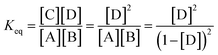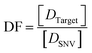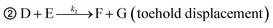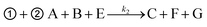Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Systematic comparison between toehold exchange and toehold displacement: exploration for highly specific and sensitive DNA detection†
Wen Yu‡
a,
Lan Tang‡b,
Ju-Hui Qiuc,
Zhang Zhangd,
Li-Li Zhoua,
Jun-Long Lia and
Guo-Ming Xie *a
*a
aKey Laboratory of Laboratory Medical Diagnostics of Education, Department of Laboratory Medicine, Chongqing Medical University, Chongqing 400016, P. R. China. E-mail: guomingxie@cqmu.edu.cn
bThe Public Health Center, The First Affiliated Hospital of Chongqing Medical University, Chongqing 400016, P. R. China
cState Key Laboratory of Membrane Biology, Tsinghua-Peking Center for Life Sciences, School of Life Sciences, Tsinghua University, Beijing 100084, China
dDepartment of Laboratory Medicine, Affiliated Hospital of Southwest Medical University, Luzhou, Sichuan, P. R. China
First published on 22nd August 2017
Abstract
The detection of nucleic acid variations with high specificity and sensitivity is essential for the good practice of precision medicine. Herein, we explore improving the sensitivity while ensuring the specificity of nucleic acid detection at single base resolution. “Toehold exchange” and “toehold displacement” are the two most widely used strategies in nucleic acid analysis due to their high theoretical specificity. However, to the best of our knowledge, there is no direct comparison between the two strategies to determine which one is more specific for nucleic acid analysis. First, a systematic comparison of the specificity of SNV detection based on toehold exchange and toehold displacement is performed, which shows that toehold exchange exhibits higher discrimination than toehold displacement, both experimentally and theoretically. Second, “plug the reaction”, which is a new principle to improve sensitivity without sacrificing specificity is proposed for the first time and demonstrated. This is achieved by adding a “plugging probe” at the equilibrium of toehold exchange. The plugging probe reacts with the product of toehold exchange, thus breaking the equilibrium of the reaction and successfully amplifying the signal. In summary, this work provides universal guidelines for nucleic acid analysis such as the detection of SNVs and rare mutations, which are required for high specificity, and the principle of plug the reaction will present new avenues for sensitive nucleic acid analysis.
1. Introduction
Variations in nucleic acid sequences, including DNA, mRNA, and miRNA, have been demonstrated to play crucial roles in many diseases, such as cancers and cardiovascular disease.1–3 Even a single base mutation in RNA or DNA sequences may cause the occurrence, development, and drug resistance of diseases, and has great impact on their prognosis and therapy.4–6 With the development of precision medicine, detection of sequence variants at base–pair resolution is of great importance particularly in early cancer diagnosis, in which tumor samples may contain very little target variants and have many interferential sequences. A wide variety of technologies have been developed for nucleic acid analysis, including hybridization, PCR, isothermal amplification, sequencing, as well as their combinations.7–9 Although these technologies have distinct advantages when used in nucleic acid analysis, there are still some drawbacks that limit the scope of their application, for example, PCR often results in false positives and artifactual mutations, while sequencing technology suffers from a definite intrinsic error rate (5 × 10−4 to 10−2 per nucleotide).10,11 Due to these limitations, the detection of sequence variants such as single nucleotide variants (SNVs) and rare mutations requires high specificity, which is still a challenge. Even next generation sequencing cannot easily detect rare mutations and single molecule mutations.12,13To detect SNVs and rare mutations, nucleic acid testing is required to maximize specificity with sufficient sensitivity. There is a myriad of research focused on improving the sensitivity of nucleic acid analysis,7,8 such as binding-induced DNA assembly and incorporating isothermal amplification using DNA nanotechnology and other metal nanomaterials.14–16 Although these methods achieved high sensitivity and low detection limitation (as low as fM), they usually have the disadvantage of low sequence selectivity or specificity, which causes them to be rarely used in clinical diagnosis. There is scarce research on specificity which is in fact no less important than sensitivity in clinical detection. Most detection methods just use toehold displacement or toehold exchange to guarantee specificity; however, no research has compared the two strategies to determine which one exhibits higher discrimination ability for nucleic acid analysis.
Both toehold displacement and toehold exchange are competitive hybridization reactions, where an incoming DNA strand outcompetes the other strand (usually known as a blocker or protector) from a DNA or RNA duplex to form a better matched duplex. This process is controlled by the Gibbs free energy of hybridization. Toehold displacement which was first introduced by Yurke is a highly specific process in which an input strand displaces a blocker strand to bind to its complementary partner.17 Toehold exchange, which was developed from toehold displacement, possesses fast reaction kinetics, high specificity and has the potential to be used in large reaction networks.18–21 David optimized the specificity of nucleic acid hybridization based on toehold exchange,20 but there is still confusion about the specificity of toehold exchange and toehold displacement. On one hand, toehold exchange has been demonstrated to possess high selectivity for the detection of single base mutations in double-stranded DNA,22–24 as well as for the enhanced specific detection of other mutations.25–27 On the other hand, there are more studies based on toehold displacement. For example, combined with materials such as gold nanoparticles, graphene, and nanodendrites, toehold displacement has been used to detect mRNA and DNA.28–31 Based on toehold displacement, highly selective detection of DNA and miRNA has been achieved,32–37 and also in situ molecular visualization.38–40 Herein, the specificity of SNV detection based on toehold exchange and toehold displacement is systematically compared in a more obvious manner. We believe this will provide universal guidelines for nucleic acid analysis, which are required for high specificity.
Also, we intended to improve sensitivity without sacrificing specificity by adding another “plugging probe” to plug the reaction. By performing the signal amplification step after the discrimination reaction, the specificity is not influenced.41 Differing from target sequence amplification which may amplify both unwanted sequences and target sequences and may also introduce artifactual mutations, our signal amplification strategy is energy-efficient. The process of the “plug the reaction” is illustrated as follows. Once the discrimination reaction reaches equilibrium based on toehold exchange, a fluorescent reporter strand is produced. Then an additional plugging probe is added to react with the fluorescent reporter strand based on toehold displacement, which consumes the product of the toehold exchange reaction, thus breaking the equilibrium of the reaction and making the reversible reaction move forward and producing more fluorescence signals. Additionally, to demonstrate the potential applications of plug the reaction, the probes are slightly modified. For example, labelling the plugging probe with the same fluorophore can further improve sensitivity.
Since the sequence specificity can be well represented by the discrimination of SNVs, herein, we first systematically compare the performance of SNVs detection based on toehold exchange and toehold displacement in three different buffers (20 mM Tris–HCL, 1× PBS and 1× TAE), and using the results provide guidelines for highly specific nucleic acid analysis. Besides, we analyze the discrimination factor (DF, which represents the discrimination ability) of the above two strategies from both theoretical and experimental results. Finally, a new principle (plug the reaction), which is aimed at improving sensitivity without sacrificing specificity, is proposed and its potential application also demonstrated.
2. Experimental
2.1 Materials and chemicals
All chemicals were purchased from Sigma-Aldrich Chemical Co (St. Louis, USA). All oligonucleotides were synthesized and further purified via HPLC by Sangon (Shanghai, China). 6-Carboxylfluorescein (FAM) was used as a fluorophore and 4-((4-(dimethylamino) phenyl)azo) benzoic acid (Dabcyl) was used as a quencher according to fluorescence resonance energy transfer. The sequences are listed in Scheme S1.†2.2 Preparation of the probes
To prepare 200 nM double-stranded probes (probe1/probe1(2) and probe2/probe2(2)), 400 nM of each fluorophore (protector) or quencher (compliment) labeled strand was separately dissolved in the same buffer, Tris–HCl (pH 8.0, containing 1 M Na+), 1× TAE (containing 12.5 mM Mg2+) or 1× PBS (pH 7.4). Then the two solutions (fluorophore and quencher) were mixed and allowed to equilibrate for over 30 min. All the other probes (probe3/probe3(2) and probe4) and oligonucleotides were dissolved in the same Tris–HCl buffer (pH 8.0, containing 1 M Na+) unless specifically mentioned. All experiments and purifications were performed at room temperature (25 ± 2 °C) unless specifically mentioned.2.3 Fluorescence measurement
Kinetics were recorded using a Cary Eclipse Fluorescence spectrophotometer and the following parameters: excitation wavelength: 480 nm, emission wavelength: 520 nm, excitation slit: 10 nm and emission slit: 10 nm (note: all experiments were performed in 20 mM Tris–HCl with 1 M Na+ at room temperature (25 ± 2 °C) unless specifically mentioned).For the comparison of toehold displacement and toehold exchange, first, 100 μL 200 nM labeled probe (probe1 or probe2) was injected to a quartz fluorescence cuvette (45 × 12.5 × 7.5 mm3, path length 5 mm, inside width 1 mm). Subsequently, 100 μL 200 nM of target/SNVs was injected into the cuvette.
To evaluate the ΔGsep of the modified fluorophore and quencher, first, 100 μL 200 nM FAM labeled strand X (containing 200 nM catalyst C) was injected into a cuvette, and then 100 μL 200 nM duplex YZ (probe4) was injected into the cuvette.
For the plug the reaction experiments, following the toehold exchange experiment, 100 μL 200 nM probe3/probe3(2) was injected into a cuvette. To demonstrate the potential use of plug the reaction, the procedure was the same as that for plug the reaction above, except that probe3(2) was intentionally modified.
3. Results and discussion
3.1 Performance of toehold displacement and toehold exchange
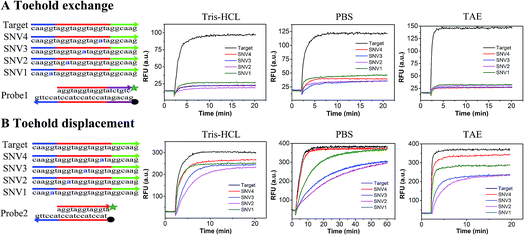
In order to compare the performance of SNV detection based on toehold exchange and toehold displacement, two types of probes were designed. Probe1 was designed based on toehold exchange and probe2 was designed based on toehold displacement. Probe1 and probe2 were both pre-formed duplexes that contained a fluorophore labeled strand and a quencher labeled protector strand. In order to compare their performances on the same basis, the sequences were designed to have base groups in series so that the four SNVs could have the same mutation and neighbors even though they occur at different positions (SNV1 in toehold, SNV2 in place of the double-strand region, SNV3 in the middle position and SNV4 in the end of branch migration region). When the probes were incubated with target or SNVs, the perfectly matched target displaced the protector strand with high efficiency and released high fluorescence, whereas the SNVs hardly displaced the protector strand and generated low fluorescence. Fig. 1 shows the relative fluorescence intensity (RFU) of the two detection systems (probe1 with target/SNVs and probe2 with target/SNVs) in three different buffers (20 mM Tris–HCl, 1× PBS, and 1× TAE containing 12.5 mM Mg2+). The fluorescence intensity at equilibrium in the target group was dramatically higher than that in the SNVs group through toehold exchange, but was only a little higher through toehold displacement. Moreover, in all three buffers, toehold exchange only needed 2–3 minutes to reach equilibrium for both target and SNVs. In contrast, toehold displacement needed 2–3 minutes to reach equilibrium for the target, but much longer for SNVs, especially for SNV2 and SNV3 in PBS which need more than 40 minutes to reach equilibrium. Three different buffers (20 mM Tris–HCl, 1× PBS, and 1× TAE containing 12.5 mM Mg2+) were used to compare toehold exchange with toehold displacement. As shown in Fig. 1, the performances of toehold exchange in the three buffers were almost the same, but the performance of toehold displacement differed from each other. For example, the performance of toehold displacement for SNVs in PBS was much slower than in Tris–HCl and TAE. This may have been caused by the different ions in different buffers. However, in all the buffers, the specificity of toehold exchange was better than toehold displacement (as shown in Table S1,† the value of DF for toehold exchange was about 6–8 and that for toehold displacement was only 1–2).Based on these results, it can be concluded that: (1) toehold exchange exhibits higher discrimination than toehold displacement at reaction equilibrium in any buffer system. Toehold exchange can distinguish SNVs no matter where the single base mutation is. These results are consistent with the theory of thermodynamics, as predicted by NUPACK. The Gibbs free energy (ΔG) for the toehold exchange reaction was almost 0 kcal mol−1 (Table S3†) which permits a high level of discrimination, whereas the ΔG for toehold displacement was −8.59 kcal mol−1 (Table S4†). Since ΔG determines the occurrence and productivity of a reaction, when ΔG ≈ 0 kcal mol−1, a slight change in ΔG can cause a huge difference in the reaction and thus optimal specificity. (2) The kinetics of toehold exchange is faster than that for toehold displacement, especially for SNVs. (3) Although the toehold displacement exhibits low discrimination at reaction saturation, it can achieve good discrimination in an appropriate buffer (e.g. 1× PBS) early in reaction, which relies on the kinetic difference, especially when the mutation occurs at the first or middle position of the migration strand, such as SNV2 or SNV3. This may be due to the fact that when mutations occur in the region of the double strand near the toehold, the reaction needs to break the pre-formed base-pairing and form a mismatch bubble after the reaction. However, when mutation occurs at the toehold region, there is no need to break the pre-formed base-pairing. When mutation occurs at the end of the branch migration region, there are already many matched bases before the mismatch bubble. This makes it difficult for the mismatched strands to be separated and therefore reduces the specificity. (4) The yield of toehold displacement is higher than toehold exchange because toehold displacement is thermodynamically more favorable owing to its relatively lower ΔGrea. To further demonstrate the universality of the methods, we prolonged the sequences of the target and SNVs and found that the results were similar to the short sequence except that the equilibrium time was longer (Fig. S1†).
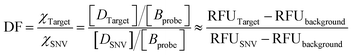
The discrimination factor (DF), which represents the discrimination ability, can be defined as the signal of the target to the signal of the SNVs. The calculated experimental DF is shown in Tables S1 and S2.†
The theoretical DF calculated using the thermodynamic equation is shown in Tables S3 and S4.† In a simplistic view, both toehold displacement and toehold exchange can be described as a reversible reaction as follows: A + B ![[left over right harpoons]](https://www.rsc.org/images/entities/char_21cb.gif) C + D, where A is the target or SNV, B is the probe, C is the waste and D is the fluorescent product. When the equilibrium of the reaction A + B
C + D, where A is the target or SNV, B is the probe, C is the waste and D is the fluorescent product. When the equilibrium of the reaction A + B ![[left over right harpoons]](https://www.rsc.org/images/entities/char_21cb.gif) C + D is achieved:
C + D is achieved:
ΔG = −RT![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ln(Keq) = (ΔGC + ΔGD) − (ΔGA + ΔGB) ln(Keq) = (ΔGC + ΔGD) − (ΔGA + ΔGB) |
where, ΔG is the Gibbs energy change, T is the Kelvin temperature, R is the gas constant and Keq is the equilibrium rate constant of reaction.
Both the experimental DF and theoretical DF for toehold exchange were higher than that for toehold displacement (Tables S1, S3 and S4†), which demonstrates that the specificity of toehold exchange is much better than toehold displacement.
However, there was difference between the theoretical DF and experimental DF values especially in toehold exchange. For example, the theoretical DFexh (DF of exchange) was 10.7 and the experimental DFexh was 6.02. This difference may due to the temperature, buffer condition, error from yield and prediction error of the NUPACK software because it disregards the modification of the fluorophore and quencher.
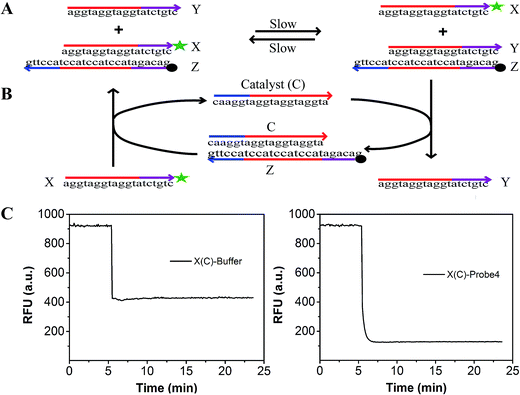
To evaluate the effect of the modification of the fluorophore and quencher on the reaction, we introduced a fluorescence strategy based on a generalized method for measuring the separation standard free energy (ΔGsep) of the modified FAM fluorophore and Dabcyl quencher (Fig. 2). The principle of this method is described in detail in Text S3.† Briefly, the absolute value of ΔGsep was equal to the gather standard free energy (ΔGgath) of the modified FAM fluorophore and Dabcyl quencher (Fig. 2A). The reaction can be described as: X + YZ ![[left over right harpoons]](https://www.rsc.org/images/entities/char_21cb.gif) Y + XZ, where X is the strand modified with FAM, Y is the strand without modification and Z is the strand modified with Dabcyl quencher.42 By designing strand X, which differs from the protector strand Y, to consist of probe4 by only the FAM modification, the reaction standard free energy (ΔGrea) was close to ΔGgath. A catalyst strand C was added to accelerate the reaction (Fig. 2B). When 100 μL buffer was added to 100 μL strand X (containing catalyst strand C), the fluorescence of X decreased because of the dilution effect (left of Fig. 2C). When 100 μL probe4 was added to 100 μL strand X (containing catalyst strand C), the fluorescence of X dramatically decreased because of the dilution effect plus the effect of the strand displacement reaction (right of Fig. 2C). Thus, based on Fig. 2C, we can evaluate the efficiency of the strand displacement reaction. Fig. S2† shows the relationship between the fluorescence intensities and the concentration of the FAM modified strand X. Based on Fig. S2† and 2, the ΔGsep was calculated to be 0.995 kcal mol−1 (Text S3†).
Y + XZ, where X is the strand modified with FAM, Y is the strand without modification and Z is the strand modified with Dabcyl quencher.42 By designing strand X, which differs from the protector strand Y, to consist of probe4 by only the FAM modification, the reaction standard free energy (ΔGrea) was close to ΔGgath. A catalyst strand C was added to accelerate the reaction (Fig. 2B). When 100 μL buffer was added to 100 μL strand X (containing catalyst strand C), the fluorescence of X decreased because of the dilution effect (left of Fig. 2C). When 100 μL probe4 was added to 100 μL strand X (containing catalyst strand C), the fluorescence of X dramatically decreased because of the dilution effect plus the effect of the strand displacement reaction (right of Fig. 2C). Thus, based on Fig. 2C, we can evaluate the efficiency of the strand displacement reaction. Fig. S2† shows the relationship between the fluorescence intensities and the concentration of the FAM modified strand X. Based on Fig. S2† and 2, the ΔGsep was calculated to be 0.995 kcal mol−1 (Text S3†).
Discrimination based on reaction kinetics is not very practical in most diagnostic applications since it usually requires advanced instrumentation and control over the concentrations of the SNVs and target, equilibrium discrimination is much more practical and preferable in real clinical diagnosis. Both the experimental DF and theoretical DF reveal that the toehold exchange exhibits higher equilibrium discrimination than toehold displacement no matter where the single base mutation is. These results will provide universal guidelines for nucleic acid analysis, which are required for high specificity. It can be combined with other strategies such as the use of blocking strands, sinkers and clutch probes, which have been used in specific nucleic acid detection to further improve specificity. For example, when toehold exchange was combined with blocking probes, the DF improved by about two times (Text S4 and Fig. S3†). Thus, toehold exchange was selected for use in our further experiments. Additionally, since the specificity of toehold exchange is position independent, we only chose SNV2 to represent SNVs for the following experiments.
3.2 Plug the reaction
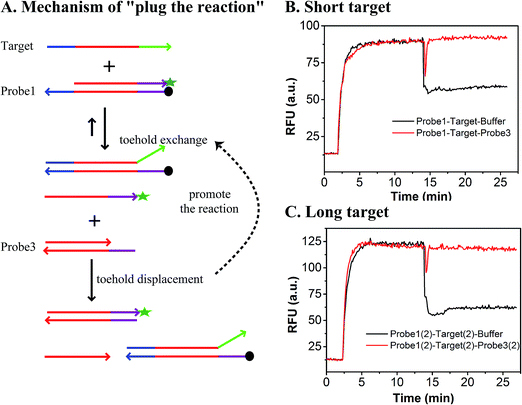
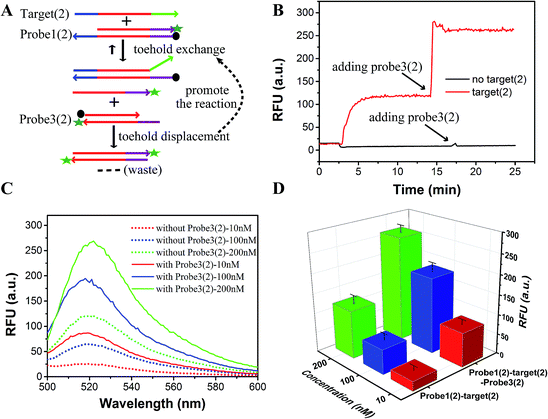
As shown in Fig. 1, the yield of toehold displacement was almost three times higher than toehold exchange, thus we expected that the combination of toehold exchange with toehold displacement would achieve both high specificity and sensitive detection.A plugging probe (probe3) was designed to react with the fluorescence reporter strand (the product of toehold exchange), thus plugging the former toehold exchange reaction and causing more FAM labeled strand to be released (Fig. 3A). The two-step reaction can be described as:
➀ A + B ![[left over right harpoons]](https://www.rsc.org/images/entities/char_21cb.gif) C + D (toehold exchange) C + D (toehold exchange) |
where, A is the target or SNVs, B is probe1, C is the waste, D is the intermediate fluorescent product, E is probe3, G is the waste and F is the final fluorescent product.
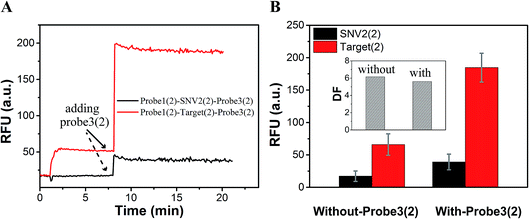
As shown in Fig. 3B and C, the RFU dropped immediately when probe3 was added to the system at equilibrium because of the dilution effect. Also, the RFU increased immediately, which was caused by plug the reaction. However, there was only a dilution effect without plug the reaction once buffer was added to the system.
In order to demonstrate that specificity would not be influenced through plug the reaction, we detected SNV2 and target based on this system and calculated the DF with time (Fig. 4). By adding probe3, the RFU of both the target and SNV2 decreased and increased (Fig. 4A), but the DF was not reduced by adding probe3 (Fig. 4B). These results show that the DF will not be reduced by plug the reaction through the addition of probe3. Furthermore, we designed a series of probe3 with different lengths of toehold to maximize the efficiency of plugging the reaction. Fig. 5 shows the RFU change over time by adding probe3 with different lengths of toehold in the second step. The black line represents the basic line upon the addition buffer after the first step of the reaction. The other colorful lines represent the RFU over time with the addition of probe3 with different lengths of toehold in the second step. The results showed that when the length of toehold of probe3 was seven, the RFU was the highest (green line) compared with the buffer (black line). This was caused by the combined effects of thermodynamics and kinetics. Thus, when the length of toehold of probe3 was seven, the efficiency of plugging the reaction was best.
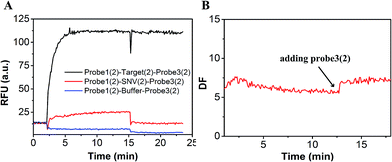 | ||
| Fig. 4 Evaluation of the DF based on plug the reaction. (A) Fluorescence of SNV2 and target detection responses over time. (B) DF responses over time. | ||
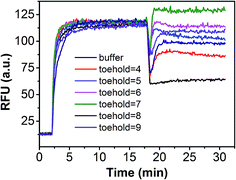 | ||
| Fig. 5 Performance of plug the reaction based on different probe3 with different lengths of toehold. | ||
As a proof-of-concept, detection systems based on plug the reaction were designed to demonstrate the potential use of plug the reaction. In one system, probe3 and probe1 were labeled with the same fluorophore FAM, which successfully amplified the fluorescence signal (Fig. 6). As shown in Fig. 6A, the DNA target reacted with probe1 based on toehold exchange, generating a fluorescent strand. Then the generated fluorescent strand reacted with probe3 based on toehold displacement, which broke the equilibrium of toehold exchange and produced an increased fluorescent signal. As shown in Fig. 6B, the fluorescent intensity increased more than two times when probe3 was added to the reaction. Fig. 6C shows the fluorescence intensity before the addition of probe3 (dotted line) and after the addition of probe3 (solid line) at different concentrations of target (10 nM, 100 nM and 200 nM). This shows that the signal can be amplified 2–3 times by adding probe3 to plug the reaction (Fig. 6C and D). Using 100 nM target and SNV2 as an example, we can see that both the RFUs of SNV and target increased by adding probe3 (Fig. 7A), but the DFs before and after the addition of probe3 were almost the same (Fig. 7B).
In summary, we combined toehold displacement with toehold exchange and formed the plug the reaction system, which increased their sensitivity by two times while ensuring specificity. The sensitivity can be further enhanced if other amplification steps are incorporated, such as PCR and binding-induced DNA assembly. We expect that the principle of consuming or taking away the product of toehold exchange to plug the reaction and amplify the signal will be useful in further applications.
4. Conclusions
Herein, we systematically compared the performance of SNV detection based on toehold exchange and toehold displacement, and found that the equilibrium discrimination of toehold exchange is better than toehold displacement, which is more practical in clinical applications. Also, the calculated DF of toehold exchange is higher than that of toehold displacement both theoretically and experimentally. Second, we introduced a new concept, plugging the reaction, to improve sensitivity without sacrificing specificity. We believe that this work will be of great significance in the development of both specific and sensitive nucleic acid analysis. It has the potential to be used in chips, next-generation sequencing, PCR and other detection strategies for accomplishing the ultra-specific detection of SNVs, rare mutations and other nucleic acid analyses.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by the Natural Science Research Foundation of China (81672112, 81171415) and the Chongqing Yuzhong District Science and Technology Project (20140108).Notes and references
- S. Misale, R. Yaeger, S. Hobor, E. Scala, M. Janakiraman, D. Liska, E. Valtorta, R. Schiavo, M. Buscarino, G. Siravegna, K. Bencardino, A. Cercek, C. T. Chen, S. Veronese, C. Zanon, A. Sartore-Bianchi, M. Gambacorta, M. Gallicchio, E. Vakiani, V. Boscaro, E. Medico, M. Weiser, S. Siena, F. Di Nicolantonio, D. Solit and A. Bardelli, Nature, 2012, 486, 532–536 CAS.
- M. Peifer, L. Fernandez-Cuesta, M. L. Sos, J. George, D. Seidel, L. H. Kasper, D. Plenker, F. Leenders, R. Sun, T. Zander, R. Menon, M. Koker, I. Dahmen, C. Muller, V. Di Cerbo, H. U. Schildhaus, J. Altmuller, I. Baessmann, C. Becker, B. de Wilde, J. Vandesompele, D. Bohm, S. Ansen, F. Gabler, I. Wilkening, S. Heynck, J. M. Heuckmann, X. Lu, S. L. Carter, K. Cibulskis, S. Banerji, G. Getz, K. S. Park, D. Rauh, C. Grutter, M. Fischer, L. Pasqualucci, G. Wright, Z. Wainer, P. Russell, I. Petersen, Y. Chen, E. Stoelben, C. Ludwig, P. Schnabel, H. Hoffmann, T. Muley, M. Brockmann, W. Engel-Riedel, L. A. Muscarella, V. M. Fazio, H. Groen, W. Timens, H. Sietsma, E. Thunnissen, E. Smit, D. A. Heideman, P. J. Snijders, F. Cappuzzo, C. Ligorio, S. Damiani, J. Field, S. Solberg, O. T. Brustugun, M. Lund-Iversen, J. Sanger, J. H. Clement, A. Soltermann, H. Moch, W. Weder, B. Solomon, J. C. Soria, P. Validire, B. Besse, E. Brambilla, C. Brambilla, S. Lantuejoul, P. Lorimier, P. M. Schneider, M. Hallek, W. Pao, M. Meyerson, J. Sage, J. Shendure, R. Schneider, R. Buttner, J. Wolf, P. Nurnberg, S. Perner, L. C. Heukamp, P. K. Brindle, S. Haas and R. K. Thomas, Nat. Genet., 2012, 44, 1104–1110 CrossRef CAS PubMed.
- B. Vogelstein, N. Papadopoulos, V. E. Velculescu, S. Zhou, L. A. Diaz Jr and K. W. Kinzler, Science, 2013, 339, 1546–1558 CrossRef CAS PubMed.
- N. Kolevzon, D. Hashoul, S. Naik, A. Rubinstein and E. Yavin, Chem. Commun., 2016, 52, 2405–2407 RSC.
- M. Liong, A. N. Hoang, J. Chung, N. Gural, C. B. Ford, C. Min, R. R. Shah, R. Ahmad, M. Fernandez-Suarez, S. M. Fortune, M. Toner, H. Lee and R. Weissleder, Nat. Commun., 2013, 4, 1752 CrossRef PubMed.
- S. Soverini, S. Branford, F. E. Nicolini, M. Talpaz, M. W. Deininger, G. Martinelli, M. C. Muller, J. P. Radich and N. P. Shah, Leuk. Res., 2014, 38, 10–20 CrossRef CAS PubMed.
- H. Zhang, F. Li, B. Dever, X. F. Li and X. C. Le, Chem. Rev., 2013, 113, 2812–2841 CrossRef CAS PubMed.
- Y. Zhao, F. Chen, Q. Li, L. Wang and C. Fan, Chem. Rev., 2015, 115, 12491–12545 CrossRef CAS PubMed.
- D. Khodakov, C. Wang and D. Y. Zhang, Adv. Drug Delivery Rev., 2016, 105, 3–19 CrossRef CAS PubMed.
- Y. He, J. Wu, D. C. Dressman, C. Iacobuzio-Donahue, S. D. Markowitz, V. E. Velculescu, L. A. Diaz Jr, K. W. Kinzler, B. Vogelstein and N. Papadopoulos, Nature, 2010, 464, 610–614 CrossRef CAS PubMed.
- A. Gore, Z. Li, H. L. Fung, J. Young, S. Agarwal, J. Antosiewicz-Bourget, I. Canto, A. Giorgetti, M. A. Israel, E. Kiskinis, J. H. Lee, Y. H. Loh, P. D. Manos, N. Montserrat, A. D. Panopoulos, S. Ruiz, M. L. Wilbert, J. Yu, E. F. Kirkness, J. C. Izpisua Belmonte, D. J. Rossi, J. A. Thomson, K. Eggan, G. Q. Daley, L. S. Goldstein and K. Zhang, Nature, 2011, 471, 63–67 CrossRef CAS PubMed.
- M. T. Gregory, J. A. Bertout, N. G. Ericson, S. D. Taylor, R. Mukherjee, H. S. Robins, C. W. Drescher and J. H. Bielas, Nucleic Acids Res., 2016, 44, e22 CrossRef PubMed.
- M. W. Schmitt, E. J. Fox, M. J. Prindle, K. S. Reid-Bayliss, L. D. True, J. P. Radich and L. A. Loeb, Nat. Methods, 2015, 12, 423–425 CrossRef CAS PubMed.
- L. Liu, Q. Yang, J. Lei, N. Xu and H. Ju, Chem. Commun., 2014, 50, 13698–13701 RSC.
- K. Knez, K. P. Janssen, J. Pollet, D. Spasic and J. Lammertyn, Small, 2012, 8, 868–872 CrossRef CAS PubMed.
- S. Su, Y. Wu, D. Zhu, J. Chao, X. Liu, Y. Wan, Y. Su, X. Zuo, C. Fan and L. Wang, Small, 2016, 12, 3794–3801 CrossRef CAS PubMed.
- B. Yurke, A. J. Turberfield, A. P. Mills Jr, F. C. Simmel and J. L. Neumann, Nature, 2000, 406, 605–608 CrossRef CAS PubMed.
- D. Y. Zhang, A. J. Turberfield, B. Yurke and E. Winfree, Science, 2007, 318, 1121–1125 CrossRef CAS PubMed.
- D. Y. Zhang and E. Winfree, J. Am. Chem. Soc., 2009, 131, 17303–17314 CrossRef CAS PubMed.
- D. Y. Zhang, S. X. Chen and P. Yin, Nat. Chem., 2012, 4, 208–214 CrossRef CAS PubMed.
- X. Song, A. Eshra, C. Dwyer and J. Reif, RSC Adv., 2017, 7, 28130–28144 RSC.
- S. X. Chen, D. Y. Zhang and G. Seelig, Nat. Chem., 2013, 5, 782–789 CrossRef CAS PubMed.
- J. S. Wang and D. Y. Zhang, Nat. Chem., 2015, 7, 545–553 CrossRef CAS PubMed.
- Z. Zhang, J. L. Li, J. Yao, T. Wang, D. Yin, Y. Xiang, Z. Chen and G. Xie, Biosens. Bioelectron., 2016, 79, 488–494 CrossRef CAS PubMed.
- T. Wang, L. Zhou, S. Bai, Z. Zhang, J. Li, X. Jing and G. Xie, Biosens. Bioelectron., 2016, 78, 464–470 CrossRef CAS PubMed.
- Z. Wu, T. Ma, J. L. Coll, F. Liu, H. Zhang, Y. Ma, Z. Wang, Q. Jin, H. Mao and J. Zhao, Biosens. Bioelectron., 2016, 80, 175–181 CrossRef CAS PubMed.
- H. Xu, W. Deng, F. Huang, S. Xiao, G. Liu and H. Liang, Chem. Commun., 2014, 50, 14171–14174 RSC.
- A. E. Prigodich, P. S. Randeria, W. E. Briley, N. J. Kim, W. L. Daniel, D. A. Giljohann and C. A. Mirkin, Anal. Chem., 2012, 84, 2062–2066 CrossRef CAS PubMed.
- R. E. Paliwoda, F. Li, M. S. Reid, Y. Lin and X. C. Le, Anal. Chem., 2014, 86, 6138–6143 CrossRef CAS PubMed.
- T. Miyahata, Y. Kitamura, A. Futamura, H. Matsuura, K. Hatakeyama, M. Koinuma, Y. Matsumoto and T. Ihara, Chem. Commun., 2013, 49, 10139–10141 RSC.
- M. T. Hwang, P. B. Landon, J. Lee, D. Choi, A. H. Mo, G. Glinsky and R. Lal, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 7088–7093 CrossRef CAS PubMed.
- C. Li, Y. Li, Y. Chen, R. Lin, T. Li, F. Liu and N. Li, RSC Adv., 2016, 6(78), 74913–74916 RSC.
- Y. Yu, T. Wu, A. Johnson-Buck, L. Li and X. Su, Biosens. Bioelectron., 2016, 82, 248–254 CrossRef CAS PubMed.
- F. Yang, S. Wang, Y. Zhang, L. Tang, D. Jin, Y. Ning and G. J. Zhang, Biosens. Bioelectron., 2016, 82, 32–39 CrossRef CAS PubMed.
- D. A. Khodakov, A. S. Khodakova, A. Linacre and A. V. Ellis, J. Am. Chem. Soc., 2013, 135, 5612–5619 CrossRef CAS PubMed.
- D. A. Khodakov, A. S. Khodakova, D. M. Huang, A. Linacre and A. V. Ellis, Sci. Rep., 2015, 5, 8721 CrossRef CAS PubMed.
- S. X. Chen and G. Seelig, J. Am. Chem. Soc., 2016, 138, 5076–5086 CrossRef CAS PubMed.
- R. Deng, L. Tang, Q. Tian, Y. Wang, L. Lin and J. Li, Angew. Chem., Int. Ed., 2014, 53, 2389–2393 CrossRef CAS PubMed.
- M. J. Levesque, P. Ginart, Y. Wei and A. Raj, Nat. Methods, 2013, 10, 865–867 CrossRef CAS PubMed.
- D. Y. Duose, R. M. Schweller, J. Zimak, A. R. Rogers, W. N. Hittelman and M. R. Diehl, Nucleic Acids Res., 2012, 40, 3289–3298 CrossRef CAS PubMed.
- T. H. Ho, K. X. Dang, S. Lintula, K. Hotakainen, L. Feng, V. M. Olkkonen, E. W. Verschuren, T. Tenkanen, C. Haglund, K. L. Kolho, U. H. Stenman and J. Stenman, Nucleic Acids Res., 2015, 43, e4 CrossRef PubMed.
- C. Wang, J. H. Bae and D. Y. Zhang, Nat. Commun., 2016, 7, 10319 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental section, additional data, sequence data. See DOI: 10.1039/c7ra07481f |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2017 |