O⋯Cl, etc.). In vitro interaction studies of [Cu2(valdien)2⋯2Cl2CHCOOH] with ct-DNA revealed good binding propensity which was reflected by its higher Kb, K and KSV values. The nuclease activity of cocrystal [Cu2(valdien)2⋯2Cl2CHCOOH] with pBR322 plasmid DNA was ascertained by the gel electrophoretic mobility assay which revealed an efficient cleaving ability via the oxidative pathway involving singlet oxygen and the superoxide anion as the ROS responsible for mediating the double stranded DNA break reactions. The in vitro antiproliferative activity was ascertained via the MTT assay, and the IC50 values were found to be 15.0 ± 0.15 (treatment upon 24 h) for MCF-7 cancer cell lines. Furthermore, on exposure of cocrystal [Cu2(valdien)2⋯2Cl2CHCOOH] 1 to MCF-7 cancer cell lines, ROS level was significantly increased. Thus, the results of in vitro antiproliferative activity validated that cocrystal [Cu2(valdien)2⋯2Cl2CHCOOH] 1 is a potent chemotherapeutic molecular drug entity for MCF-7 carcinomas, and it warrants further in vivo investigations.4. Experimental section
4.1. Materials and measurements
Cu(CH3COO)2·H2O (Merck), o-vanillin (Sigma-Aldrich), dichloroacetic acid (fluka), 6X loading dye (Fermentas Life Science), and supercoiled pBR322 plasmid DNA (Sigma-Aldrich) were utilized as received. The disodium salt of CT-DNA was purchased from Sigma Chem. Co. and was stored at 4 °C.The diagnostic kits, reagents, and other specified chemicals, for cytotoxic studies, were procured from Sigma Chemical Company Pvt. Ltd, St Louis, MO, USA. DMEM, antibiotics/antimycotics solution and FBS were purchased from Invitrogen, Life Technologies (USA). Plastic and Culture wares consumables used in this study were procured from Nunc, Denmark.
Carbon, hydrogen, and nitrogen contents were carried out on CHN Elemental Analyzer (model: Elementar Vario EL III). The Fourier-transform infrared (FT-IR) spectra were done on Spectrum Two Perkin-Elmer FT-IR spectrometers. The ESI-MS spectrum was recorded on Micromass Quattro II triple quadrupole mass spectrometer. Electronic spectra were recorded on PerkinElmer Lambda 35 UV-vis spectrometer in MeOH using 1 cm path length cuvette, and data were reported in λmax/nm. Fluorescence measurements were determined on an RF-5301 PC spectrofluorophotometer (Shimadzu).
4.2. Synthesis of ligand (H2valdien)
The pro-ligand H2valdien was synthesized from the reaction of o-vanillin (2 mmol, 0.304 g) and diethylenetriamine (1 mmol, 0.108 ml) according to the procedure reported earlier.504.3. Synthesis of the cocrystal [Cu2(valdien)2⋯2Cl2CHCOOH] (1)
To a stirred solution of H2valdien (1 mmol, 0.371 g) and Cu(CH3COO)2·H2O (1 mmol, 0.199 g) in 20 ml of MeOH, triethylamine (2 mmol, 0.278 ml) was added. The resulting clear dark green solution was stirred for 30 min then filtered. To the filtered solution dichloroacetic acid (2 mmol, 0.164 ml) in 5 ml MeCN was added. Then, the solution was placed in a refrigerator for slow evaporation at 4 °C. After 2–3 month, rectangular-shaped brown crystals were collected. Yields = 65–70%. EA, IR, UV-vis, Fluorescence and ESI-mass data are given in the ESI.†4.4. Computational details
The computational programme “ORCA” package was used to carry out density functional theory (DFT) calculations51,52 using the B3LYP functionals53 with Aldrich's def2-TZVP basis set for copper atoms and def2-SVP basis set for C, H, O, N atoms for the geometry optimization and single-point calculations.54–56 Starting geometry was obtained from single crystal X-ray data. Atomic coordinates of optimized structure (Fig. S13†) of cocrystal [Cu2(valdien)2⋯2Cl2CHCOOH] 1 is given in Table S4,† in good agreement with single crystal X-ray structure. To calculate the density of the electron for all atoms to get NCI index measurements, def2-TZVP basis set were used for the structure of 1.57–59 To speed up the calculations we employed the resolution of identity (RI) approximation with the decontracted auxiliary def2-SVP/J and def2-TZV/J Coulomb fitting basis sets and the chain-of-spheres approximation to exact exchange with empirical van der Waals correction as executed in ORCA.60,61 The molecular docking studies were carried out by using Autodock Vina version 1.1.2.62,63 All rotatable bonds inside the ligand were allowed to rotate freely, and receptor was considered rigid. From the http://www.rcsb.org./pdb (protein data bank), the B-DNA dodecamer d(CGCGAATTCGCG)2 (PDB ID: 1BNA) crystal structure was retrieved The Discovery Studio 4.1, and PyMol64,65 were used to visualize minimum energy favorable docked poses. Hirshfeld surface was mapped using Crystal Explorer66 software using crystal structure coordinates of CIF files.4.5. Crystal structure determination
Bruker SMART APEX CCD diffractometer was used to collect single crystal X-ray data of cocrystal [Cu2(valdien)2····2Cl2CHCOOH] 1 at 100 K on a using graphite monochromatic MoKα radiation (λ= 0.71073 Å). The linear absorption coefficients, scattering factors for the atoms and the anomalous dispersion corrections were referred for X-ray crystallography from the International Tables.67 The data integration and reduction were analyzed by using SAINT software.68 Empirical absorption correction was applied to the collected reflections with SADABS,69 and the space group was determined using XPREP.70By using the direct methods SHELXTL-2016, the structure was solved and refined on F2 by full-matrix least-squares using the SIR-97 program package.71 Only a few H atoms could be located in the difference Fourier maps in the structure. The remaining were positioned in calculated positions using idealized geometries (riding model) and assigned fixed isotropic displacement parameters. All non-H atoms were refined anisotropically. The refinement and crystal data are presented in Table 1.
4.6. Methodology for biological studies
Nucleic acid binding experiments were performed in Tris–HCl/NaCl (5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50) buffer at pH 7.2 which included absorption spectral traces, emission spectroscopy and circular dichroism measurements confronted to the standard methods and practices previously adopted by our laboratory.72–76 While measuring the absorption spectra an equal amount of DNA was added to the reference solution to eliminate the absorption of the CT-DNA itself, and the absorbance of the Tris buffer was subtracted through baseline correction. The cleavage experiments of supercoiled pBR322 DNA (300 ng) in Tris–HCl/NaCl (5
50) buffer at pH 7.2 which included absorption spectral traces, emission spectroscopy and circular dichroism measurements confronted to the standard methods and practices previously adopted by our laboratory.72–76 While measuring the absorption spectra an equal amount of DNA was added to the reference solution to eliminate the absorption of the CT-DNA itself, and the absorbance of the Tris buffer was subtracted through baseline correction. The cleavage experiments of supercoiled pBR322 DNA (300 ng) in Tris–HCl/NaCl (5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 mM) buffer at pH 7.2 was studied by agarose gel electrophoresis. The samples were incubated for 45 min at 37 °C, and the cleavage process with and without ROS was monitored using agarose gel electrophoresis. A loading buffer comprising of 25% bromophenol blue, 0.25% xylene cyanol, and 30% glycerol was added, and electrophoresis was done at 30 V for 1 h in Tris–HCl buffer 1% agarose gel containing 1.0 mg ml−1 ethidium bromide. In vitro experiment on MCF7 (human, mammary gland, metastatic site, epithelial cell, breast cancer) cell line viz., cell proliferation, cell culture, MTT assay, AO/EB staining and ROS generation assays were carried by adopting standard methods77–79 with slight modification as reported earlier by us and references therein.80–85
50 mM) buffer at pH 7.2 was studied by agarose gel electrophoresis. The samples were incubated for 45 min at 37 °C, and the cleavage process with and without ROS was monitored using agarose gel electrophoresis. A loading buffer comprising of 25% bromophenol blue, 0.25% xylene cyanol, and 30% glycerol was added, and electrophoresis was done at 30 V for 1 h in Tris–HCl buffer 1% agarose gel containing 1.0 mg ml−1 ethidium bromide. In vitro experiment on MCF7 (human, mammary gland, metastatic site, epithelial cell, breast cancer) cell line viz., cell proliferation, cell culture, MTT assay, AO/EB staining and ROS generation assays were carried by adopting standard methods77–79 with slight modification as reported earlier by us and references therein.80–85
The cell lines was purchased from ATCC.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
We gratefully acknowledge the DST–PURSE program, DST–FIST and DRS–1 (SAP) from UGC, New Delhi, India. The authors extend their appreciation to the Deanship of Scientific Research at King Saud University for funding this work through research group No. RG-1438-006. Authors are thankful to the staff of IIT KANPUR for the assistance in Single Crystal X-ray studies.References
- Ö. Almarsson and M. J. Zaworotko, Chem. Commun., 2004, 1889–1896 RSC.
- J. W. Steed, Trends Pharmacol. Sci., 2013, 34, 185–193 CrossRef CAS PubMed.
- P. Sanphui, Mol. Pharmaceutics, 2015, 12, 889–897 CrossRef CAS PubMed.
- H. D. Clarke, M. B. Hickey, B. Moulton, J. A. Perman, M. L. Peterson, Ł. Wojtas, Ö. Almarsson and M. J. Zaworotko, Cryst. Growth Des., 2012, 12, 4194–4201 CAS.
- N. Schultheiss and A. Newman, Cryst. Growth Des., 2009, 9, 2950–2967 CAS.
- M. Khan, V. Enkelmann and G. J. Brunklaus, J. Am. Chem. Soc., 2010, 132, 5254–5263 CrossRef CAS PubMed.
- S. Aitipamula, R. Banerjee, A. K. Bansal, K. Biradha, M. L. Cheney, A. R. Choudhury, G. R. Desiraju, A. G. Dikundwar, R. Dubey, N. Duggirala, P. P. Ghogale, S. Ghosh, P. K. Goswami, N. R. Goud, R. K. R. Jetti, P. Karpinski, P. Kaushik, D. Kumar, V. Kumar, B. Moulton, A. Mukherjee, G. Mukherjee, A. S. Myerson, V. Puri, A. Ramanan, T. Rajamannar, C. M. Reddy, N. Rodriguez-Hornedo, R. D. Rogers, T. N. Guru Row, P. Sanphui, N. Shan, G. Shete, A. Singh, C. C. Sun, J. A. Swift, R. Thaimattam, T. S. Thakur, R. K. Thaper, S. P. Thomas, S. Tothadi, V. R. Vangala, P. Vishweshwar, D. R. Weyna and M. J. Zaworotko, Cryst. Growth Des., 2012, 12, 2147–2152 CAS.
- M. Nayak, R. Koner, H. H. Lin, U. Florke, H. H. Wei and S. Mohanta, Inorg. Chem., 2006, 45, 10764–10773 CrossRef CAS PubMed.
- S. Hazra, R. Koner, M. Nayak, H. A. Sparkes, J. A. K. Howard and S. Mohanta, Cryst. Growth Des., 2009, 9, 3603–3608 CAS.
- S. Sarkar, M. Nayak, M. Fleck, U. Florke, S. Dutta, R. Koner and S. Mohanta, Eur. J. Inorg. Chem., 2010, 735–743 CrossRef CAS.
- S. Hazra, S. Sasmal, M. Nayak, H. A. Sparkes, J. A. K. Howard and S. Mohanta, CrystEngComm, 2010, 12, 470–477 RSC.
- C. C. Chou, C. C. Su, H. L. Tsai and K. H. Lii, Inorg. Chem., 2005, 44, 628–632 CrossRef CAS PubMed.
- M. Palaniandavar, R. J. Butcher and A. W. Addison, Inorg. Chem., 1996, 35, 467–471 CrossRef CAS PubMed.
- R. C. Holz and L. C. Thompson, Inorg. Chem., 1993, 32, 5251–5256 CrossRef CAS.
- P. Jones, R. S. Vagg and P. A. Williams, Inorg. Chem., 1984, 23, 4110–4111 CrossRef CAS.
- W. J. Evans, T. J. Boyle and J. W. Ziller, Inorg. Chem., 1992, 31, 1120–1122 CrossRef CAS.
- J. R. Wang, Q. Yu, W. Dai and X. Mei, Chem. Commun., 2016, 52, 3572–3575 RSC.
- S. Aitipamula, P. S. Chow and R. B. H. Tan, CrystEngComm, 2009, 11, 1823–1827 RSC.
- P. Grobelny, A. Mukherjee and G. R. Desiraju, CrystEngComm, 2011, 13, 4358–4364 RSC.
- M. L. Cheney, D. R. Weyna, N. Shan, M. Hanna, L. Wojtas and M. J. Zaworotko, J. Pharm. Sci., 2011, 100, 2172–2181 CrossRef CAS PubMed.
- I. Nugrahani, S. Asyarie, S. N. Soewandhi and S. Ibrahim, Int. J. Pharmacol., 2007, 3, 475–481 CAS.
- D. A. Sica, Drugs, 2002, 62, 443–462 CrossRef CAS PubMed.
- S. Bangalore, G. Kamalakkannan, S. Parkar and F. H. Messerli, Am. J. Med., 2007, 120, 713–719 CrossRef PubMed.
- O. D. Putra, T. Furuishi, E. Yonemochi, K. Terada and H. Uekusa, Cryst. Growth Des., 2016, 16, 3577–3581 CAS.
- Y. Suh, I. Amelio, T. G. Urbano and M. Tavassoli, Cell Death Dis., 2014, 5, 1018 CrossRef PubMed.
- Q. S. C. Chu, R. Sangha, J. Spratlin, L. J. Vos, J. R. Mackey, A. J. B. McEwan, P. Venner and E. D. Michelakis, Invest. New Drugs, 2015, 33, 603–610 CrossRef CAS PubMed.
- G. Lin, D. K. Hill, G. Andrejeva, J. K. R. Boult, H. Troy, A. C. L. F. W. T. Fong, M. R. Orton, R. Panek, H. G. Parkes, M. Jafar, D. M. Koh, S. P. Robinson, I. R. Judson, J. R. Griffiths, M. O. Leach, T. R. Eykyn and Y. L. Chung, Br. J. Cancer, 2014, 111, 375–385 CrossRef CAS PubMed.
- L. H. Stockwin, S. X. Yu, S. Borgel, C. Hancock, T. L. Wolfe, L. R. Phillips, M. G. Hollingshead and D. L. Newton, Int. J. Cancer, 2010, 127, 2510–2519 CrossRef CAS PubMed.
- P. Kaufmann, K. Engelstad, Y. Wei, S. Jhung, M. C. Sano, D. C. Shungu, W. S. Millar, X. Hong, C. L. Gooch, X. Mao, J. M. Pascual, M. Hirano, P. W. Stacpoole, S. DiMauro and D. C. De Vivo, Neurology, 2006, 66(3), 324–330 CrossRef CAS PubMed.
- (a) K. D. Karlin and Z. Tyeklar, Bioinorganic Chemistry of Copper, Chapman & Hill, New York, 1993 Search PubMed; (b) W. Kaim and J. Rall, Angew. Chem., Int. Ed., 1996, 35, 43–60 CrossRef CAS; (c) I. Papazoglou, P. J. Cox, A. G. Hatzidimitriou, C. Kokotidou, T. Choli-Papadopoulou and P. Aslanidis, Eur. J. Med. Chem., 2014, 78, 383–391 CrossRef CAS PubMed; (d) C. Belle, C. Beguin, I. Gautier-Luneau, S. Hamman, C. Philouze, J. L. Pierre, F. Thomas and S. Torelli, Inorg. Chem., 2002, 41, 479–491 CrossRef CAS PubMed.
- (a) E. K. Van den Beuken and B. L. Feringa, Tetrahedron, 1998, 54, 12985–13011 CrossRef CAS; (b) C. J. Cairns and D. H. Busch, Coord. Chem. Rev., 1986, 69, 1–55 CrossRef; (c) P. Akilan, M. Thirumavalavan and M. Kandaswamy, Polyhedron, 2003, 22, 1407–1413 CrossRef CAS; (d) L. K. Thompson, F. L. Lee and E. J. Gabe, Inorg. Chem., 1988, 27, 39–46 CrossRef CAS.
- (a) D. S. Rajaa, N. S. P. Bhuvaneshb and K. Natarajan, Eur. J. Med. Chem., 2011, 46, 4584–4594 CrossRef PubMed; (b) M. Alagesan, N. S. Bhuvanesh and N. Dharmaraj, Eur. J. Med. Chem., 2014, 78, 281–293 CrossRef CAS PubMed; (c) Y. Gou, Y. Zhang, J. X. Qi, Z. P. Zhou, F. Yang and H. Liang, J. Inorg. Biochem., 2015, 144, 47–55 CrossRef CAS PubMed; (d) Balewski, F. Sązewski, P. J. Bednarski, M. Gdaniec, E. Borys and A. Makowska, Molecules, 2014, 19, 17026–17051 CrossRef PubMed; (e) N. Chitrapriya, W. Wang, Y. J. Jang, S. K. Kim and J. H. Kim, J. Inorg. Biochem., 2014, 140, 153–159 CrossRef CAS PubMed.
- (a) E. Halevas, O. Tsave, M. P. Yavropoulou, A. Hatzidimitriou, J. G. Yovos, V. Psycharis, C. Gabriel and A. Salifoglou, J. Inorg. Biochem., 2015, 147, 99 CrossRef CAS PubMed; (b) K. Lirdprapamongkol, J. Peterkramb, T. Suthiphongchai, R. Surarit, C. Srisomsap, G. Dannhardt and J. Svasti, J. Agric. Food Chem., 2009, 57, 3055 CrossRef CAS PubMed; (c) J. Vanco, J. Marek, Z. Travnicek, E. Racanska, J. Muselik and O. Svajlenova, J. Inorg. Biochem., 2008, 102, 595 CrossRef CAS PubMed; (d) C. Patra, A. K. Bhanja, C. Sen, D. Ojha, D. Chattopadhyay, A. Mahapatra and C. Sinha, Sens. Actuators, B, 2016, 228, 287 CrossRef CAS; (e) A. K. Bhanja, C. Patra, S. Mondal, D. Ojha, D. Chattopadhyay and C. Sinha, RSC Adv., 2015, 5, 48997 RSC.
- P. C. Bruijnincx and P. J. Sadler, Curr. Opin. Chem. Biol., 2008, 12, 197–206 CrossRef CAS PubMed.
- T. W. Hambley, Dalton Trans., 2007, 4929–4937 RSC.
- C. Santini, M. Pellei, V. Gandin, M. Porchia, F. Tisato and C. Marzano, Chem. Rev., 2014, 114, 815–862 CrossRef CAS PubMed.
- T. Storr, K. H. Thompson and C. Orvig, Chem. Soc. Rev., 2006, 35, 534–544 RSC.
- S. Sarkar, T. Mukherjee, S. Sen, E. Zangrando and P. Chattopadhyay, J. Mol. Struct., 2010, 980, 117–123 CrossRef CAS.
- M. Usman, F. Arjmand, M. Ahmad, M. S. Khan, I. Ahmad and S. Tabassum, Inorg. Chim. Acta, 2016, 453, 193–201 CrossRef CAS.
- J. X. Song, J. M. Chen and T. B. Lu, Cryst. Growth Des., 2015, 15, 4869–4875 CAS.
- R. Chaudret, B. de Courcy, J. Contreras-Garcia, E. Gloaguen, A. Zehnacker-Rentien, M. Mons and J. P. Piquemal, Phys. Chem. Chem. Phys., 2014, 16, 9876 RSC.
- J. A. Harrison, M. A. Sajjad, P. Schwerdtfeger and A. J. Nielson, Cryst. Growth Des., 2016, 16, 4934–4942 CAS.
- N. Han, Y. Zeng, X. Li, S. Zheng and L. Meng, J. Phys. Chem. A, 2013, 117, 12959–12968 CrossRef CAS PubMed.
- M. Ganeshpandian, R. Loganathan, E. Suresh, A. Riyasdeen, M. A. Akbarsha and M. Palaniandavar, Dalton Trans., 2014, 43, 1203–1219 RSC.
- R. Manikandan, N. Chitrapriya, Y. J. Jang and P. Viswanathamurthi, RSC Adv., 2013, 3, 11647–11657 RSC.
- M. Cory, D. D. McKee, J. Kagan, D. W. Henry and J. A. Miller, J. Am. Chem. Soc., 1985, 107, 2528–2536 CrossRef CAS.
- J. R. Lakowicz and G. Weber, Biochemistry, 1973, 12, 4161–4170 CrossRef CAS PubMed.
- R. Loganathan, S. Ramakrishnan, E. Suresh, M. Palaniandavar, A. Riyasdeen and M. A. Akbarsha, Dalton Trans., 2014, 43, 6177–6194 RSC.
- V. A. Kawade, A. A. Kumbhar, A. S. Kumbhar, C. Nather, A. Erxleben, U. B. Sonawane and R. R. Joshi, Dalton Trans., 2011, 40, 639–650 RSC.
- J. Long, F. Habib, P. H. Lin, I. Korobkov, G. Enright, L. Ungur, W. Wernsdorfer, L. F. Chibotaru and M. Murugesu, J. Am. Chem. Soc., 2011, 133, 5319–5328 CrossRef CAS PubMed.
- F. Neese, The ORCA program system, Wiley Interdiscip. Rev.: Comput. Mol. Sci., 2012, 2, 73–78 CrossRef CAS.
- F. Neese, Orca. An ab Initio, Density Functional and Semiempirical Program Package version, 2009 Search PubMed.
- C. Lee, W. Yang and R. G. Parr, Phys. Rev. B: Condens. Matter Mater. Phys., 1988, 37, 785–789 CrossRef CAS.
- F. Weigend and R. Ahlrichs, Phys. Chem. Chem. Phys., 2005, 7, 3297–3305 RSC.
- A. Schaefer, C. Huber and R. Ahlrichs, J. Chem. Phys., 1994, 100, 5829–5835 CrossRef CAS.
- A. Schaefer, H. Horn and R. Ahlrichs, J. Chem. Phys., 1992, 97, 2571–2577 CrossRef CAS.
- E. R. Johnson, S. Keinan, P. Mori-Sánchez, J. Contreras-Garcıa, A. J. Cohen and W. Yang, J. Am. Chem. Soc., 2010, 132, 6498–6506 CrossRef CAS PubMed.
- J. Contreras-Garcia, E. R. Johnson, S. Keinan, R. Chaudret, J. P. Piquemal, D. N. Beratan and W. Yang, J. Chem. Theory Comput., 2011, 7, 625–632 CrossRef CAS PubMed.
- A. R. Allouche, J. Comput. Chem., 2011, 32, 174 CrossRef CAS PubMed.
- S. Grimme, J. Antony, S. Ehrlich and H. Krieg, J. Chem. Phys., 2010, 132, 154104–154119 CrossRef PubMed.
- C. Steffen, K. Thomas, U. Huniar, A. Hellweg, O. Rubner and A. Schroer, J. Comput. Chem., 2010, 31, 2967–2970 CAS.
- O. Trott and A. J. Olson, J. Comput. Chem., 2010, 31, 455–461 CAS.
- M. F. Sanner, J. Mol. Graphics Modell., 1999, 17, 57–61 CAS.
- Accelrys Software Inc., Discovery Studio Modeling Environment, Release 4.0, San Diego, Accelrys Software Inc., 2013 Search PubMed.
- The PyMOL Molecular Graphics System, Version 1.5.0.4 Schrödinger, LLC Search PubMed.
- M. A. Spackman and D. Jayatilaka, CrystEngComm, 2009, 11, 19–32 RSC.
- C. H. MacGillavry, G. D. Rieck and K. Lonsdale, International Tables for X-Ray Crystallography, Kynoch Press, Birmingham, England, 1952, vol. III, pp. 257–269 Search PubMed.
- SAINT, version 6.02, Bruker AXS, Madison, WI, 1999 Search PubMed.
- G. M. Sheldrick, SADABS, Empirical Absorption Correction Program, University of Göttingen, Göttingen, Germany, 1997 Search PubMed.
- XPREP, version 5.1, Siemens Industrial Automation Inc., Madison, WI, 1995 Search PubMed.
- G. M. Sheldrick, SHELXTL Version 2016/6, http://shelx.uniac.gwdg.de/SHELX/index.php Search PubMed.
- J. Marmur, J. Mol. Biol., 1961, 3, 208–218 CrossRef CAS.
- M. E. Reicmann, S. A. Rice, C. A. Thomas and P. A. Doty, J. Am. Chem. Soc., 1954, 76, 3047–3053 CrossRef.
- A. Wolfe, G. H. Shimer and T. Meehan, Biochemistry, 1987, 26, 6392–6396 CrossRef CAS PubMed.
- F. Arjmand, M. Muddassir and R. H. Khan, Eur. J. Med. Chem., 2010, 45, 3549–3557 CrossRef CAS PubMed.
- S. Yadav, I. Yousuf, M. Usman, M. Ahmad, F. Arjmand and S. Tabassum, RSC Adv., 2015, 5, 50673–50690 RSC.
- D. Chandra, K. V. Ramana, L. Wang, B. N. Christensen, A. Bhatnagar and S. K. Srivastava, Invest. Ophthalmol. Visual Sci., 2002, 43, 2285–2292 Search PubMed.
- J. A. Buege and S. D. Aust, Methods Enzymol., 1978, 52, 302–310 CAS.
- S. A. S. Sakinah, S. T. Handayani and L. P. A. Hawariah, Cancer Cell Int., 2007, 7, 1 CrossRef PubMed.
- I. Yousuf, F. Arjmand, S. Tabassum, L. Toupet, R. A. Khan and M. A. Siddiqui, Dalton Trans., 2015, 44, 10330–10342 RSC.
- S. Tabassum, A. Asim, R. A. Khan, F. Arjmand, D. Rajakumar, P. Balaji and M. A. Akbarsha, RSC Adv., 2015, 5, 47439–47450 RSC.
- A. Sarkar, A. R. Paital, R. A. Khan, F. Arjmand, V. Bertolasi, C. Mathonière, R. Clérac and D. Ray, Dalton Trans., 2013, 42, 12495–12506 RSC.
- R. A. Khan, M. Usman, D. Rajakumar, P. Balaji, A. Alsalme, F. Arjmand, K. AlFarhan, M. A. Akbarsha, F. Marchetti, C. Pettinari and S. Tabassum, Sci. Rep., 2017, 7, 45229 CrossRef CAS PubMed.
- R. A. Khan, A. de Almeida, K. Al-Farhan, A. Alsalme, A. Casini, M. Ghazzali and J. Reedijk, J. Inorg. Biochem., 2016, 165, 128–135 CrossRef CAS PubMed.
- R. A. Khan, K. Al-Farhan, A. de Almeida, A. Alsalme, A. Casini, M. Ghazzali and J. Reedijk, J. Inorg. Biochem., 2014, 140, 1–5 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. CCDC 1402001. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c7ra08262b |
| This journal is © The Royal Society of Chemistry 2017 |