DOI:
10.1039/C7RA07520K
(Paper)
RSC Adv., 2017,
7, 41779-41786
Endocytosis mechanism of a novel proteoglycan, extracted from Ganoderma lucidum, in HepG2 cells
Received
8th July 2017
, Accepted 11th August 2017
First published on 29th August 2017
Abstract
A novel protein tyrosine phosphatase 1B (PTP1B) inhibitor, FYGL, extracted from Ganoderma lucidum, was first reported to have an efficient hypoglycemic effect and high safety in vivo in our previous study. However, the underlying mechanism of the anti-diabetes activity was still unclear. The dominant effective component of FYGL was demonstrated to be a neutral hyperbranched proteoglycan macromolecule. Therefore, it is necessary to first elucidate how the biomacromolecule works at the cellular level, in order to understand its possible functional mechanism in vivo. Herein, we focused on demonstrating the cell internalization and endocytosis mechanism of FYGL in HepG2 cells. A series of cellular uptake pathways was explored, including clathrin-mediated endocytosis, caveolae/lipid-mediated endocytosis, and macropinocytosis. Confocal laser scanning microscopy and flow cytometry were used to demonstrate the absorption of FYGL by HepG2 cells. Furthermore, pharmacologically selective inhibitors and Western blot methods were used to demonstrate that macropinocytosis mediated by c-Src/PI3K cascades was the preferred route for the uptake of FYGL in cells. Our study provided the basis of uptake of FYGL for an efficient hypoglycemic effect. The functional mechanism of the signaling pathway in vivo will be reported in the future.
1. Introduction
The protein tyrosine phosphatase 1B (PTP1B), a member of the protein tyrosine phosphatase (PTPs) family, is overexpressed in insulin-sensitive peripheral tissues, such as liver and muscle tissues, in type 2 diabetes mellitus (T2DM) patients; it negatively regulates the insulin signaling pathway by dephosphorylating insulin receptor substrate (IRS) on tyrosine residues.1,2 Inhibiting PTP1B activity is considered an efficient strategy for the management of T2DM. Therefore, identifying PTP1B inhibitors is a topic of current interest; however, no ideal inhibitor has been found for clinical use thus far, because of either poor bioavailability or low safety.3,4 Interestingly, our previous study found a novel PTP1B inhibitor, FYGL, which was extracted from Ganderma lucidum, a well-known Chinese medicinal fungus that has long been used as a traditional Chinese medicine.5,6 The molecular weight of FYGL is 2.6 × 105, and the structure of the dominant component is shown in Fig. 1. FYGL is an amphiphilic hyperbranched proteoglycan consisting of hydrophilic polysaccharides (4 saccharide units) and lipophilic protein moieties (17 amino acids) with a ratio of 77![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 17 for saccharide
17 for saccharide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) protein.7 We first demonstrated that FYGL has an efficient hypoglycemic effect and high safety in vivo,5 but mechanism was still unclear. In order to understand the possible functional mechanism of FYGL, it is necessary to first reveal how the biomacromolecule works at the cellular level. Therefore, herein, we focused on demonstrating the cell internalization and endocytosis mechanism of FYGL in HepG2 cells, which are engaged in glycogen synthesis.
protein.7 We first demonstrated that FYGL has an efficient hypoglycemic effect and high safety in vivo,5 but mechanism was still unclear. In order to understand the possible functional mechanism of FYGL, it is necessary to first reveal how the biomacromolecule works at the cellular level. Therefore, herein, we focused on demonstrating the cell internalization and endocytosis mechanism of FYGL in HepG2 cells, which are engaged in glycogen synthesis.
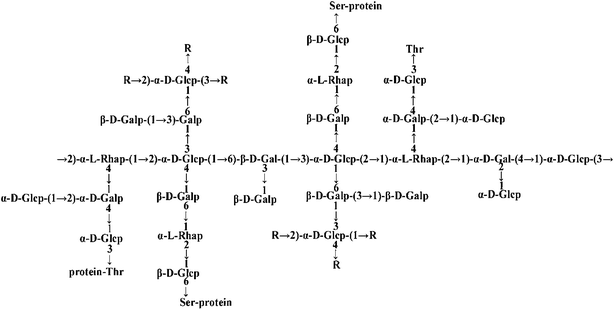 |
| | Fig. 1 The structure of FYGL (Ara: arabinose, Gal: galactose, Glc: glucose, Rha: rhamnose, Thr: threonine, Ser: serine). The suffixes p and f represent pyranose and furanose, respectively. | |
Cell internalization of a macromolecular drug involves specific or multiple endocytosis processes.8 Endocytosis is a process involving the uptake of particles, fluids and macromolecules from the outside to the inside of a cell.9 The endocytosis mechanisms mainly include clathrin-mediated endocytosis (CME) and clathrin-independent endocytosis (CIE); the latter involves caveolae/lipid-mediated endocytosis and macropinocytosis.9 Endocytic pathways are key to current drug delivery strategies and functional materials used in the field of biomedicine.10 Macropinocytosis was first observed in 1931, representing a distinct endocytic pathway in mammalian cell lines,11,12 and can contribute to the efficiency by which numerous cell-penetrating peptides translocate to the cytoplasm.13 This process is an actin-dependent endocytic pathway and occurs through large vesicles of variable sizes (diameter 0.2–5 μm) called macropinosomes.14 As a typical macropinocytosis tracer, dextran is often used to co-localize with the studied drug in cells for proving the macropinocytosis mechanism.15 A protein tyrosine kinase implicated in cancer and some intracellular signaling cascades,16 c-sarcomagene (c-Src), can promote cell proliferation and inhibit cell apoptosis via PI3K/Akt cascades.17 In the initial stage of macropinocytosis, c-Src induces cell ruffling and influences the cell shape, following which it closes the outermost margin to form a vesicle.18
Pharmacologically selective inhibitors are generally used to study the mechanism of endocytosis.9 Among the inhibitors, chlorpromazine can inhibit clathrin-mediated endocytosis but not specifically during the process.19 Nystatin can decompose cholesterol, and thus inhibit caveolae/lipid-mediated endocytosis.20 Amiloride negatively influences macropinocytosis.21 Genistein mediates some kinases involved in endocytosis,22 and LY294002 is a common inhibitor of phosphatidylinositol-3 kinase (PI3K), which regulates macropinocytosis and RhoA-mediated endocytosis, a member of the Ras homolog gene family.23
In the present study, we attempt to reveal the specific endocytosis process through which FYGL enters the HepG2 cells, for drug uptake. A clear understanding of the endocytosis mechanism of FYGL at the cellular level will shed light on the roles of traditional natural macromolecular products in medicine.
2. Materials and methods
2.1 Materials
FYGL was extracted from Ganderma lucidum in our laboratory.5 Dulbecco's modified Eagle's medium (DMEM), opti-minimal essential medium (opti-MEM) and fetal bovine serum (FBS) were purchased from Gibco Co. Ltd. (USA). Paraformaldehyde (4%), penicillin and streptomycin were purchased from Sangon Co. Ltd. (Shanghai, China). Cell counting kit-8 (CCK-8) was acquired from Dojindo Co. Ltd. (Shanghai, China). Fluorescein isothiocyanate (FITC), 4′,6-diamidino-2-phenylindole (DAPI), chlorpromazine, nystatin, genistein and amiloride were obtained from Aladdin Co. Ltd. (Shanghai, China). Rhodamine-phalloidin, a protein marker of FBS and polylysine were sourced from Yeasen Co. Ltd. (Shanghai, China). RIPA lysis buffer, lipofectamine 6000 and LY294002 were purchased from Beyotime Co. Ltd. (Shanghai, China). Tetramethylrhodamine isothiocyanate (TRITC) labeled-dextran was purchased from Xibao Co. Ltd. (Shanghai, China). HepG2 cell lines were provided by Fuxiang Co. Ltd. (Shanghai, China).
2.2 Oil/water (O/W) partition coefficient determination
Octanol and water were mixed and shaken vigorously to form two phases of water-saturated octanol (oil phase, phase-O) and octanol-saturated water (water phase, phase-W). The concentration of FYGL in phase-W was measured by the bicinchoninic acid (BCA) assay for protein analysis. FYGL was dissolved in phase-O to form 250 μg mL−1 solution. A given volume of phase-O was mixed with phase-W, and the mixture was shaken at 30 °C until the distribution of FYGL attained equilibrium in the two phases. Finally, the O/W partition coefficient (KOW) was calculated based on eqn (1):24| |
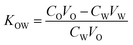 | (1) |
where CO is the original concentration of FYGL in phase-O, VO is the volume of phase-O, CW is the equilibrium concentration of FYGL in phase-W, and VW is the volume of phase-W.
2.3 Measurement of FYGL particle size
A solution of FYGL (2 mg mL−1) was prepared in phosphate buffered solution (PBS), and the particle size measured by Zetasizer Nano (Malvern, UK).
2.4 Cell culture
HepG2 cells were cultured in DMEM supplement with 10% FBS and 1% penicillin–streptomycin at 37 °C, in 5% CO2 atmosphere.
2.5 CCK-8 test
HepG2 cells were seeded into 96-well plates, and then endocytic selective inhibitors (with different concentrations in DMEM with 2% FBS) were added into the wells and incubated for 4.5 h at 37 °C. CCK-8 was added and cultured for another 0.5–2 h. Each sample was replicated in three wells. Finally the optical density (OD) was measured at 450 nm by a microplate reader (Bio-Tek, USA) and the cell viability was calculated.
2.6 Intracellular uptake of FYGL
FYGL was labeled by a green fluorescence marker, FITC, to form FITC-FYGL by the following procedure. FITC (1 mg) was dissolved in 1 mL DMSO, and the solution was added into sodium buffer containing 1 mg mL−1 FYGL at a volume ratio of 1 to 10. This mixture was stirred for 24 h at 4 °C, away from light, to form the FITC-FYGL solution. HepG2 cells were transferred into 12-well plates and incubated with the FITC-FYGL solution for 6 h, then washed with PBS, fixed with 4% paraformaldehyde, permeated with 0.2% Triton X-100/PBS, and stained with DAPI for the cellular nucleus and rhodamine-phalloidin for the cytoskeleton. The HepG2 cells were finally mounted on the glass slide for observation of the uptake of FYGL in by a confocal laser scanning microscope (C2+, Nikon, Japan). In addition, the HepG2 cells were also transferred into 6-well plates at a density of 5 × 105 to 1 × 106 cells per well and incubated with FITC-FYGL at different concentrations for 6 h; the cell suspensions were then analyzed by flow cytometry (Gallios, Beckman coulter, USA) at channel FL1 (excitation wavelength at 488 nm and emission wavelength at 525 nm) to demonstrate the absorption of FYGL by the HepG2 cells.
2.7 Endocytic inhibition of FYGL in HepG2 cells
HepG2 cells were transferred into 6-well plates with a density of 5 × 105 to 1 × 106 cells per well, pre-incubated with endocytic selective inhibitors at safe concentrations for 0.5 h, and then incubated with FITC-FYGL at the indicated concentration for 4 h. Finally, the cell suspensions were analyzed by flow cytometry at channel FL1 and observed by confocal laser scanning microscope with the same protocol as described in part 2.6.
2.8 FYGL in HepG2 cells transfected by c-Src siRNA
HepG2 cells were seeded into 6-well plates. A mixture solution of 20 μM c-Src siRNA (synthesized by Sangon Bio. Co, sequence shown in Table 1) and 5 μL lipofectamine 6000 were premixed in opti-MEM and added into the wells. The solution was removed 6 h later and the cells were washed thrice with PBS. A solution of FITC-FYGL at the indicated concentration in DMEM with 2% FBS was added into the wells and incubated for 4 h at 37 °C, followed by analysis of FCM and confocal laser scanning microscopy.
Table 1 The c-Src siRNA sequence
| Sense (5′ to 3′) |
CAAGAGCAAGCCCAAGGAU |
| Anti-sense (5′ to 3′) |
AUCCUUGGGCUUGCUCUUG |
2.9 c-Src protein extraction and Western blot
HepG2 cells were lysed in RIPA lysis buffer (Beyotime Co. Ltd) and clarified by centrifugation (12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g, 10 min, 4 °C). The cell lysates were separated by 8% SDS-PAGE, transferred to a polyvinylidene fluoride (PVDF) membrane, and then immunoblotted with rabbit polyclonal c-Src antibody (Sangon Co, Shanghai, China) and secondary goat anti-rabbit antibody (Cell Signaling Technology, USA). Bands were visualized with enhanced chemiluminescence solution (ECL, Biotanon) and detected by Image Lab camera (Bio-Rad, Germany); they were then quantified by densitometry scanning by the Image J software.
000 × g, 10 min, 4 °C). The cell lysates were separated by 8% SDS-PAGE, transferred to a polyvinylidene fluoride (PVDF) membrane, and then immunoblotted with rabbit polyclonal c-Src antibody (Sangon Co, Shanghai, China) and secondary goat anti-rabbit antibody (Cell Signaling Technology, USA). Bands were visualized with enhanced chemiluminescence solution (ECL, Biotanon) and detected by Image Lab camera (Bio-Rad, Germany); they were then quantified by densitometry scanning by the Image J software.
2.10 Intracellular co-localization of FYGL and dextran
HepG2 cells were seeded at a density of 2 × 104 cells per well; to these cells 100 μg mL−1 FITC-FYGL was added and incubated for 4 h. Following this, the medium was removed, TRITC-dextran was added and incubated for 10 min, and then the cells were fixed with 4% paraformaldehyde and observed by a confocal laser scanning microscope.
2.11 Statistical analysis
All data were presented as mean ± SD. One-way ANOVA test was performed to analyze the statistical significance between the two groups. A difference is considered to be statistically significant when the p value was <0.05.
3. Results and discussion
3.1 Oil/water (O/W) partition coefficient of FYGL
A working drug should possess amphiphilic properties in order to be transported in the human body and penetrate into the cell membrane. The oil/water partition coefficient is an important index for the amphiphilic property of a compound. The equilibrium concentration of FYGL in phase-W was obtained according to the standard curve plotted by the BCA assay. The partition coefficient KOW of FYGL, calculated from eqn (1), was 1.29 ± 0.08, larger than 1.0, suggesting that FYGL is soluble in lipids to some extent, in addition to water. It is thus possibly capable of penetrating through cell membranes and entering cells,25 and the structure of hydrophilic polysaccharide and lipophilic protein moieties including 17 amino acid residuals play an important role in cell penetration, especially the side chain of particular serine/threonine or tyrosine residues which are related to phosphorylation.26
3.2 Particle size of FYGL
Fig. 2 shows the particle size distribution of FYGL in water at room temperature. The particle size of FYGL is mainly distributed in the range 400–1000 nm, and polydispersity index (PDI) is 0.57 ± 0.03, suggesting that FYGL exists as aggregated particles in aqueous solution. Thus, the uptake of FYGL in cells maybe based on the mechanism of macropinocytosis in which the macropinocytic vesicles can endocytose the particles in the size range 200–5000 nm.
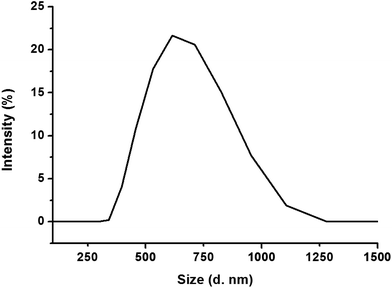 |
| | Fig. 2 The particle size distribution of FYGL in water at room temperature. Data are presented as mean ± SD (n = 3). | |
3.3 Intracellular uptake of FYGL
Fig. 3 shows the relative fluorescent intensity of green FITC-labeled FYGL, FITC-FYGL, at different concentrations by FCM analysis. The peak position in the x-axis moved towards right as the FITC-FYGL concentration increased from 25 to 200 μg mL−1 (Fig. 3A); the fluorescent intensity significantly increased in a dose-dependent manner (Fig. 3B). The confocal laser scanning microscopy images in Fig. 3C depict the intracellular uptake of FITC-FYGL in HepG2 cells.
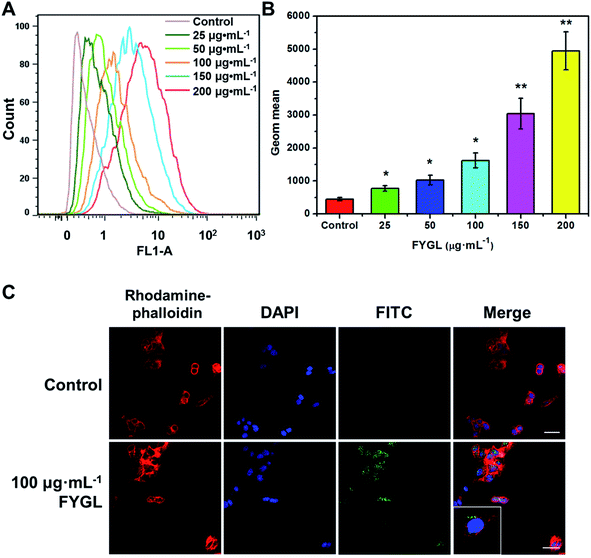 |
| | Fig. 3 (A) FCM shows the peak position of FITC-FYGL at the designated concentrations (incubation for 6 h at 37 °C) in channel FL1. (B) The Geom. mean reflects the relative fluorescent intensity of FITC-FYGL at different concentrations in HepG2 cells. (C) Confocal laser scanning microscopy images of the HepG2 cells after incubation with FITC-FYGL for 4 h. The scale bar represents 50 μm. The blue (DAPI labelled), red (rhodamine labelled) and green (FITC labelled) colors represent the cell nucleus, cell cytoskeleton, and FYGL, respectively. The inset shows the enlarged cell containing FYGL (green). Data are presented as mean ± SD (n = 3). *p < 0.05, **p < 0.01 versus the former group. | |
3.4 The safe concentrations of FITC-FYGL and endocytic selective inhibitors in HepG2 cells
In order to investigate the endocytic mechanism, the safety of FITC-FYGL and the applied endocytic selective inhibitors for cells is necessary. The safety of FITC-FYGL and the selective inhibitors, genistein, amiloride, chlorpromazine, nystatin and LY294002, for HepG2 cells was tested using CCK-8. From Fig. 4, it is found that FITC-FYGL is safe for HepG2 cells in a wide concentration range; even at concentrations higher than 300 μg mL−1, the cell is still viable up to 85%. The safety of the applied endocytic inhibitors was also tested (data not shown). Table 2 lists the safe concentrations of FITC-FYGL and the applied endocytic selective inhibitors in HepG2 cells, which will be used in the subsequent experiments.
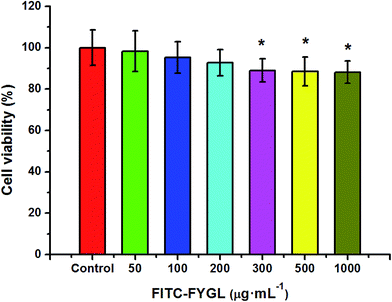 |
| | Fig. 4 In vitro cell viability of HepG2 cells incubated with different concentrations of FITC-FYGL for 24 h at 37 °C. Data are presented as mean ± SD (n = 6). *p < 0.05 versus the control group. | |
Table 2 CCK-8 test of safe concentrations on HepG2 cells
| Agent |
Safe concentration (μg mL−1) |
| Chlorpromazine |
50 |
| Nystatin |
50 |
| Amiloride |
100 |
| LY294002 |
10 |
| Genistein |
500 |
| FITC-FYGL |
200 |
3.5 Influence of endocytic selective inhibitors on FYGL uptake in HepG2 cells
Fig. 5A shows the FCM data of HepG2 cells (blank), the cells incubated with 100 μg mL−1 FITC-FYGL (control), and 100 μg mL−1 FITC-FYGL addition to preincubated 50 μg mL−1 chlorpromazine, 50 μg mL−1 nystatin, 100 μg mL−1 amiloride, 10 μg mL−1 LY294002, and 500 μg mL−1 genistein. From Fig. 5B, the fluorescence at λ = 488 nm was referred to that of blank group, and the FITC-FYGL fluorescence at λ = 488 nm in control group was normalized to 100. It was found that neither chlorpromazine nor nystatin had a significant inhibitory effect on endocytosis of FYGL, so the possibilities of clathrin-mediated endocytosis and caveolae/lipid-mediated endocytosis are excluded. However, the FITC-FYGL fluorescence in the groups with selective inhibitors of amiloride, LY294002 and genistein were all remarkably attenuated to different extents, compared with that in the control group, indicating that the uptake of FITC-FYGL in the cells was inhibited by amiloride, LY294002 and genistein. The results were also demonstrated by confocal laser scanning microscopy (Fig. 5C), which revealed that almost no green FITC-FYGL was observed in the cells with those inhibitors. Amiloride was reported to specifically inhibit macropinocytosis by lowering the submembraneous pH;27 herein, the significant inhibition of the endocytic process of FYGL by amiloride indicated that the cellular uptake of FYGL was regulated by macropinocytosis. Macropinocytosis requires several kinases to activate numerous signaling pathways, which coordinate the formation of a macropinosome.28 Genistein can inhibit either tyrosine kinases or caveolae pinching to regulate endocytosis.22 In this study, the caveolae/lipid-mediated endocytosis of FYGL was excluded because nystatin did not inhibit FYGL entering the cells (Fig. 5); thus genistein may work on the inhibition of tyrosine kinase activity to block the uptake of FYGL.
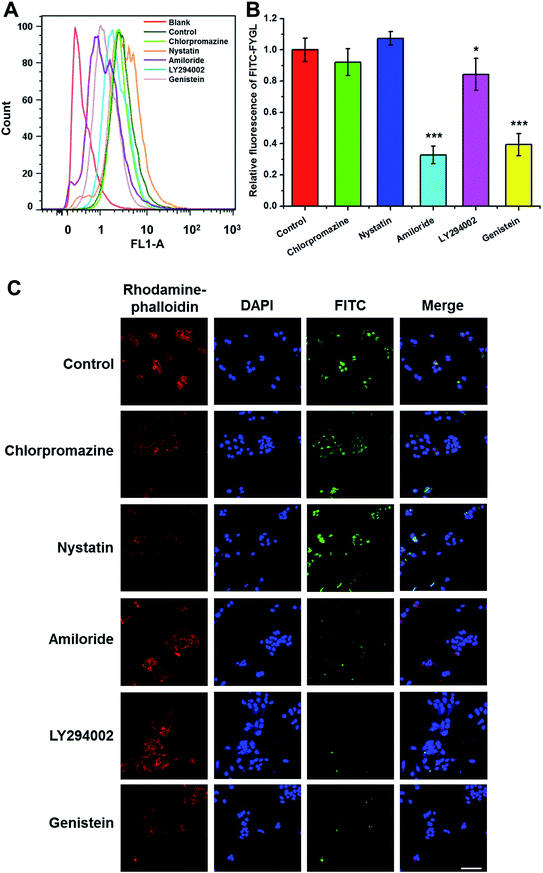 |
| | Fig. 5 (A) FCM shows the peak position of 100 μg mL−1 FITC-FYGL with the selective inhibitor (chlorpromazine, nystatin, amiloride, LY294002, and genistein) in channel FL1. (B) The relative fluorescent intensity of FITC-FYGL with the selective inhibitors in HepG2 cells. Data are presented as mean ± SD (n = 3). *p < 0.05, ***p < 0.001 versus the control group. (C) Confocal laser scanning microscopy images of HepG2 cells incubated with FITC-FYGL for 4 h in the absence or presence of inhibitor, chlorpromazine, nystatin, amiloride, LY294002, and genistein. The scale bar represents 50 μm. | |
As a quercetin analogue, LY294002 is widely used to specifically inhibit PI3K activity and macropinosome formation.29,30 PI3K activity in the process of membrane ruffling affects the formation of macropinosomes and phagosomes, especially the closure of ruffles and pseudopodia, in addition to the influence on the roles of insulin and insulin growth factor-1 in signal transduction.31–33 Herein, LY294002 down-regulated the uptake of FYGL in HepG2 cells, indicating that the uptake process might involve the PI3K cascade.
3.6 Endocytosis of FYGL mediated by c-Src protein
c-Src is a non-receptor tyrosine kinase and the upstream protein of PI3K, modulating ruffling in the early stages of phagocytosis.18,34,35 Thus, inhibition of c-Src activity blocks the macropinocytosis process.33 From Fig. 6A, HepG2 cells transfected with siRNA inhibited the expression of c-Src to a remarkable extent, compared with control group. The uptake of FYGL in HepG2 cells was also attenuated, which was demonstrated by FCM and the confocal laser scanning microscopy images in Fig. 6B–D. The results indicate that the endocytosis of FYGL in HepG2 cells is possibly on the c-Src/PI3K cascade involved in the macropinocytosis process.
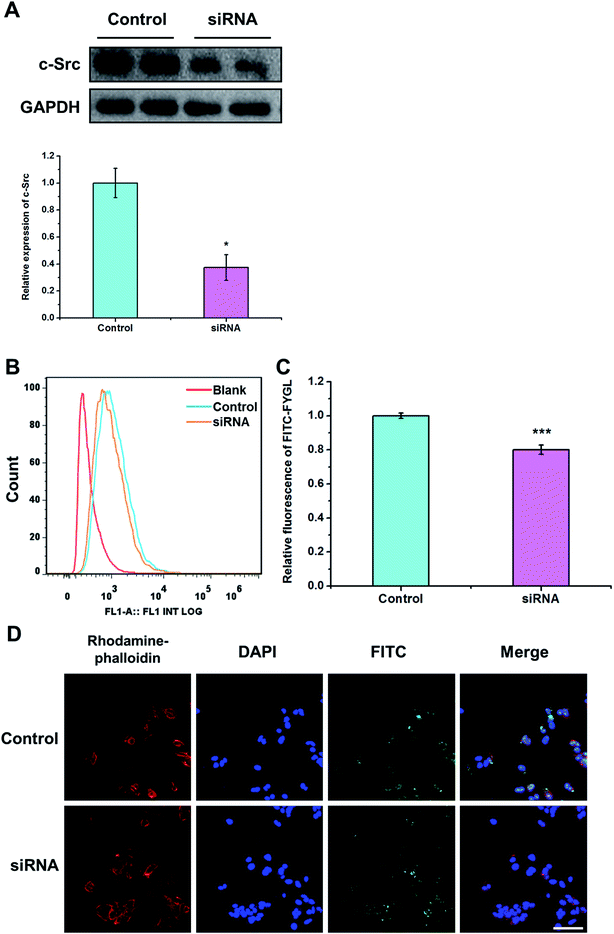 |
| | Fig. 6 (A) Western blot of c-Src protein in the control group and in siRNA transfected group. Data are presented as mean ± SD (n = 3). *p < 0.05 versus the control group. (B) FCM shows the peak position of 100 μg mL−1 FITC-FYGL in siRNA transfected HepG2 cells in channel FL1. (C) The relative fluorescent intensity of FITC-FYGL in siRNA transfected HepG2 cells. (D) Confocal laser scanning microscopy images of siRNA transfected HepG2 cells incubated with FITC-FYGL for 4 h. The scale bar represents 50 μm. Data are presented as mean ± SD (n = 3). ***p < 0.001 versus the control group. | |
3.7 Intracellular co-localization of FYGL and dextran
Dextran is used as a macropinocytosis tracer in endocytosis mechanism research.15 The method of co-localization was used to prove that FYGL and dextran enter the cell via the same endocytosis pathway, if both compounds are overlaid in the cell. From the images in Fig. 7, we can observe that when FYGL and dextran were cultured in HepG2 cells, they were co-localized in the cells, and the merged image of FYGL (green, FITC labelled) and dextran (red, TRITC labelled) showed orange cells. The results demonstrated again that the uptake of FYGL in HepG2 cells was mainly through macropinocytosis and also identified the result of Fig. 3C.
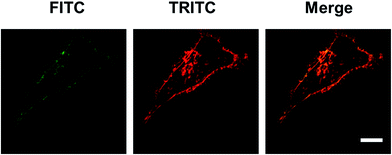 |
| | Fig. 7 Confocal laser scanning microscopy images of HepG2 cells incubated with FITC-FYGL for 4 h and TRITC-dextran for 10 min. The scale bar represents 10 μm. | |
4. Conclusions
In conclusion, we demonstrated that FYGL in aqueous solutions existed as aggregates with particle sizes in the range 400–1000 nm, and could be absorbed by HepG2 cells via the macropinocytosis mechanism. Moreover, we excluded clathrin-mediated endocytosis and caveolae/lipid-mediated endocytosis of FYGL by the pharmacologically selective inhibitor approach and proved that macropinocytosis mediated by c-Src/PI3K cascades was the preferred endocytic route. FYGL is a multidomain peptide that contains a polysaccharide and peptide domain and can potentially play multiple roles in the treatment of diseases. The specific structure of FYGL is an ideal case of bottom-up design where the modulation of chemical structure leads to controlled self-assembly and nanostructure, and eventually control of biological functions.36 Our study provided the uptake basis of FYGL for efficient hypoglycemic action. The functional mechanism of the signaling pathway in vivo will be reported in the future.
Authors' contribution
ZY and FW performed the experiments and statistical analysis, ZY and PZ designed the experiments and wrote the manuscript, and HJY and PZ provided the funds.
Conflicts of interest
The authors declare the researchers in this study have no conflict of interest.
Acknowledgements
This study was supported by the Natural Science Foundation of China (No. 21374022 and 81374032).
References
- C. Taghibiglou, F. Rashid-Kolvear, S. C. Van Iderstine, H. Le-Tien, I. G. Fantus, G. F. Lewis and K. Adeli, J. Biol. Chem., 2002, 277, 793–803 CrossRef CAS PubMed.
- M. Elchebly, P. Payette, E. Michaliszyn, W. Cromlish, S. Collins, A. L. Loy, D. Normandin, A. Cheng, J. Himms-Hagen, C. C. Chan, C. Ramachandran, M. J. Gresser, M. L. Tremblay and B. P. Kennedy, Science, 1999, 283, 1544–1548 CrossRef CAS PubMed.
- T. O. Johnson, J. Ermolieff and M. R. Jirousek, Nat. Rev. Drug Discovery, 2002, 1, 696–709 CrossRef CAS PubMed.
- M. Zaklos-Szyda, I. Majewska, M. Redzynia and M. Koziolkiewicz, Curr. Top. Med. Chem., 2015, 15, 2431–2444 CrossRef PubMed.
- B. S. Teng, C. D. Wang, H. J. Yang, J. S. Wu, D. Zhang, M. Zheng, Z. H. Fan, D. Pan and P. Zhou, J. Agric. Food Chem., 2011, 59, 6492–6500 CrossRef CAS PubMed.
- H. T. Ma, J. F. Hsieh and S. T. Chen, Phytochemistry, 2015, 114, 109–113 CrossRef CAS PubMed.
- D. Pan, L. Q. Wang, C. H. Chen, B. W. Hu and P. Zhou, Carbohydr. Polym., 2015, 117, 106–114 CrossRef CAS PubMed.
- C. Orellana-Tavra, S. A. Mercado and D. Fairen-Jimenez, Adv. Healthcare Mater., 2016, 5, 2261–2270 CrossRef CAS PubMed.
- T.-G. Iversen, T. Skotland and K. Sandvig, Nano Today, 2011, 6, 176–185 CrossRef CAS.
- P. Zhao, M. Cao, L. N. Song, H. Wu, K. Hu, B. Chen, Q. W. Wang and N. Gu, RSC Adv., 2016, 6, 96635–96643 RSC.
- W. H. Lewis, Bull. Johns Hopkins Hosp., 1931, 49, 17–27 Search PubMed.
- M. C. Kerr and R. D. Teasdale, Traffic, 2009, 10, 364–371 CrossRef CAS PubMed.
- A. T. Jones, J. Cell. Mol. Med., 2007, 11, 670–684 CrossRef CAS PubMed.
- J. A. Swanson and C. Watts, Trends Cell Biol., 1995, 5, 424–428 CrossRef CAS PubMed.
- S. Elmquist, R. Libelius, G. Lawoko and S. Tagerud, Muscle Nerve, 1992, 15, 876–884 CrossRef CAS PubMed.
- R. J. Jones, V. G. Brunton and M. C. Frame, Eur. J. Cancer, 2000, 36, 1595–1606 CrossRef CAS PubMed.
- B. R. Wong, D. Besser, N. Kim, J. R. Arron, M. Vologodskaia, H. Hanafusa and Y. Choi, Mol. Cell, 1999, 4, 1041–1049 CrossRef CAS PubMed.
- A. Aderem and D. M. Underhill, Annu. Rev. Immunol., 1999, 17, 593–623 CrossRef CAS PubMed.
- L. H. Wang, K. G. Rothberg and R. G. W. Anderson, J. Cell Biol., 1993, 123, 1107–1117 CrossRef CAS PubMed.
- C. K. Payne, S. A. Jones, C. Chen and X. Zhuang, Traffic, 2007, 8, 389–401 CrossRef CAS PubMed.
- M. Koivusalo, C. Welch, H. Hayashi, C. C. Scott, M. Kim, T. Alexander, N. Touret, K. M. Hahn and S. Grinstein, J. Cell Biol., 2010, 188, 547–563 CrossRef CAS PubMed.
- L. Pelkmans, D. Puntener and A. Helenius, Science, 2002, 296, 535–539 CrossRef CAS PubMed.
- P. Khandelwal, W. G. Ruiz and G. Apodaca, EMBO J., 2010, 29, 1961–1975 CrossRef CAS PubMed.
- Y. B. Tewari, M. M. Miller, S. P. Wasik and D. E. Martire, J. Chem. Eng. Data, 1982, 27, 451–454 CrossRef CAS.
- L. Jiang, L. Wang, M. Guo, G. Yin and R.-Y. Wang, Sens. Actuators, B, 2011, 156, 825–831 CrossRef CAS.
- Y. Katayama, Bull. Chem. Soc. Jpn., 2017, 90, 12–21 CrossRef CAS.
- M. A. West, M. S. Bretscher and C. Watts, J. Cell Biol., 1989, 109, 2731–2739 CrossRef CAS PubMed.
- Y. Egami, T. Taguchi, M. Maekawa, H. Arai and N. Araki, Front. Physiol., 2014, 5, 374 Search PubMed.
- C. J. Vlahos, W. F. Matter, K. Y. Hui and R. F. Brown, J. Biol. Chem., 1994, 269, 5241–5248 CAS.
- N. Araki, M. Hamasaki, Y. Egami and T. Hatae, Cell Struct. Funct., 2006, 31, 145–157 CrossRef CAS PubMed.
- N. Araki, M. T. Johnson and J. A. Swanson, J. Cell Biol., 1996, 135, 1249–1260 CrossRef CAS PubMed.
- K. Kotani, K. Yonezawa, K. Hara, H. Ueda, Y. Kitamura, H. Sakaue, A. Ando, A. Chavanieu, B. Calas, F. Grigorescu, M. Nishiyama, M. D. Waterfield and M. Kasuga, EMBO J., 1994, 13, 2313–2321 CAS.
- S. Wennstrom, P. Hawkins, F. Cooke, K. Hara, K. Yonezawa, M. Kasuga, T. Jackson, L. Claessonwelsh and L. Stephens, Curr. Biol., 1994, 4, 385–393 CrossRef CAS PubMed.
- M. Mettlen, A. Platek, P. Van Der Smissen, S. Carpentier, M. Amyere, L. Lanzetti, P. de Diesbach, D. Tyteca and P. J. Courtoy, Traffic, 2006, 7, 589–603 CrossRef CAS PubMed.
- M. Okada and H. Nakagawa, J. Biol. Chem., 1989, 264, 20886–20893 CAS.
- A. N. Moore and J. D. Hartgerink, Acc. Chem. Res., 2017, 50, 714–722 CrossRef CAS PubMed.
|
| This journal is © The Royal Society of Chemistry 2017 |
Click here to see how this site uses Cookies. View our privacy policy here.  Open Access Article
Open Access Article *a
*a
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 17 for saccharide
17 for saccharide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) protein.7 We first demonstrated that FYGL has an efficient hypoglycemic effect and high safety in vivo,5 but mechanism was still unclear. In order to understand the possible functional mechanism of FYGL, it is necessary to first reveal how the biomacromolecule works at the cellular level. Therefore, herein, we focused on demonstrating the cell internalization and endocytosis mechanism of FYGL in HepG2 cells, which are engaged in glycogen synthesis.
protein.7 We first demonstrated that FYGL has an efficient hypoglycemic effect and high safety in vivo,5 but mechanism was still unclear. In order to understand the possible functional mechanism of FYGL, it is necessary to first reveal how the biomacromolecule works at the cellular level. Therefore, herein, we focused on demonstrating the cell internalization and endocytosis mechanism of FYGL in HepG2 cells, which are engaged in glycogen synthesis.

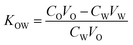
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g, 10 min, 4 °C). The cell lysates were separated by 8% SDS-PAGE, transferred to a polyvinylidene fluoride (PVDF) membrane, and then immunoblotted with rabbit polyclonal c-Src antibody (Sangon Co, Shanghai, China) and secondary goat anti-rabbit antibody (Cell Signaling Technology, USA). Bands were visualized with enhanced chemiluminescence solution (ECL, Biotanon) and detected by Image Lab camera (Bio-Rad, Germany); they were then quantified by densitometry scanning by the Image J software.
000 × g, 10 min, 4 °C). The cell lysates were separated by 8% SDS-PAGE, transferred to a polyvinylidene fluoride (PVDF) membrane, and then immunoblotted with rabbit polyclonal c-Src antibody (Sangon Co, Shanghai, China) and secondary goat anti-rabbit antibody (Cell Signaling Technology, USA). Bands were visualized with enhanced chemiluminescence solution (ECL, Biotanon) and detected by Image Lab camera (Bio-Rad, Germany); they were then quantified by densitometry scanning by the Image J software.






