Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Evaluation of the efficacy and safety of Ganoderma lucidum mycelium-fermented liquid on gut microbiota and its impact on cardiovascular risk factors in human†
Qizheng Wua,
Houcheng Zhanga,
Peng Gorge Wang*ab and
Min Chen *a
*a
aThe State Key Laboratory of Microbial Technology, National Glycoengineering Research Center, School of Life Science, Shandong University, Jinan, Shandong 250100, China. E-mail: chenmin@sdu.edu.cn
bDepartment of Chemistry, Georgia State University, Atlanta, Georgia 30303, USA
First published on 21st September 2017
Abstract
Ganoderma lucidum is a Chinese traditional medicine with various bioactivities. However, the impacts of a Ganoderma lucidum mycelium-fermented liquid (GLFL), as a promising alternative product, on the gut microbes and cardiovascular risk factors have not been explored in humans to date. In this study, the composition of polysaccharides in GLFL was analyzed, and eight volunteers were fed with GLFL to investigate its influence on gut microbes and cardiovascular risk factors. High-throughput 16S rRNA gene sequencing technique was utilized to investigate the effects of GLFL on the diversity of the bacterial communities and the composition of gut microbiota. The results demonstrated that GLFL significantly altered β diversity and several phyla of gut microbiota in volunteers. Moreover, GLFL provided protection to humans by promoting the growth of probiotics, incorporating genus Lactobacillus (p < 0.05), and reducing pathogens containing genus Campylobacter and Aggregatibacter (p < 0.05); however, GLFL also had detrimental effects as it increased the population of Firmicutes/Bacteroidetes and opportunistic pathogens including genus Acinetobacter, Pseudomonas, Serratia, Stenotrophomonas, Peptococcus and S24-7 (p < 0.05) and reduced that of probiotics genus Lactococcus. Finally, we found that GLFL reduced plasma low-density lipoprotein cholesterol (LDL-c) in volunteers. This is helpful for further understanding the effect of Ganoderma lucidum or its related products on human body.
Introduction
Ganoderma lucidum, which is a traditional medical mushroom, has been used to promote health and longevity.1 Since G. lucidum is rare in nature, as an alternative method, submerged fermentation of Ganoderma lucidum mycelium is carried out as it requires shorter time for cultivation.2–4 Prior assays indicated that the G. lucidum mycelium-fermented liquid (GLFL) included ganoderic acid and extracellular and intracellular polysaccharides5 as effective constituents.The Ganoderma lucidum mycelium-fermented liquid was verified to strengthen health in host. Previous report suggested that components of GLFL had antitumor activity,6 and recent assays also found that GLFL induced human peripheral blood mononuclear cells (PBMC) to synthesize the tumor necrosis factor α (TNF-α)7–9 as a supporting therapy in cancer patients. Moreover, another assay manifested that GLFL had hepatoprotective properties in rat.10 However, the biological activities and functions of GLFL have not been extensively studied especially in the fields of regulation of gut microbiota and cardiovascular diseases.
The gut microbiota and cardiovascular risk factors play a crucial role in host health.11,12 Several reports have found that Ganoderma lucidum-related products could regulate the gut microbiota and cardiovascular risk factors. Previous reports suggested that a water extract of Ganoderma lucidum mycelium reversed the composition of gut microbiota in obese mice.13 GLFL was proven to reduce plasma low-density lipoprotein cholesterol (LDL-c) and triglycerides, total cholesterol, and increased high-density lipoprotein cholesterol (HDL-c) in rat.14 However, it is unclear whether the Ganoderma lucidum mycelium-fermented liquid produces any effect on gut microbiota and cardiovascular risk factors in human.
In this study, to evaluate the efficacy and safety of Ganoderma lucidum mycelium-fermented liquid, which contained extracellular and intracellular biopolymers, we examined the composition of polysaccharides of GLFL purchased from market, performed a systematic analysis of gut microbiota following oral administration of GLFL by the 16S rDNA sequencing technique, as well as analyzed the plasma cardiovascular risk factors. Our study will provide the foundation of how GLFL influences the human body.
Materials and methods
Volunteers and administration
A cohort of eight healthy volunteers, consisting of equal number of females and males, participated in this study. Volunteers drank 50 mL GLFL every day for one month from April 1 to 30, 2016. The administration was random in a day. Volunteers were needed to conform to all the following conditions: (1) females or males should be ≥20 years of age; (2) they should have normal body mass index (BMI = 18.5–24.9 kg m−2); (3) they should have no smoking history, (4) no severe drinking history, and (5) no medical history; (6) they should not be vegetarians; (7) they should not be taking probiotics, prebiotics or antibiotics one month before commencement of the study; and (8) they should not be taking probiotics, prebiotics or antibiotics within one month of enrollment. All procedures of this study involving human participants were performed in compliance with the guidelines of Biomedical research ethics review method involving people (China). The study was approved by the Ethics Committee of Shandong University, and written informed consent was obtained from each volunteer before commencement of the study.Experimental design
The fresh stool and serum samples were obtained before GLFL administration (named pre-feeding group), and the fresh stool and serum samples were also obtained at the end of the experiment (named post-feeding group). The gut microbiota was extracted from stool and then analyzed by the high-throughput 16S rRNA gene sequencing technique; the plasma lipids and lipoproteins were analyzed by an automatic biochemical analyzer. Finally, the data of the pre-feeding group as a baseline (control) was compared with the data of the post-feeding group. Moreover, monosaccharides of GLFL were analyzed by gas chromatography/mass spectrometry.The components of GLFL
In our study, the Ganoderma lucidum mycelium-fermented liquid was purchased from a commercial producer (Hongshun Biological Technology Co., Ltd, Weihai, China), which comprised extracellular and intracellular biopolymers, containing 2.9% (w/w) carbohydrate, 0.19% (w/w) protein, <0.1% (w/w) lipids, 96.5% (w/w) water, 0.2% (w/w) dietary fiber, and 0.25% (w/w) other components. The dry biomass of GLFL was ≥3.5% (w/v), whereas the percentage of polysaccharides in GLFL was ≥0.7% (w/v).Extraction of the GLFL crude polysaccharides
GLFL was mixed with four-fold volume of 100% EtOH at 4 °C for 12 hours to precipitate the GLFL crude polysaccharides. The precipitate was pelleted (15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 ×0g, 4 °C, 10 minutes), and then, the supernatant was drained and lyophilized.
000 ×0g, 4 °C, 10 minutes), and then, the supernatant was drained and lyophilized.
Gas chromatography/mass spectrometry analysis for monosaccharides
Collection of stool specimens and gut microbiota
The fresh stool specimens were obtained at the start and end of the experiment. Fresh stool specimens were processed immediately after defecation as follows: in brief, a stool specimen weighing 500 mg was suspended in 10 mL 0.01 M pH = 7.2–7.4 sterile phosphate buffered solution (PBS). Then, the mixture was stirred for 10 minutes. The supernatant was obtained by centrifugation at 100g for 3 minutes. The supernatant was centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 10 minutes to obtain the precipitates containing gut microbiota. The precipitate was resuspended in 10 mL 0.01 M pH = 7.2–7.4 PBS, and then was centrifuged at 12000g for 10 minutes to obtain the precipitate. The abovementioned process was repeated 3 times. The bacterial pellets were resuspended in 1 mL sterile PBS and saved at 4 °C for DNA extraction.
000g for 10 minutes to obtain the precipitates containing gut microbiota. The precipitate was resuspended in 10 mL 0.01 M pH = 7.2–7.4 PBS, and then was centrifuged at 12000g for 10 minutes to obtain the precipitate. The abovementioned process was repeated 3 times. The bacterial pellets were resuspended in 1 mL sterile PBS and saved at 4 °C for DNA extraction.
DNA extraction from gut microbiota and sequencing
Gut bacterial DNA was extracted using the TIANGEN® TIANamp Bacteria DNA Kit (TIANGEN Biotech Co., Ltd, Beijing, China) according to the manufacturer's instructions.The gut bacterial V4 hypervariable region of 16S rDNA was amplified with the universal primer pair B341F (5′-CCTACGGGNGGCWGCAG-3′) and B785R (5′-GACTACHVGGGTATCTAATCC-3′) using the following conditions: initial denaturation at 95 °C for 3 min, 25 cycles at 95 °C for 30 s, 55 °C for 30 s, 72 °C for 30 s, and 72 °C for 5 min. The PCR amplification procedure was conducted using a GeneAmp PCR System 9700 (Life Technologies, Carlsbad, CA, USA). PCR amplification was performed using the KAPA HiFi HotStart ReadyMix PCR Kit (Kapa Biosystems, Woburn, MA, USA). PCR amplicon libraries were built and purified by the QiaQuick Gel Extraction Kit (Qiagen, USA). Then, the concentration of the PCR amplicon libraries was detected using a Qubit Fluorometer 3.0 (Invitrogen). Amplicon sequencing was performed using an Illumina Miseq platform at Beijing Ori-Gene Science and Technology Corp., LTD. (Beijing, China).
Bioinformatic analysis
After sequence assembly and sequence filtration, high-quality sequences were clustered into OTUs based on 97% sequence similarity according to USEARCH.15 OTUs were used for analysis of α diversity (nseqs, sobs, chao, ace, coverage, shannon, npshannon, and simpson) by the mothur method. Rarefaction curves were calculated for all the samples. For β diversity, the OTU sequences were aligned with the SILVA database using PyNAST,16 and the phylogenetic tree was built with the FastTree software.17 The unweighed and weighed distance matrix between communities was generated with the UniFrac software,18 which was the basis of principal coordinates analysis (PCoA).Analysis of the major cardiovascular risk factors
Herein, 3 mL blood was obtained from 8 volunteers before feeding and after feeding, and then, it was put into the Automatic Biochemical Analyzer, Hitachi 7100 (Hitachi, Ltd, Japan), to analyze the major cardiovascular risk factors.Statistical analysis
The data of the pre-feeding group as a baseline (control) was compared with the data of the post-feeding group. The Metastats statistical software was used to test the differences of species abundance between two groups. Student's t-test was performed to test the differences. Differences were considered significant at P < 0.05. Data are denoted as mean ± standard deviation (SD).Results and discussion
Composition of the polysaccharides of GLFL
Ganoderma polysaccharide is considered as one of the beneficial ingredients. However, there is a wide diversity in the chemical composition among different Ganoderma polysaccharides; this results in various bioactivities. Thus, characterization of the composition of polysaccharide is crucial. We analyzed the composition of the polysaccharides by gas chromatography/mass spectrometry. The carbohydrate portion was found to comprise glucose (52.01%), galactose (19.36%), arabinose (6.41%), fucose (4.67%), xylose (7.45%), and mannose (10.11%). Our results supported the data obtained by other researchers indicating that the polysaccharides isolated from Ganoderma consisted of mannose, glucose, arabinose, fucose, xylose, and galactose with different combinations.19GLFL changed the diversity of gut bacteria
To address the changes in the gut bacterial structure, we characterized the gut bacterial community diversity in the pre-feeding and post-feeding GLFL group by pyrosequencing of 16S rDNA covering the V4 hypervariable regions. To exclude the influence of host adiposity, age, and geographical location on the gut microbiome, the eight volunteers selected for this study were healthy adults, living in northern China, and had a normal body mass index (BMI = 18.5–24.9 kg m−2) (ESI Table 1†).After selecting the high-quality sequences, we obtained 120![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 259 sequences (mean ± S.D., 15
259 sequences (mean ± S.D., 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 032 ± 3829) from the pre-feeding group and 89
032 ± 3829) from the pre-feeding group and 89![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 760 sequences (mean ± S.D., 11
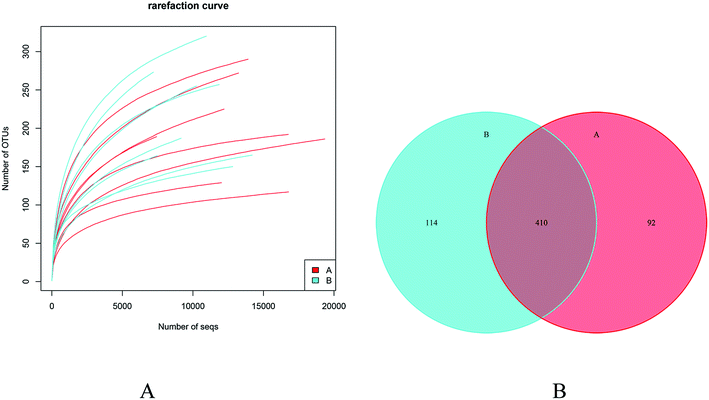
760 sequences (mean ± S.D., 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 220 ± 2571) from the post-feeding group (Table 1). Moreover, rarefaction curve analysis indicated that sufficient sequences were obtained to conduct the pyrosequencing analysis (Fig. 1A). More OTUs (524 OTUs) were detected in the post-feeding GLFL group as compared to those in the pre-feeding GLFL group (502 OTUs), but both groups shared 410 OTUs (Fig. 1B). Therefore, GLFL assisted in the generation of 22 OTUs.
220 ± 2571) from the post-feeding group (Table 1). Moreover, rarefaction curve analysis indicated that sufficient sequences were obtained to conduct the pyrosequencing analysis (Fig. 1A). More OTUs (524 OTUs) were detected in the post-feeding GLFL group as compared to those in the pre-feeding GLFL group (502 OTUs), but both groups shared 410 OTUs (Fig. 1B). Therefore, GLFL assisted in the generation of 22 OTUs.
| Sample | Clean_seqs | Filter_seqs | Percent |
|---|---|---|---|
| a A and B correspond to pre-feeding group and post-feeding group, respectively. | |||
| A1 | 9055 | 7859 | 86.79% |
| A2 | 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 972 972 |
13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 004 004 |
86.86% |
| A3 | 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 141 141 |
20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 295 295 |
87.70% |
| A4 | 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 065 065 |
14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 424 424 |
79.85% |
| A5 | 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 886 886 |
17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 741 741 |
77.52% |
| A6 | 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 045 045 |
13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 115 115 |
56.91% |
| A7 | 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 112 112 |
16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 238 238 |
70.26% |
| A8 | 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 209 209 |
17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 583 583 |
75.76% |
| B1 | 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 162 162 |
12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 562 562 |
82.85% |
| B2 | 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 492 492 |
12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 031 031 |
77.66% |
| B3 | 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 784 784 |
13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 871 871 |
60.88% |
| B4 | 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 728 728 |
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 956 956 |
74.39% |
| B5 | 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 338 338 |
9724 | 72.90% |
| B6 | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 350 350 |
8022 | 77.51% |
| B7 | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 251 251 |
7817 | 76.26% |
| B8 | 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 152 152 |
14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 777 777 |
63.83% |
Although no significant differences in the α diversity were detected between two groups (ESI Table 2†), the β diversity between two groups had evident difference, as observed by calculation of unweighed unifrac. Principal coordinates analysis (PCoA) suggested that the second principal component PC2, which accounted for 20.3% of variance in the data, could completely differentiate the post-feeding group from the pre-feeding groups (Fig. 2). This indicated that the composition of gut microbiota changed greatly after the administration of GLFL.
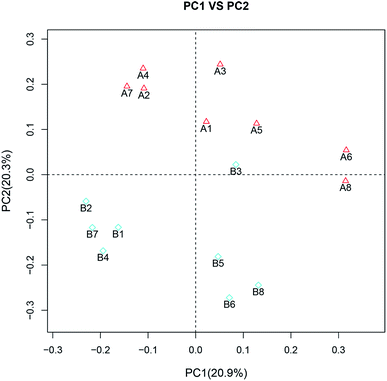 | ||
| Fig. 2 Principal coordinate analysis (PCoA). A and B correspond to the pre-feeding group and post-feeding group, respectively. | ||
The composition of fecal microbiota was changed after feeding the GLFL
To address how GLFL altered the gut microbiome, we compared the microbiota composition in the pre-feeding and post-feeding GLFL groups. Taxonomic profiling demonstrated that Firmicutes and Bacteroidetes were the main gut bacteria in two groups (Fig. 3). At the phylum level, in the pre-feeding group, the percentage of Firmicutes (F) was 62.98% and that of Bacteroidetes (B) was 23.91%, and F/B = 2.63. However, in the post-feeding group, the percentage of Firmicutes was 68.54% and that of Bacteroidetes was 19.23%, and F/B = 3.56. It was noted that F/B increased 1.35 times, which was consistent with the previous result obtained after feeding Ganoderma lucidum polysaccharides in mice4 and contrary to the result obtained after feeding water extracts of cultured Ganoderma lucidum mycelium in mice.13 A previous report suggested that the gut microbiota of obese human and obese mice had a significantly greater F/B ratio.20 Moreover, GLFL administration increased the population of Acidobacteria (0 vs. 0.0520%, p < 0.05), Lentisphaerae (0 vs. 0.0057%, p < 0.05), Planctomycetes (0.0034% vs. 0.0424%, p < 0 0.05), and Tenericutes (0 vs. 0.0049%, p < 0.05) at the phylum level. Another feature was that the population of phylum Proteobacteria reduced from 8.822% to 6.758% (p > 0.05) (Fig. 4). This result was contrary to previous result, which suggested that mushroom polysaccharide induced elevation of Proteobacteria.21 In murine models, recent result suggested that less Firmicutes and more Bacteroidetes and Proteobacteria were associated with low grade intestinal inflammation.22 Thus, GLFL might reduce the probability of low-grade intestinal inflammation.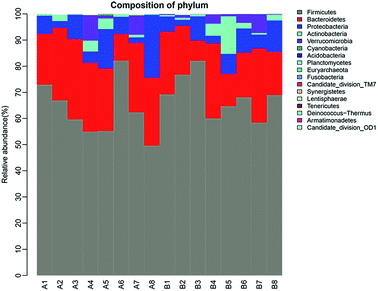 | ||
| Fig. 3 The composition of gut bacteria at the phylum level. Different colors represent different gut microbiota, and the height of the column represents the proportion of gut microbiota. | ||
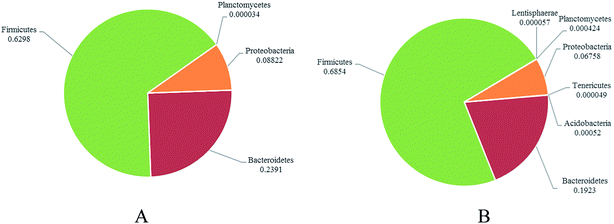 | ||
| Fig. 4 The distribution of dominated gut bacteria at the phylum level between the pre-feeding group (A) and post-feeding group (B). | ||
Focusing on the genus level allowed us to further understand how GLFL altered the gut microbiome. In this study, there were 39 genera, which were significantly altered (P < 0.05) (ESI Table 3†). Herein, we mainly analyzed numerous genera, which had relation with the health.
There were 12 genera that significantly decreased (Fig. 5A). The population of the Campylobacter genus decreased significantly (p = 0.045) after administration of GLFL. Campylobacter genus is associated with diarrheal disease, and it has been viewed as a crucial risk factor for the development of inflammatory bowel disease.23 The population of Aggregatibacter also decreased significantly (p = 0.000999) from 0.0105% to 0. Aggregatibacter is a genus of the phylum Proteobacteria, which contains three species, namely A. actinomycetemcomitans, A. aphrophilus, and A. segnis. A. actinomycetemcomitans is a causative agent of periodontal disease,24 and a recent study suggested that A. aphrophilus is associated with cerebral abscess.25 The population of Succinivibrio genus decreased from 0.0274% to 0. Previous study indicated that the decreased abundance of Succinivibrio was related to parasite infection in pigs.26 However, the population of two beneficial gut bacteria also decreased in this study. The population of Lactococcus genus decreased from 0.0057% to 0 and that of the Turicibacter genus decreased from 0.2556 to 0.0041. Lactococcus genus, which is used on a large scale by the dairy industry, is generally recognized as safe for human consumption. Recent study has suggested that dogs with inflammatory bowel disease have lower abundance of Turicibacter,27 which may be a beneficial gut microbiota. The other genera whose population decreased are shown in Fig. 5A.
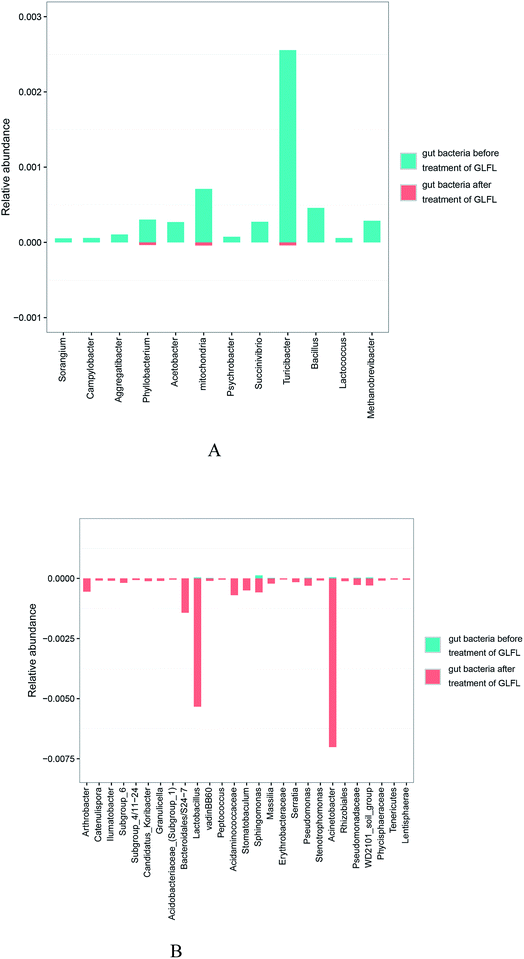 | ||
| Fig. 5 The population of genera shown have been decreased significantly (A) and that of the genera shown have been increased significantly (B). | ||
There were 27 genera, whose population significantly increased (Fig. 5B). The population of Lactobacillus genus increased from 0.0036% to 0.5335%. For many decades, Lactobacillus genus has been known as a beneficial bacteria, and adequate amount of assays have verified that Lactobacillus genus had antitumor activity, antitoxic activity, cholesterol-lowering activity, and antioxidant activity.28 The population of Acinetobacter genus increased mostly from 0.0042% to 0.7015%. This genus is the main reason for infection in fragile patients in the hospital, particularly the species Acinetobacter baumannii, causing skin infections.29 As is known, Acinetobacter species are found frequently in natural environment such as water and soil, as well as in hospital environment, human skin, and foods.30 Interestingly, it was also found in gut microbiota in this study. The population of Pseudomonas genus increased from 0.0022% to 0.0304%. Previous study indicated that infectious species, such as Pseudomonas aeruginosa, of this genus was a key conditioned pathogen in immunocompromised patients and caused life-threatening lung infections in individuals with cystic fibrosis.31 The population of the Serratia genus increased from 0 to 0.0155%, and this bacterium was an opportunistic human pathogen, causative agent of severe nosocomial infections.32 The population of the Stenotrophomonas genus increased from 0 to 0.086%. The numerous recent literatures reported to date have shown that Stenotrophomonas maltophilia, belonging to Stenotrophomonas genus, is an opportunistic pathogen that causes nosocomial infections.33 The population of the Peptococcus genus also increased from 0 to 0.0053%. Species in this genus are a normal part of the human gut microbiome, but Peptococcus magnus is a human pathogen.34 Interestingly, the population of the S24-7, belonging to the Bacteroidales family, increased from 0 to 0.1428%. The recent study indicated that this Bacteroidales family was related to the degradation of particular carbohydrates such as α-glucan, host glycan, and plant glycan.35 In our study, GLFL could contain analogous carbohydrates; thus, the abovementioned family consumed them and then proliferated acutely. Another study suggested that mice fed with a high-fat carbohydrate-free diet had more S24-7;36 thus, S24-7 might be bad gut microbiota.
Water extracts of the cultured Ganoderma lucidum mycelium (WEGL) have been proven to alter the gut microbiota significantly (P < 0.05). Chang et al. proved that a water extract of Ganoderma lucidum mycelium decreased Firmicutes-to-Bacteroidetes ratios and Lactobacillus genus level in mice.13 However, our results suggested that GLFL elevated the Firmicutes-to-Bacteroidetes ratios and Lactobacillus genus level in humans. However, both results suggested that WEGL and GLFL decreased Proteobacteria phylum level and Lactococcus genus level. As GLFL is a complex containing WEGL, extracellular biopolymers, and other compounds, the discrepancy and similarity of the effect of GLFL and WEGL on gut microbiota can be explained by their components. Moreover, the difference of animal model was another reason for discrepancy.
GLFL decreased plasma low-density lipoprotein in humans
Major cardiovascular risk factors, including triglycerides (TG), total cholesterol (TC), glucose, low-density lipoprotein cholesterol (LDL-c), and high-density lipoprotein cholesterol (HDL-c), were analyzed between pre-and post-feeding groups. There was no significant difference between two groups in terms of these factors, but every volunteer after being administered with GLFL exhibited a lower plasma LDL-c (Table 2). This effect may be attributed to the elevation of cholesterol-lowering Lactobacillus genus, as abovementioned. Yang et al. found that the extracts of submerged mycelium culture of Ganoderma lucidum not only reduced low-density lipoprotein cholesterol, but also decreased total cholesterol and triglyceride, and increased high-density lipoprotein cholesterol in rats.14 Moreover, Tong et al. revealed that Ganoderma lucidum without fermentation decreased the LDL-c, TG, and TC level significantly (P < 0.05), whereas further increased the plasma concentration of HDL-c in rats.37 However, we found that GLFL only decreased low-density lipoprotein cholesterol in humans. There were two reasons for the discrepancy. First, their animal model was diet-induced hyperlipidemic rat, whereas our model was a healthy human. Second, they utilized the extracts of submerged mycelium culture of Ganoderma lucidum or G. lucidum fruiting body, whereas we used the submerged mycelium culture of Ganoderma lucidum containing all extracts and other contents. Therefore, difference of model and aliment may result in different consequences.| Groups | Glucose (mmol L−1) | TC (mmol L−1) | TG (mmol L−1) | HDL-c (mmol L−1) | LDL-c (mmol L−1) |
|---|---|---|---|---|---|
| a A and B correspond to pre-feeding group and post-feeding group, respectively. TC, TG, HDL-c and LDL-c correspond to total cholesterol, triglycerides, high density lipoprotein cholesterol and low density lipoprotein cholesterol, respectively. | |||||
| A | 4.99 ± 0.28 | 3.98 ± 0.83 | 0.86 ± 0.42 | 1.60 ± 0.38 | 2.34 ± 0.73 |
| B | 4.88 ± 0.15 | 3.90 ± 0.87 | 0.88 ± 0.28 | 1.52 ± 0.45 | 1.93 ± 0.70 |
Conclusion
When Ganoderma lucidum mycelium-fermented liquid was fed to humans, it profoundly altered the gut microbiota. In the post-feeding group, there was evident difference in β diversity as compared to the case of the pre-feeding group; this suggested that GLFL had altered the composition of gut microbiota. At the phylum level, GLFL increased the population of Firmicutes, whereas it decreased the population of Bacteroidetes and Proteobacteria. At the genus level, there were 39 genera, which were significantly altered (P < 0.05). As we expected, GLFL promoted the growth of probiotics such as Lactobacillus genus, whereas it suppressed the growth of pathogens such as Campylobacter genus. However, we also found that GLFL was unsafe as it increased the population of opportunistic pathogens, such as Acinetobacter genus, and reduced the population of probiotics such as Lactococcus genus. In addition, there was no doubt that GLFL could reduce plasma LDL-c. These results were not identical with previous consequences that were produced from Ganoderma lucidum without fermentation, WEGL or GLFL, which were fed to murine. Based on our results, we appeal to re-examine the effect of Ganoderma lucidum or its derived products on humans and the consequences may be different from previous successful results in murine.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This work was financially supported by the State Key Laboratory of Microbial Technology Projects Fund and the National Natural Science Foundation of China (No. 31270983).References
- B. S. Sanodiya, G. S. Thakur, R. K. Baghel, G. B. Prasad and P. S. Bisen, Ganoderma lucidum: a potent pharmacological macrofungus, Curr. Pharm. Biotechnol., 2009, 10, 717 CAS.
- C. W. Huie and D. Xin, Chromatographic and electrophoretic methods for Lingzhi pharmacologically active components, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2004, 812, 241–257 CrossRef CAS.
- S. Q. Huang, S. Ding and L. Fan, Antioxidant activities of five polysaccharides from Inonotus obliquus, Int. J. Biol. Macromol., 2012, 50, 1183–1187 CrossRef CAS PubMed.
- K. Li, C. Zhuo, C. Teng, S. Yu, X. Wang, Y. Hu, G. Ren, M. Yu and J. Qu, Effects of Ganoderma lucidum polysaccharides on chronic pancreatitis and intestinal microbiota in mice, Int. J. Biol. Macromol., 2016, 93, 904–912 CrossRef CAS PubMed.
- Y. J. Tang and J. J. Zhong, Fed-batch fermentation of Ganoderma lucidum for hyperproduction of polysaccharide and ganoderic acid, Enzyme Microb. Technol., 2002, 31, 20–28 CrossRef CAS.
- S. Y. Lee, T. S. Kang, S. O. Moon, I. D. Lew and M. Y. Lee, Fractionation and antitumor activity of the water soluble exo-polysaccharide by submerged cultivation of Ganoderma lucidum mycelium, Korean J. Appl. Microbiol. Biotechnol., 1996, 24, 459–464 CAS.
- J. Habijanič, M. Berovič, B. Wraber, D. Hodzar and B. Boh, Immunostimulatory effects of fungal polysaccharides from Ganoderma lucidum submerged biomass cultivation, Food Technol. Biotechnol., 2001, 39, 327–331 Search PubMed.
- M. Berovic, J. Habijanic, I. Zore, B. Wraber, D. Hodzar, B. Boh and F. Pohleven, Submerged cultivation of Ganoderma lucidum biomass and immunostimulatory effects of fungal polysaccharides, J. Biotechnol., 2003, 103, 77 CrossRef CAS PubMed.
- J. Habijanic, M. Berovic, B. Boh, M. Plankl and B. Wraber, Submerged cultivation of Ganoderma lucidum and the effects of its polysaccharides on the production of human cytokines TNF-α, IL-12, IFN-γ, IL-2, IL-4, IL-10 and IL-17, New Biotechnol., 2015, 32, 85 CrossRef CAS PubMed.
- C. H. Song, B. K. Yang, K. S. Ra, D. H. Shon, E. J. Park, G. I. Go and Y. H. Kim, Hepatoprotective effect of extracellular polymer produced by submerged culture of Ganoderma lucidum WK-003, J. Microbiol. Biotechnol., 1998, 8, 277–279 Search PubMed.
- J. C. Clemente, L. K. Ursell, L. W. Parfrey and R. Knight, The impact of the gut microbiota on human health: an integrative view, Cell, 2012, 148, 1258–1270 CrossRef CAS PubMed.
- W. H. Organization, WHO|Cardiovascular diseases (CVDs) factsheet, 2013 Search PubMed.
- C. J. Chang, C. S. Lin, C. C. Lu, J. Martel, Y. F. Ko, D. M. Ojcius, S. F. Tseng, T. R. Wu, Y. Y. M. Chen and J. D. Young, Ganoderma lucidum reduces obesity in mice by modulating the composition of the gut microbiota, Nat. Commun., 2015, 6, 7489 CrossRef CAS PubMed.
- B. K. Yang, S. C. Jeong and C. H. Song, Hypolipidemic effect of exo- and endo-biopolymers produced from submerged mycelial culture of Ganoderma lucidum in rats, J. Microbiol. Biotechnol., 2002, 12, 872–877 CAS.
- R. C. Edgar, UPARSE: highly accurate OTU sequences from microbial amplicon reads, Nat. Methods, 2013, 10, 996 CrossRef CAS PubMed.
- J. G. Caporaso, K. Bittinger, F. D. Bushman, T. Z. DeSantis, G. L. Andersen and R. Knight, PyNAST: a flexible tool for aligning sequences to a template alignment, Bioinformatics, 2010, 26, 266–267 CrossRef CAS PubMed.
- M. N. Price, P. S. Dehal and A. P. Arkin, FastTree: computing large minimum evolution trees with profiles instead of a distance matrix, Mol. Biol. Evol., 2009, 26, 1641–1650 CrossRef CAS PubMed.
- C. Lozupone and R. Knight, UniFrac: a new phylogenetic method for comparing microbial communities, Appl. Environ. Microbiol., 2005, 71, 8228–8235 CrossRef CAS PubMed.
- I. C. Ferreira, S. A. Heleno, F. S. Reis, D. Stojkovic, M. J. Queiroz, M. H. Vasconcelos and M. Sokovic, Chemical features of Ganoderma polysaccharides with antioxidant, antitumor and antimicrobial activities, Phytochemistry, 2015, 114, 38 CrossRef CAS PubMed.
- L. Zhao, The gut microbiota and obesity: from correlation to causality, Nat. Rev. Microbiol., 2013, 11, 639 CrossRef CAS PubMed.
- X. Xu and X. Zhang, Lentinula edodes-Derived Polysaccharide Alters the Spatial Structure of Gut Microbiota in Mice, PLoS One, 2015, 10, e0115037 Search PubMed.
- F. Hildebrand, T. L. A. Nguyen, B. Brinkman, R. G. Yunta, B. Cauwe, P. Vandenabeele, A. Liston and J. Raes, Inflammation-associated enterotypes, host genotype, cage and inter-individual effects drive gut microbiota variation in common laboratory mice, Genome Biol., 2013, 14, R4 CrossRef PubMed.
- R. C. Spiller, Role of infection in irritable bowel syndrome, J. Gastroenterol., 2007, 17(42 suppl), 41 CrossRef PubMed.
- B. Henderson, J. M. Ward and D. Ready, Aggregatibacter (Actinobacillus) actinomycetemcomitans: a triple A* periodontopathogen?, Periodontology, 2010, 54, 78–105 CrossRef PubMed.
- S. P. Ahamed, S. Lath, G. J. Degabriele and V. T. Mathew, Cerebral abscess caused by Aggregatibacter aphrophilus, Neurosciences, 2010, 15, 40 Search PubMed.
- R. W. Li, S. Wu, W. Li, K. Navarro, R. D. Couch, D. Hill and J. F. Urban Jr, Alterations in the porcine colon microbiota induced by the gastrointestinal nematode Trichuris suis, Infect. Immun., 2012, 80, 2150–2157 CrossRef CAS PubMed.
- G. Rossi, G. Pengo, M. Caldin, P. A. Palumbo, J. M. Steiner, N. D. Cohen, A. E. Jergens and J. S. Suchodolski, Comparison of microbiological, histological, and immunomodulatory parameters in response to treatment with either combination therapy with prednisone and metronidazole or probiotic VSL#3 strains in dogs with idiopathic inflammatory bowel disease, PLoS One, 2014, 9, e94699 Search PubMed.
- A. D. Cerbo, B. Palmieri, M. Aponte, J. C. Moralesmedina and T. Iannitti, Mechanisms and therapeutic effectiveness of lactobacilli, J. Clin. Pathol., 2015, 69, 187–203 CrossRef PubMed.
- A. Y. Peleg, H. Seifert and D. L. Paterson, Acinetobacter baumannii: emergence of a successful pathogen, Clin. Microbiol. Rev., 2008, 21, 538–582 CrossRef CAS PubMed.
- H. Seifert, L. Dijkshoorn, P. Gerner-Smidt, N. Pelzer, I. Tjernberg and M. Vaneechoutte, Distribution of Acinetobacter species on human skin: comparison of phenotypic and genotypic identification methods, J. Clin. Microbiol., 1997, 35, 2819–2825 CAS.
- E. Kintz and J. B. Goldberg, Regulation of lipopolysaccharide O antigen expression in Pseudomonas aeruginosa, Future Microbiol., 2008, 3, 191–203 CrossRef CAS PubMed.
- L. Vetter, G. Schuepfer, S. P. Kuster and M. Rossi, A Hospital-wide Outbreak of Serratia marcescens, and Ishikawa's “Fishbone” Analysis to Support Outbreak Control, Qual. Manag. Health Care, 2016, 25, 1 CrossRef PubMed.
- L. Hauben, L. Vauterin, E. Moore, B. Hoste and J. Swings, Genomic diversity of the genus Stenotrophomonas, Int. J. Syst. Evol. Microbiol., 1999, 49, 1749–1760 CAS.
- A.-M. Bourgault, J. E. Rosenblatt and R. H. Fitzgerald, Peptococcus magnus: a significant human pathogen, Ann. Intern. Med., 1980, 93, 244–248 CrossRef CAS PubMed.
- K. L. Ormerod, D. L. A. Wood, N. Lachner, S. L. Gellatly, J. N. Daly, J. D. Parsons, C. G. O. Dal'Molin, R. W. Palfreyman, L. K. Nielsen and M. A. Cooper, Genomic characterization of the uncultured Bacteroidales family S24-7 inhabiting the guts of homeothermic animals, Microbiome, 2016, 4, 36 CrossRef PubMed.
- M. Serino, E. Luche, S. Gres, A. Baylac, M. Bergé, C. Cenac, A. Waget, P. Klopp, J. Iacovoni and C. Klopp, Metabolic adaptation to a high-fat diet is associated with a change in the gut microbiota, Gut, 2012, 61, 543–553 CrossRef CAS PubMed.
- C. C. Tong, Y. K. Choong, S. Mohamed, N. M. Mustapha and N. A. Umar, Efficacy of Ganoderma lucidum on plasma lipids and lipoproteins in rats fed with high cholesterol diet, Nutr. Food Sci., 2008, 38, 229–238 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra08087e |
| This journal is © The Royal Society of Chemistry 2017 |