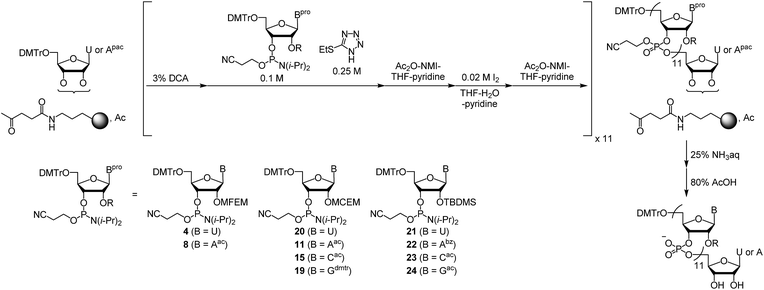
Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Synthesis of 2′-O-monohaloethoxymethyl-modified RNAs and their duplex formation ability†
Rintaro Iwata Haraa,
Masayuki Kageyamab,
Koichiro Araib,
Naoki Uchiyamab and
Takeshi Wada *a
*a
aFaculty of Pharmaceutical Sciences, Tokyo University of Science, 2641 Yamazaki, Noda, Chiba 278-8510, Japan. E-mail: twada@rs.tus.ac.jp
bGraduate School of Frontier Sciences, The University of Tokyo, 5-1-5 Kashiwanoha, Kashiwa, Chiba 277-8562, Japan
First published on 23rd August 2017
Abstract
We synthesized 2′-O-monohaloethoxymethyl-modified RNAs and evaluated their duplex formation ability. The effects of 2-chloroethoxymethyl (MCEM) and 2-fluoroethoxymethyl groups on the RNA/RNA duplex stability was found to depend on both base sequences and halogen atoms. Only the 2′-O-MCEM-rU12/rA12 duplex was found to be significantly more stable than the unmodified duplex. In this study, it is proposed through UV melting analyses, isothermal titration calorimetry measurements, and molecular mechanics calculations that this stabilization might result from enthalpic stabilization due to interactions between the MCEM groups and nucleobases in the complementary strand.
Introduction
Nucleic acid drugs, which function as therapeutics by suppressing the expression of disease-causing genes or by other mechanisms, are generally required to be chemically modified, as natural DNA and RNA molecules are degraded by endogenous nucleases in vivo.1 For several decades, numerous chemically modified oligonucleotides have been synthesized to improve not only their stability in vivo but also the affinity for target RNAs and pharmacokinetics and cytotoxicity of nucleic acid drugs.2 Chemically modified oligonucleotide derivatives are broadly classified into three types according to the site where chemical modifications are introduced: phosphodiester backbones, sugars, and nucleobases. Of these modifications, the first two modifications are seen in the structure of all approved nucleic acid drugs. For example, phosphorothioate linkages, in which one of the non-bridging oxygen atoms are replaced with a sulfur atom, are seen in mipomersen (Kynamuro®),3 formivirsen (Vitravene®),4 and nusinersen (Spinraza®).5 Mipomersen and nusinersen also contain 2′-O-2-methoxyethyl groups, whereas eteplirsen (exondys 51™) is based on the phosphordiamidate morpholino oligomer.6Among chemically modified oligonucleotides, 2′-modified nucleic acids are generally used to improve the nuclease resistance of siRNA and other RNA-based drugs.7 This improvement is significant in that RNA molecules are much more susceptible to degradation than DNA molecules. Furthermore, chimeric oligomers containing natural DNA and 2′-modified RNA, known as gapmers,8 from part of recent trends in antisense therapeutics, of which mipomersen is a successful example. Although many 2′-modified RNA molecules have been synthesized, the number of 2′-modified RNA molecules that can be easily prepared is still limited.9 Although 2′-O-alkylation is one of the simplest 2′-modifications, even such modifications are limited in some cases, partly because alkylation of nucleobases competes with 2′-O-alkylation as a significant side reaction in the synthesis of monomer units for oligonucleotide synthesis.10 As a result, 2′-O-alkoxymethyl groups has attracted much attention, because they can be introduced onto any nucleosides without such side reactions.11
Previous investigation reported on 2′-O-haloethoxymethyl groups as a potential 2′-O-modification,12 and the introduction of 2-chloroethoxymethyl (MCEM) and dichloroethoxymethyl modification of the 2′-OH group on rU12 have been shown to significantly increase the stability of RNA/RNA duplex whereas the 2′-O-ethoxymethyl (EOM) modification did not show such stabilization effect. Other than the duplex stabilization, easily accessible 2′-O-haloethoxymethyl modified RNAs might be useful for the heavy atom isomorphous replacement method13 in X-ray crystallography of nucleic acids, and the introduction of fluorine atoms is standard strategy for altering pharmacokinetics.14
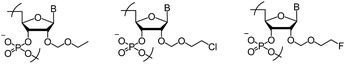
In the present study, the 2-fluoroethoxymethyl (MFEM) group as a novel 2′-O-haloethoxymethyl modification (Fig. 1) and the properties of 2′-O-MCEM and 2′-O-MFEM rA12 and rU12 are compared. The synthesis and properties of 2′-MCEM-modified RNAs containing all the four nucleobases is then reported.
Results and discussion
Synthesis of monomers and oligo RNAs
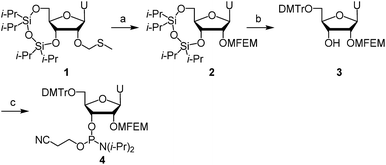
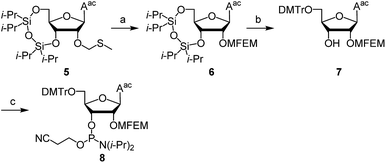
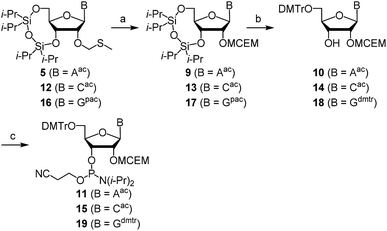
First, we describe the synthesis of ribonucleoside phosphoramidites bearing a 2′-O-haloethoxymethyl group. We previously reported the synthesis of 2′-O-MCEM and other haloethoxymethyl modified rU phosphoramidites,12 and other groups reported the synthesis of 2′-O-cyanoethoxymethyl (2′-O-CEM) modified ribonucleoside phosphoramidites,15 the synthesis of monomers employed in this study was therefore based on these procedures. The synthesis of 2′-O-MFEM-modified rU phosphoramidite is shown in Scheme 1. Compound 1 was prepared according to the literature.12 The introduction of an MFEM group was accomplished using N-iodosuccinimide (NIS) and trifluoromethanesulfonic acid (TfOH) as activators to produce 2′-O-MFEM modified compound 2. The silyl group on 2 was removed by treatment with fluoride ion, and the 5′-OH group was protected by a DMTr group to give compound 3. Finally, the 3′-OH group was phosphitylated and the 2′-O-MFEM-modified rU phosphoramidite 4 was obtained with good yield. Other phosphoramidite monomers were synthesized in a similar manner. Scheme 2 shows the synthesis of 2′-O-MFEM-modified rA phosphoramidite 8 and Scheme 3 shows the synthesis of 2′-O-MCEM-modified rA phosphoramidite 11. In the synthesis of these rA monomers, the amino group on the nucleobase was protected with an acetyl group. As shown in Scheme 3, the synthesis of 2′-O-MCEM-modified rC and rG phosphoramidite, indicates an acetyl group and a phenoxyacetyl (pac) group were used for the protection of amino groups in the nucleobases. Starting materials 5, 12, and 16 are known compounds and their synthetic procedures are described in the literature.11 Both 2′-O-MFEM- and 2′-O-MCEM-rAac monomers 8 and 11 and the 2′-O-MCEM-rCac monomer 15 were obtained with good yields. However, in the synthesis of the 2′-O-MCEM-rGpac monomer, unintended removal of the pac group occurred and the subsequent tritylation resulted in N-, 5′-O-di-DMTr compound 18 being obtained as the main product. Although the DMTr group on the guanine base is removed during the oligomer synthesis, the unprotected guanine is reported to be less reactive in the oligomer synthesis by the phosphoramidite method.16 Therefore, 18 was phosphitylated and 2′-O-MCEM-rGdmtr phosphoramidite 19 was used in the oligomer synthesis.Next, RNA oligomers containing 2′-O-monohaloethoxymethyl ribonucleosides were produced through the use of the synthesized phosphoramidites. Automated solid-phase synthesis was applied to produce RNA oligomers using the appropriate monomer units as shown in Scheme 4. Both rU- or rApac-anchored CPG were used and the DMTr group was removed by treatment with dichloroacetic acid (DCA). Each condensation reaction was conducted using phosphoramidites 4, 8, 11, 15, 19, or 2′-O-MCEM rU phosphoramidite 20 whose synthesis was previously reported,12 or commercially available 2′-O-TBDMS protected phosphoramidites, and 5-(ethylthio)-1H-tetrazole (ETT) acting as an acidic activator followed by the subsequent oxidation by iodine after capping resulted in the phosphotriester. By repeating these reactions, 2′-O-haloethoxymethyl-modified RNA 12mers were synthesized. The sequences, site of modifications and the yields of them were shown in Table 1. As indicated by entries 4–8, rGUCAGUCAGUCA was selected as a sequence containing all four nucleobases. All 2′-O-modified RNA 12mers were successfully synthesized in this method, and identified by mass spectrometry after purification with reverse phase HPLC (RP-HPLC).
| Entry | Sequence | Yield | m/z (calcd) | m/z (found) |
|---|---|---|---|---|
| a Bold nucleotides are 2′-O-MCEM modified, and underlined nucleotides are 2′-O-MFEM-modified RNA oligomers were identified with ESI-MS in entry 1, and with MALD-TOF MS in entry 2–8. | ||||
| 1 | rAAAAAAAAAAAA | 41% | 980.149([M − 5H]5−) | 980.13 |
| 2 | 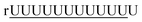 |
15% | 4446.70([M − H]−) | 4446.85 |
| 3 | rAAAAAAAAAAAA | 16% | 4723.03([M − H]−) | 4721.49 |
| 4 | rGUCAGUCAGUCA | 5% | 4163.55([M − H]−) | 4164.81 |
| 5 | rGUCAGUCAGUCA | 6% | 4163.55([M − H]−) | 4163.17 |
| 6 | rGUCAGUCAGUCA | 4% | 4067.57([M − H]−) | 4069.17 |
| 7 | rGUCAGUCAGUCA | 9% | 4439.56([M − H]−) | 4440.33 |
| 8 | rGUCAGUCAGUCA | 23% | 4809.57([M − H]−) | 4808.98 |
UV melting analysis
The melting temperature (Tm) of the duplex of each synthetic RNA 12mer and the complementary RNA 12mer was measured. As shown in Table 2, the Tm value of unmodified rU12/rA12 was 16.4 °C. 2′-O-MCEM-rU12/rA12 formed a more stable duplex (26.8 °C, entry 2) than the unmodified one as previously reported.12 As shown in entry 4, Tm of 2′-O-MFEM rU12/rA12 was similar to that of the unmodified RNA duplex. These results indicate that a fluorine atom on the ethoxymethyl group does not contribute to the stabilization of the RNA/RNA duplex unlike the chlorine atom. Entries 3 and 5 show Tm values of rU12/2′-O-modified-rA12. Contrary to expectation, the introduction of each monohaloethoxy group destabilized the duplex. Furthermore, rU12/2′-O-MFEM-rA12 was too unstable for the calculation of Tm.| Entry | 2'-O-modified RNA | Complementary RNA | Tm/°C | ΔTm/°C | ΔTm per mod/°C |
|---|---|---|---|---|---|
| a Bold nucleotides are 2′-O-MCEM modified, and underlined nucleotides are 2′-O-MFEM modified. | |||||
| 1 | rUUUUUUUUUUUU | rAAAAAAAAAAAA | 16.4 | — | — |
| 2 | rUUUUUUUUUUUU | rAAAAAAAAAAAA | 26.8 | +10.4 | +0.9 |
| 3 | rAAAAAAAAAAAA | rUUUUUUUUUUUU | 11.1 | −5.3 | −0.5 |
| 4 | 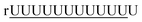 |
rAAAAAAAAAAAA | 17.3 | +0.9 | +0.1 |
| 5 | 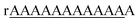 |
rUUUUUUUUUUUU | <10 | — | — |
| 6 | rGUCAGUCAGUCA | rUGACUGACUGAC | 61.6 | — | — |
| 7 | rGUCAGUCAGUCA | rUGACUGACUGAC | 58.7 | −2.9 | −0.7 |
| 8 | rGUCAGUCAGUCA | rUGACUGACUGAC | 58.3 | −3.3 | −0.8 |
| 9 | rGUCAGUCAGUCA | rUGACUGACUGAC | 58.6 | −3.0 | −1.0 |
| 10 | rGUCAGUCAGUCA | rUGACUGACUGAC | 57.8 | −3.8 | −0.5 |
| 11 | rGUCAGUCAGUCA | rUGACUGACUGAC | 56.0 | −5.6 | −0.5 |
Entries 6–11 in Table 2 are the Tm values for the 2′-O-MCEM-modified-RNA duplexes containing four nucleobases. In entries 7–9, consecutive three or four ribonucleosides in rGUCAGUCAGUCA were modified. Regardless of the sites of modification, Tm values decreased to a similar extent compared with the unmodified RNA duplex (−0.7 °C, −0.8 °C, and −1.0 °C per modification in entries 7, 8, and 9, respectively). In entries 10 and 11, although RNA 12mers bearing seven and eleven MCEM modifications showed lower Tm values, each ΔTm per mod was relatively small compared with those in entries 7–9 (−0.5 °C per modification in both entries 10 and 11). The Tm value (56 °C) of 2′-O-MCEM-modified 12mer RNA in entry 11 was high enough to a form duplex under physiological conditions (neutral pH, 37 °C) although the value was less than that of the unmodified RNA/RNA duplex.
Resistance against SVPD
To evaluate nuclease resistance of the 2′-O-MFEM modified RNA, we carried out the nuclease assay by using snake venom phosphodiesterase (SVPD). 2′-O-Me-rU12, 2′-O-MEM-rU12, and 2′-O-MFEM-rU12 were treated with SVPD for 2 h, and the degradation ratio was estimated by RP-HPLC analysis. As shown in Table 3, 2′-O-MFEM-rU12 showed a certain extent of nuclease resistance and its degradation ratio was between those of 2′-O-Me-rU12 and 2′-O-MCEM-rU12.| Entry | Modification | Degradation ratio/% |
|---|---|---|
| 1 | Me | 42 |
| 2 | MCEM | 86 |
| 3 | MFEM | 62 |
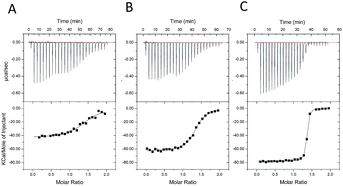
Study for the stability of 2′-O-MCEM-rU12/rA12 duplexes
To study the difference in thermal duplex stability between unmodified rU12/rA12 and 2′-O-MCEM-rU12/rA12, UV melting analyses of various RNA concentrations and isothermal titration calorimetry (ITC) were conducted to calculate the ΔH and ΔS values for duplex formation. Detailed Tm values in various RNA concentrations are shown in the ESI.† The relationship between Tm, ΔH, and ΔS values and total concentration of nucleic acids (Ctot) is described using the following equation (where R is the gas constant);171/Tm = (R/ΔH)ln(Ctot) + (ΔS° − R![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ln ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4)/ΔH° 4)/ΔH° |
As shown in Fig. 2, each plot of 1/Tm versus ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ctot indicates an approximately linear relationship thereby allowing ΔH and ΔS values to be calculated (parameters from Tm data in Table 4).
Ctot indicates an approximately linear relationship thereby allowing ΔH and ΔS values to be calculated (parameters from Tm data in Table 4).
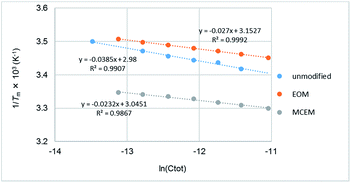 | ||
| Fig. 2 The relationship between 1/Tm and ln(Ctot) of unmodified-rU12/rA12, 2′-O-EOM-rU12/rA12, and 2′-O-MCEM-rU12/rA12. | ||
| Entry | Modification | Tm | ITC | ||
|---|---|---|---|---|---|
| ΔH (kcal mol−1) | ΔS (cal mol−1 K−1) | ΔH (kcal mol−1) | ΔS (cal mol−1 K−1) | ||
| 1 | Unmodified | −51.5 | −151 | −43.1 | −124 |
| 2 | EOM | −73.6 | −221 | −62.8 | −193 |
| 3 | MCEM | −85.6 | −258 | −77.9 | −241 |
Separately, the ITC measurement was also conducted for unmodified, 2′-O-EOM and MCEM-modified rU12/rA12 duplexes at 5 °C. The results were shown in Fig. 3, and ΔH and ΔS values are given in Table 4 (parameters from ITC data). In each method, a similar trend was observed. First, EOM modifications are enthalpically favored and entropically unfavored in the duplex formation. Second, MCEM modifications are furthermore enthalpically and less entropically favored than EOM modifications. From these results, the high stability of MCEM rU12/rA12 is postulated to be derived from enthalpic stabilization.
Next, the interaction of the MCEM groups in an RNA/RNA duplex was studied. In this investigation, the conformational analysis was conducted using molecular mechanics calculations to elucidate the most stable conformation of the MCEM groups in rU–UMCEM–UMCEM–UMCEM–U/rA5 duplex. In these calculations, RNA/RNA backbones were constrained, and the most stable conformation of MCEM groups was calculated. In the most stable conformation, each chlorine atom in the MCEM groups is located near the adenine base in the complementary RNA (Fig. 4). This location may be partly due to the fixation of an MCEM chain by electrostatic repulsion between the 2-O atom of the uracil base and oxygen atom in the MCEM group and by a gauche effect between the chlorine and oxygen atoms both in the MCEM group. Consequently, the electrostatic interactions between the chlorine atom and the electro-positive 2-H atom in the adenine base and/or the hydrophobic interactions between the MCEM group and adenine bases are expected to work effectively in the duplex. It is therefore suggested that these interactions between the MCEM groups in the rU–UMCEM–UMCEM–UMCEM–U and the adenine bases in the complementary rA5 enthalpically stabilize the RNA/RNA duplex.
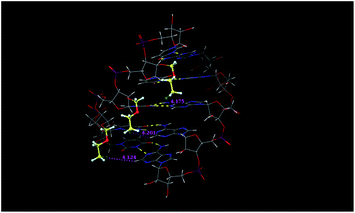 | ||
| Fig. 4 The most stable conformation of the MCEM groups in rU–UMCEM–UMCEM–UMCEM–U/rA5 duplex based on molecular mechanics calculations with a GB/SA water solvation model. | ||
Study for the thermal instability of rU12/2′-O-MCEM rA12 and rU12/2′-O-MFEM rA12 duplexes
In the case of rU12/2′-O-MCEM-rA12 and rU12/2′-O-MFEM-rA12 duplexes, UV melting analyses in a various RNA concentrations and ITC were not applicable because the Tm values were too low. Instead of these experiments, alternative experiments for the elucidation of their instability were conducted. Fig. 5 shows the temperature dependence of UV absorbance of single-stranded RNAs. As shown the Fig. 5, the UV absorbance of rA12 increases with increasing temperature, whereas the UV absorbance of rU12 is almost unchanged. This hypochromicity of rA12 at low temperatures is derived from stacking interactions of adenine bases in the single strand.18 Furthermore, 2′-O-MCEM rA12 and 2′-O-MFEM rA12 showed a significantly lower hypochromicity than that of rA12, and 2′-OMe rA12 as a reference. These results strongly suggest that MCEM and MFEM groups interacted with a nucleobase, another MCEM/MFEM group or another functional group, and subsequently disrupted the stacking interactions. The stacking interactions are advantageous in single-stranded RNAs to facilitate a preorganized structure which is advantageous for duplex formation. The collapse of base-stacking interactions caused by MCEM and MFEM groups might therefore entropically induce the significant destabilization of the duplex.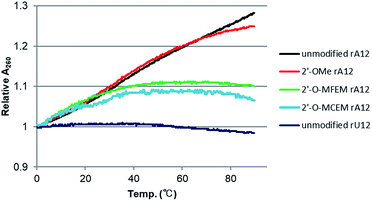 | ||
| Fig. 5 The temperature dependence of UV absorbance of 4 μM of single stranded RNAs in a 10 mM phosphate buffer containing 100 mM NaCl and 0.5 mM EDTA at pH 7.0. | ||
Conclusion
In this study, 2′-O-MFEM and 2′-O-MCEM RNA oligomers were synthesized and their duplex stability was evaluated. The thermodynamic stabilizing or destabilizing effects of these haloethoxymethyl groups for RNA/RNA duplexes were found to be dependent on base sequences and halogen atoms. The significant increase in the Tm value of 2′-O-MCEM-rU12/rA12 was postulated to be due to enthalpic stabilization as investigated by UV melting analyses, ITC measurements, and molecular mechanics calculations. Furthermore, rU12/2′-O-MCEM-rA12 and rU12/2′-O-MFEM-rA12 were found to be unstable compared with the unmodified duplex, and it was suggested that this is due to the collapse of self-stacking of nucleobases in the 2′-O-MFEM-rA12. In addition, 2′-O-MCEM-modified RNAs bearing the four nucleobases form a stable duplex with their complementary RNAs, although the duplexes were less stable than the unmodified ones. These results will prove useful in the design of siRNAs or other RNA-based nucleic acid drugs containing 2′-O-haloethoxymethyl modifications. For example, siRNAs containing many A–U base pairs at their 3′ or 5′ termini are reported to be more susceptible to nuclease digestion than those with G–C rich ones.19 This difference is considered to be attributed to partially dissociated structure in the duplexes was recognized by nucleases. In this regard, 2′-O-MCEM modification, which can stabilize rU12/rA12 when it is introduced on rU, might be useful for stabilizing the unstable termini in siRNAs.Experimental section
General information
All reactions were conducted under an argon atmosphere. 1H NMR spectra were obtained at 300 MHz on a Varian MERCURY 300 spectrometer or a JEOL AL-300 spectrometer with tetramethylsilane as an internal standard in CDCl3. 31P NMR spectra were obtained with 85% H3PO4 as an external standard (δ 0.0) in CDCl3. Reagents and solid supports were purchased from Wako Pure Chemical Industries, Tokyo Chemical Industry, Sigma-Aldrich, and Glen Research. Oligonucleotides were purchased from Hokkaido System Science. Silica gel column chromatography was performed using a silica gel 60N (63–210 μm or 40–50 μm). RP-HPLC for analysis and purification was performed using a μBondasphere 5 μm C18, 100 Å, 19 mm × 150 mm (Waters) or Source 5 RPC ST 4.6/150 (GE Healthcare). The organic solvents were purified and dried using the appropriate procedures. Mass spectra were recorded on a Voyager System 4327 (Applied Biosystem), or a 910-MS FTMS system (Varian), or an autoflex speed MALDI-TOF MS (Bruker).Conditions for UV melting analyses
The absorbance versus temperature profiles were measured using an eight-sample cell changer, in quartz cells of 1 cm path length. All experiments were performed in a 10 mM phosphate buffer containing 100 mM of NaCl and 0.1 mM EDTA at pH 7.0. The UV absorbance at 260 and 320 nm was monitored with temperature. Samples containing oligonucleotides were first rapidly heated to 95 °C followed by cooling to 0 °C at a rate of 0.5 °C min−1, and the dissociation was recorded by heating to 50 or 90 °C at a rate of 0.5 °C min−1.SVPD assay
SVPD assay was carried out under the conditions similar to those of the literature.12,20 2′-O-Me-rU12, 2′-O-MCEM-rU12, or 2′-O-MFEM-rU12 were digested with 4 × 10−4 U mL−1 of SVPD for 2 h in 50 mM Tris–HCl buffer containing 72 mM NaCl and 14 mM MgCl2 at 37 °C, pH 8.5. On the other hand, complete digestion of these RNAs were carried out using 0.1 U mL−1 of SVPD for 1 h in the same buffer. After the reaction, the products were analysed by RP-HPLC and the degradation ratios were calculated based on these experimental results.Conditions for ITC measurements
RNAs were in a 10 mM phosphate buffer containing 100 mM NaCl and 0.5 mM EDTA at pH 7.0. The unmodified or modified rU12 solution (50 μM) was titrated into a rA12 solution (5 μM) at 5 °C. Each titration consisted of a preliminary 0.5 μL injection followed by 24 subsequent 1.5 μL additions, which were performed over 3 s periods at 120–180 s intervals.Molecular mechanics calculation
Molecular mechanics calculations were performed with a GB/SA water solvation model, Amber* as a forcefield.21 RNA duplex structures were constrained, and the most stable conformation of MCEM groups was determined by minimization with the PRCG method followed by conformational searches with the mixed torsional/low mode sampling method.Automated solid-phase synthesis
The synthesis of RNA oligomers was conducted using an Expedite 8909 automated synthesizer (Applied Biosystems), based on a standard protocol for 0.2 μmol scale synthesis as below:Detritylation; 3% DCA in CH2Cl2.
Condensation: 0.1 M monomer, 0.25 M ETT in CH3CN.
Capping: cap A Ac2O–THF (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9, v/v), cap B 10% N-methylimidazole in THF–pyridine (8
9, v/v), cap B 10% N-methylimidazole in THF–pyridine (8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1).
1).
Oxidation: 0.02 M I2 in pyridine–THF–H2O (0.4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 89.6
89.6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, v/v/v).
10, v/v/v).
Capping: cap A Ac2O–THF (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9, v/v), cap B 10% N-methylimidazole in THF–pyridine (8
9, v/v), cap B 10% N-methylimidazole in THF–pyridine (8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1).
1).
After the synthesis, 25% aqueous ammonia (1 mL) was added. After 3 h, the obtained solution was concentrated and lyophilized, and then re-dissolved in water. The DMTr-on oligomers were isolated with RP-HPLC. The DMTr group was removed under the mild acidic conditions (80% aqueous acetic acid, 1 mL, 1 h) and then DMTr-off oligomers were obtained with RP-HPLC.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank professor Kohei Tsumoto (The University of Tokyo) and Satoru Nagatoishi (The University of Tokyo) for ITC measurements and helpful discussions.Notes and references
- (a) D. Bumcrot, M. Manoharan, V. Koteliansky and D. W. Y. Sah, Nat. Chem. Biol., 2006, 2, 711 CrossRef CAS PubMed; (b) T. Dowler, D. Bergeron, A. L. Tedeschi, L. Paquet, N. Ferrari and M. J. Damha, Nucleic Acids Res., 2006, 34, 1669 CrossRef CAS PubMed.
- J. Kurreck, Eur. J. Biochem., 2003, 270, 1628 CrossRef CAS PubMed.
- R. S. Geary and B. F. Baker, Clin. Pharmacokinet., 2015, 54, 133 CrossRef CAS PubMed.
- V. K. Sharma, R. K. Sharma and S. K. Singh, Med. Chem. Commun., 2014, 5, 1454 RSC.
- C. A. Chiriboga, K. J. Swoboda, B. T. Darras, S. T. Iannaccone, J. Montes, D. C. Vivo, D. A. Norris, C. F. Bennett and K. M. Bishop, Neurology, 2016, 86, 890 CrossRef CAS PubMed.
- J. R. Mendell, L. R. Rodino-Klapac, Z. Sahenk, K. Roush, L. Bird, L. P. Lowes, L. Alfano, A. M. Gomez, S. Lewis, J. Kota, V. Malik, K. Shontz, C. M. Walker, K. M. Flanigan, M. Corridore, J. R. Kean, H. D. Allen, C. Shilling, K. R. Melia, P. Sazani, J. B. Saoud, E. M. Kaye and Etepliresen study group, Ann. Neurol., 2013, 74, 637 CrossRef CAS PubMed.
- (a) M. Manoharan, Curr. Opin. Chem. Biol., 2004, 8, 570 CrossRef CAS PubMed; (b) F. Czauderna, M. Fechtner, S. Dames, H. Aygün, A. Klippel, G. J. Pronk, K. Giese and J. Kaufmann, Nucleic Acids Res., 2003, 31, 2705 CrossRef CAS PubMed.
- A. Khvorova and J. K. Watts, Nat. Biotechnol., 2017, 35, 238 CrossRef CAS PubMed.
- K. Morihiro, Y. Kasahara and S. Obika, Mol. BioSyst., 2017, 13, 235 RSC.
- (a) K. Tamauchi, T. Nakagima and M. Kinoshita, Bull. Chem. Soc. Jpn., 1986, 59, 2947 CrossRef; (b) H. Inoue, Y. Hayase, A. Imura, S. Iwai, K. Miura and E. Otsuka, Nucleic Acids Res., 1987, 15, 6131 CrossRef CAS PubMed.
- (a) T. Ohgi, Y. Matsutomi, K. Ishiyama, H. Kitagawa, Y. Shiba and J. Yano, Org. Lett., 2005, 7, 3477 CrossRef CAS PubMed; (b) C. Zhou, D. Honcharenko and J. Chattopadhyaya, Org. Biomol. Chem., 2007, 5, 333 RSC.
- K. Arai, N. Uchiyama and T. Wada, Bioorg. Med. Chem. Lett., 2011, 21, 6285 CrossRef CAS PubMed.
- W. Saenger, Principles of Nucleic Acid Structure, Springer-Verlag, New York, 1984, p. 42 Search PubMed.
- E. P. Gillis, K. J. Eastman, M. D. Hill, D. J. Donnelly and N. A. Meanwell, J. Med. Chem., 2015, 58, 8315 CrossRef CAS PubMed.
- (a) Y. Shiba, H. Matsuda, N. Watanabe, T. Ego, K. Takagaki, K. Ishiyama, T. Ohgi and J. Yano, Nucleic Acids Res., 2007, 35, 3287 CrossRef CAS PubMed; (b) J. Cieslak, A. Grajkowski, C. Ausin, A. Gapeev and S. L. Beaucage, Nucleic Acids Res., 2012, 40, 2312 CrossRef CAS PubMed.
- T. Wada and M. Sekine, Tetrahedron Lett., 1994, 35, 757 CrossRef CAS.
- (a) P. N. Borer, B. Dengler and I. Tinoco jr, J. Mol. Biol., 1974, 86, 843 CrossRef CAS PubMed; (b) N. Poklar, D. S. Pilch, S. J. Lippard, E. A. Redding, S. U. Dunham and K. J. Breslauer, Proc. Natl. Acad. Sci. U. S. A., 1996, 93, 7606 CrossRef CAS PubMed.
- (a) J. Brahms, A. M. Michelson and L. E. van Holde, J. Mol. Biol., 1966, 15, 467 CrossRef CAS PubMed; (b) H. Simpkins and E. G. Richards, J. Mol. Biol., 1967, 29, 349 CrossRef CAS.
- J. Haupenthal, C. Baehr, S. Kiermayer, S. Zeuzem and A. Piiper, Biochem. Pharmacol., 2006, 71, 702–710 CrossRef CAS PubMed.
- M. Egli, G. Minasov, V. Tareshko, P. S. Pallan, M. Teplova, G. B. Inamati, E. A. Lesnik, S. R. Owens, B. S. Ross, T. P. Prakash and M. Manoharan, Biochemistry, 2005, 44, 9045 CrossRef CAS PubMed.
- F. Mohamadi, N. G. J. Richards, W. C. Guida, R. Liskamp, M. Lipton, C. Caufield, G. Chang, T. Hendrickson and W. C. Still, J. Comput. Chem., 1990, 11, 440 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra07767j |
| This journal is © The Royal Society of Chemistry 2017 |