Open Access Article
Open Access ArticleThe mode of antimicrobial action of curcumin depends on the delivery system: monolithic nanoparticles vs. supramolecular inclusion complex†
Ilya Shlarab,
Samir Drobya,
Ruplal Choudharyc and
Victor Rodov *a
*a
aInstitute of Postharvest and Food Sciences, Agricultural Research Organization, The Volcani Center, 68 HaMaccabim Road, P.O.Box 15159, Rishon LeZion 7528809, Israel. E-mail: vrodov@agri.gov.il; Fax: +972-3-9683622; Tel: +972-3-9683609
bInstitute of Biochemistry, Food Science and Nutrition, Hebrew University of Jerusalem, Rehovot, Israel
cDepartment of Plant, Soil and Agricultural Systems, Southern Illinois University, Carbondale, USA
First published on 4th September 2017
Abstract
Curcumin has been known for a long time for its antimicrobial properties that are further increased by exposure to light. Due to the low aqueous solubility of curcumin, appropriate delivery systems are required to facilitate its implementation. In this work, we compared the antimicrobial activity toward Escherichia coli of two curcumin formulations: methyl-β-cyclodextrin supramolecular inclusion complex and polyelectrolyte-coated monolithic nanoparticles. The two formulations showed disparity both in the extent and in the mode of toxicity, highlighting the distinct properties of materials at the nanoscale. The tests showed that while curcumin–β-cyclodextrin complexes exhibited a potent bactericidal activity, the curcumin nanoparticles were bacteriostatic. Illumination with blue light significantly increased the bactericidal efficacy of curcumin–cyclodextrin complexes but had limited influence on the activity of nanoparticulate curcumin. While the antimicrobial effect of the supramolecular complex was predominantly characterized by the increase in ROS and inhibition of electron transport, the primary attributes of the nanoparticle action were membrane depolarization and reduced ATP concentrations. Interestingly, the treatment with curcumin nanoparticles induced a filamentous phenotype of the bacterium. Our results suggest that the antimicrobial properties of curcumin depend upon a delivery formulation, which may have both practical and regulatory implications on the applicability and safety of curcumin nanomaterials.
1. Introduction
The need for novel antimicrobial materials for medical, food and agricultural applications is widely acknowledged. In this regard, natural antimicrobials of plant origin are of particular interest due to their diversity, broad activity spectrum and favourable public acceptance. Curcumin, 1,7-bis-(4-hydroxy-3-methoxyphenyl)-1,6-heptadiene-3,5-dione, a natural phenylpropanoid dimer from the rhizomes of Curcuma longa L. (Zingiberaceae) and a major active constituent of the spice turmeric, used in Asia for thousands of years, has attracted attention as one of such promising compounds. Curcumin is known to exhibit antimicrobial activity against a wide range of bacterial species. Gram-positive bacteria show a significantly higher sensitivity to curcumin than the Gram-negative ones.1 The better protection of Gram-negative bacteria against antimicrobials is due to the specific structure of their cell walls. Lipopolysaccharides of the outer cell envelope represent the outermost permeability barrier for a variety of antimicrobial compounds2 responsible for the unusually slow influx of lipophilic solutes3 in Gram-negative bacteria. On the other hand, porin proteins embedded in the outer membrane represent the main channels for solute entry into the cells of Gram-negative bacteria.The antimicrobial activity of curcumin can be substantially increased by exposure to light. The broad absorption range (300–500 nm) with a maximum absorption band at 425 nm makes curcumin a potent photosensitizer, leading to the enhanced antimicrobial potency. Recent research has shown evidence that phototoxicity of curcumin towards planktonic Staphylococcus epidermidis depends on the type of nanocarrier.4 Furthermore, photosensitization allowed overcoming the limited activity of curcumin towards Gram-negative bacteria such as Salmonella typhimurium and Escherichia coli.5,6 These experiments, however, required rather high concentrations (5–10%) of dimethylsulfoxide to overcome the low solubility of curcumin and to increase its delivery and penetration through the cell walls of Gram-negative bacteria.
Despite its status of a food-grade colourant (E100), the applicability of curcumin as an antimicrobial agent is yet to be accepted by industry and by the regulatory authorities. The poor aqueous solubility of curcumin creates an obstacle for exploiting its antimicrobial potential in food systems. A substantial effort has been invested during the last two decades in developing adequate formulations and delivery systems to allow efficient delivery of biological activity of curcumin.7 These works generated numerous formulations and approaches used to solve solubility and delivery problems associated with curcumin. Among them, complexation with cyclodextrins and preparation of nanoparticles raise many expectations. Cyclodextrins are cyclic oligosaccharides produced from starch and shown to improve the solubility of hydrophobic drugs.8 Specifically, the methyl-β-cyclodextrin (MBCD) was particularly effective in increasing the delivery and antimicrobial activity of β-lactam antibiotics against a range of Gram-negative bacteria.9 It has also been shown that complexation with cyclodextrin increased curcumin solubilization and allowed the delivery of its activity to bacterial cells.10 MBCD inclusion revealed the highest antimicrobial potency of curcumin towards E. coli in comparison with a range of phenylpropanoid acids, aldehydes and alcohols and other phenolic compounds.11 Nanomaterials hold the vast promise for overcoming the delivery obstacles for antimicrobials. Numerous works have been dedicated to the preparation of curcumin nanoparticles12 as means to facilitate its aqueous solubility. In particular, ultrasound-assisted antisolvent precipitation aided by surface-stabilization with sterically and electrostatically active adsorbate layers was shown to promote curcumin dispersion stability.13 Such surface-stabilized curcumin nanoparticles significantly inhibited the growth of E. coli.14 The properties of nanomaterials may vary not only in comparison to the corresponding bulk material but also between different formulations of the same substance. Changes in physicochemical characteristics can affect chemical properties, reactivity and photostability of nanomaterials, altering their toxicity, behaviour in surrounding media and toxicokinetics. For instance, Zou et al.,15 demonstrated the effect of nanoparticle composition on encapsulation, protection, and release of curcumin.
The aim of this study was to investigate the impact of delivery system on antimicrobial activity of curcumin. The central question of the research was whether two curcumin delivery systems – surface-protected monolithic nanoparticles (i.e. solid curcumin nanoparticles stabilized with an adsorbate layer) and cyclodextrin molecular inclusion complexes – had the same mechanisms of toxicity towards E. coli. To address this question, we compared the magnitude and the mode of antimicrobial action of curcumin – methyl-β-cyclodextrin inclusion complexes and surface-protected curcumin nanoparticles in the dark and under light conditions.
2. Materials and methods
2.1. Strains and media
E. coli ATCC 25922 was used in this study. The strain was maintained at −85 °C in Lysogeny Broth (LB, pH 7.2; Difco, Sparks, MD, USA) containing 10% glycerol. To prepare fresh cultures, the bacteria were plated on LB agar and grown at 37 °C for 16 hours. These plates were stored at 4 °C for no longer than five days. Inocula for all the experiments were prepared using exponential phase cells. To avoid the selection of atypical variant clone five normally looking colonies were used for a starter culture inoculation. A sterile disposable plastic loop was used to transfer the colonies to 10 mL of LB in a sterile Erlenmeyer flask one day before the experiment. The pre-culture was incubated at 37 °C with 200 rpm shaking. After overnight growth, the pre-cultures were diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 with fresh LB medium preheated to 37 °C to resume exponential growth. The inocula were grown at 37 °C with 200 rpm shaking until reaching optical density of 0.3–0.4 at 600 nm (OD600). Cells were collected by centrifugation (3000 × g, 15 min, 20 °C), and the resulting pellet was resuspended in 5 mL of sterile phosphate buffered saline (PBS, pH 7.1). This procedure was repeated twice. The bacteria were resuspended in LB-broth to OD600 of 0.1.
400 with fresh LB medium preheated to 37 °C to resume exponential growth. The inocula were grown at 37 °C with 200 rpm shaking until reaching optical density of 0.3–0.4 at 600 nm (OD600). Cells were collected by centrifugation (3000 × g, 15 min, 20 °C), and the resulting pellet was resuspended in 5 mL of sterile phosphate buffered saline (PBS, pH 7.1). This procedure was repeated twice. The bacteria were resuspended in LB-broth to OD600 of 0.1.
2.2. Preparation of curcumin–cyclodextrin inclusion complex
The procedure of preparing curcumin–cyclodextrin inclusion complex (CCD) is schematically represented in Fig. 1. Equal volumes of 20 mM ethanolic solution of curcumin (Sigma-Aldrich, St. Lewis, MO) and 20 mM aqueous solution of MBCD (Sigma-Aldrich) were mixed and stirred for two hours on a magnetic stirrer at room temperature. Ethanol was removed from the mixture by evaporation under vacuum (Rotavapor R-124, Buchi, Flawil, Switzerland) to half-volume. The solution was then filtered through sterile 0.22 μ Millex filter with Durapore membrane (Millipore, Cork, Ireland). The concentration of the curcumin in the CCD was determined by diluting the aqueous dispersion with ethanol in a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 and measuring its absorbance with an Ultrospec 2100 pro spectrophotometer (Amersham Biosciences, Piscataway NJ) at 425 nm in comparison with the calibration curve of curcumin in 90% ethanol. The obtained filter-sterilized aqueous CCD dispersion was aliquoted and kept at −85 °C until use.
9 and measuring its absorbance with an Ultrospec 2100 pro spectrophotometer (Amersham Biosciences, Piscataway NJ) at 425 nm in comparison with the calibration curve of curcumin in 90% ethanol. The obtained filter-sterilized aqueous CCD dispersion was aliquoted and kept at −85 °C until use.
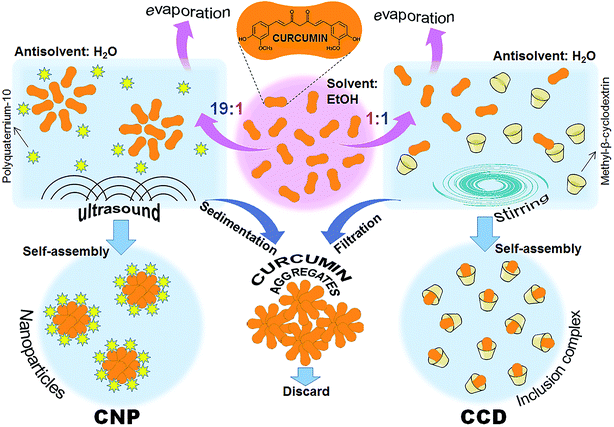 | ||
| Fig. 1 Preparation schemes of the curcumin formulations: curcumin–cyclodextrin inclusion complex (CCD) and curcumin nanoparticles (CNP). | ||
2.3. Preparation of curcumin nanoparticles
Curcumin nanoparticles (CNP) were prepared by ultrasound-assisted antisolvent precipitation using polyquaternium-10 (PQ-10) as a stabilising ligand as previously described14 (Fig. 1). Shortly, the syringe was filled with 19 mL of stabilizing ligand solution in antisolvent phase and added to 1 mL of the ethanolic curcumin solution (25 mM) under magnetic stirring at 1000 rpm. The mixture was sonicated for 20 min at 500 W, 20 kHz, using Q 500 sonicator (Qsonica LLC, Newtown, CT). The ethanol was removed from the dispersions by evaporation under vacuum. The suspensions were left to stand for 1 h, allowing sedimentation of the non-dissolved curcumin aggregates; the supernatant nanoparticle dispersions were separated by decantation and used for further investigations.2.4. Scanning electron microscopy
The water-dispersed nanoparticles were snap frozen in liquid nitrogen and freeze-dried in a model FD5508 freeze dryer (ilShin Lab Co., Yangju, Gyeonggi, South Korea). Dry nanoparticle powders were spread over a carbon tape (Ted Pella, Inc., Redding CA, USA). The samples were introduced into a chamber of a field-emission environmental scanning electron microscope XL30 ESEM-FEG (Philips, Eindhoven, The Netherlands). The chamber was flooded at pressures between 0.4 and 0.7 Torr, and samples were observed using a secondary electron detector at 20–25 kV.2.5. Dynamic light scattering (DLS) and ζ-potential measurements
The measurements of nanoparticle size and ζ-potential were taken using a Zetasizer ZS Nano (Malvern Instruments, Malvern, UK) equipped with a He–Ne laser (λ = 633 nm) at 25 °C. For the size determination, the light scattering was detected at an angle of 175°. DLS autocorrelation functions of the scattered light intensity were analysed with DTS 5.0 software provided by the manufacturer, which allowed the measurement of the distribution of the scattered intensity versus the hydrodynamic diameters and determined the polydispersity index (PDI). For the measurement of ζ-potential, the electrophoretic mobility of the particles was determined by laser Doppler velocimetry, in disposable capillary cells. The ζ-potential values were calculated by the Smoluchowski approximation of Henry's equation.162.6. Quantification of antimicrobial activity and time-kill kinetics
A final inoculum of ca. 106 CFU mL−1 was suspended in 96-well plates in LB broth supplemented with various concentrations of curcumin in the form of CCD or CNP. The plates were either exposed for 30 min to blue light as described elsewhere10 (wavelength 430–500 nm, dose 9 J cm−2), or kept under dark for the same period of time. Afterwards, the plates were incubated for 24 h at 37 °C in the dark. The doses tested were chosen based on minimum inhibitory concentrations (MIC) of curcumin determined in the previous studies: 400 μM for CNP,14 500 μM for CCD in the dark and 90 μM for CCD under light.10 CCD efficacy was tested in the concentration range of MIC to 2 × MIC with the increments of 25 and 10 μM for dark- and light-exposed samples, respectively. CNP efficacy was tested in the range of MIC to 2× MIC (maximal achievable concentration) with an increment of 100 μM for both dark- and light-exposed samples. Following the incubation at 37 °C for 24 h, the surviving bacteria were enumerated by the plate-count method on LB agar. The concentration of curcumin that produced at least 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log reduction in 24 hours was considered a minimal bactericidal concentration (MBC). For the time-kill kinetics, the bacterial cells were exposed to the both formulations in the LB medium and incubated at 37 °C. At the chosen time points, aliquots were taken, and viable CFU were enumerated by serial dilution plating on LB agar plates in triplicate. The vehicle controls, i.e. LB broth with added appropriate equivalent amounts of MBCD or PQ-10 but without curcumin, were used in all microbiological experiments and had no effect on bacterial cell proliferation.
log reduction in 24 hours was considered a minimal bactericidal concentration (MBC). For the time-kill kinetics, the bacterial cells were exposed to the both formulations in the LB medium and incubated at 37 °C. At the chosen time points, aliquots were taken, and viable CFU were enumerated by serial dilution plating on LB agar plates in triplicate. The vehicle controls, i.e. LB broth with added appropriate equivalent amounts of MBCD or PQ-10 but without curcumin, were used in all microbiological experiments and had no effect on bacterial cell proliferation.
2.7. Live/dead BacLight assay
The LIVE/DEAD BacLight Bacterial Viability Kit (Molecular Probes, Invitrogen, Eugene OR, USA) was used to assess the viability of bacteria as described by the manufacturer. The method is based on a combination of two fluorescent probes; SYTO9 is a nucleic acid stain (λex 488 nm/λemm 530 nm) that permeates the membranes of both live and dead cells, and propidium iodide (PI) (λex 488 nm/λemm 620 nm) that enters only the cells with compromised membranes. Since PI has a higher affinity for DNA than SYTO9, it can displace SYTO9 from the DNA. Thus, viable cells are stained green and dead stained red. In a spectrofluorimetric protocol, both green and red fluorescence intensities were measured using EnSpire™ multilabel plate reader (PerkinElmer, Waltham MA, USA) and the percentage of live and dead cells was calculated by comparing to a calibration curve of known ratios of live and dead cells. Each measurement consisted of three biological and four technical repetitions. Significance was calculated with a two-tailed Student's t-test relative to the respective vehicle controls. Selected samples were examined by confocal microscopy using a Leica SP8 laser scanning confocal microscope (Leica, Wetzlar, Germany), equipped with an HC PL APO 63×/1.20 water immersion objective.2.8. Nucleoid staining
For the microscopic observation of multinucleoid filaments, the bacteria were fixed with absolute ethanol at −20 °C for 15 min and then rehydrated in PBS for 15 min. The nucleoids were stained with 4′,6-diamidino-2-phenylindole (DAPI) (Sigma-Aldrich) according to manufacturer's instructions. Shortly the cells were centrifuged, the supernatant discarded and resuspended in 3 μM solution of DAPI in staining buffer (100 mM Tris, pH 7.4, 150 mM NaCl, 1 mM CaCl2, 0.5 mM MgCl2, 0.1% Nonidet P-40). After the incubation for 15 min at room temperature, the cells were examined by confocal microscopy using a Leica SP8 laser scanning confocal microscope (Leica, Wetzlar, Germany), equipped with an HC PL APO 63×/1.20 water immersion objective.2.9. Toxicity profiling
To compare between mechanisms of toxicity of CNP and CCD towards E. coli, we measured their influence on five cell vitality parameters: membrane integrity, membrane potential, cellular ROS generation, cellular ATP levels and electron transport activity as described elsewhere17 with modifications. The detailed description of the assays is available in ESI and Methods.† Shortly, E. coli cells were treated with five sub-MIC concentrations of both curcumin formulations (i.e. IC10; IC20; IC30; IC40 and IC50) which were obtained from the dose–response curve analysis (Fig. S1†). Treatments in the dark and upon illumination with blue light were followed by cell vitality assays. The results of the assays were presented in the heat map where red color indicated the results similar to the positive controls (i.e., 2-propanol for membrane integrity, carbonyl cyanide 3-chlorophenylhydrazone (CCCP) for the membrane potential assay, menadione for cellular ROS generation, carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP) for ATP levels, and sodium azide for electron transport activity), and green indicated the similarity of the results to the respective vehicle controls. The Pearson product-moment correlations (r) and their associated p-values were used to explore the relationship between the growth inhibition and cell viability parameters.2.10. SOS response assay by time-course RT-qPCR
The bacterial cells were treated either with a CCD or with CNP. As a positive control, cells were treated with 1 μg mL−1 of a known SOS inducer mitomycin C (Sigma-Aldrich).18 The appropriate vehicle control was used as a negative control. Total RNA was isolated using SV Total RNA isolation system (Promega, Madison, WI) according to manufacturers' instructions and 1 μg was used to synthesise cDNA using the Verso cDNA Synthesis kit (Thermo Scientific) and subjected to quantitative PCR (qPCR) to determine the relative expression of the selected genes. The detailed description of the qRT-PCR procedure is available in ESI and Methods.† Reactions were carried out in a total volume of 20 μL containing 1 μL cDNA, 5 μL Fast SYBR Green master mix (Applied Biosystems) and 0.3 μM forward and reverse primers (ESI Table S1†). Amplifications were conducted in MicroAmp 96-well plates (Applied Biosystems) and consisted of 20 s at 95 °C; followed by 40 cycles of 95 °C for 3 s, 60 °C for 30 s. The efficiency and critical threshold values obtained from the amplification data were used to calculate the relative amounts of cDNA according to the method described by Pfaffl.192.11. Fluorescence spectroscopy
Fluorescence emission spectra of CNP, CCD and of the bacterial cells treated with both formulations were recorded at room temperature using a EnSpire™ multilabel plate reader (PerkinElmer). The samples were excited at 350 nm, and emission was recorded in 370–650 nm range. The spectra were normalised to the same maximum value of emission intensity.2.12. Statistical methods
The statistical significance of measured biochemical parameters was evaluated with a two-tailed Student's t-test, relative to the control. The Pearson product-moment correlations and their associated p-values were used to explore the relationship between the growth inhibition and cell viability parameters. In both cases, P values of <0.05 were regarded as significant. All curve fittings were performed using GraphPad Prism software, version 7.0 (GraphPad, San Diego, CA, USA). All statistical analyses were carried out using Statistica™ 8.0 (Statsoft Inc., Tulsa, USA).3. Results and discussion
3.1. The characteristics of curcumin formulations
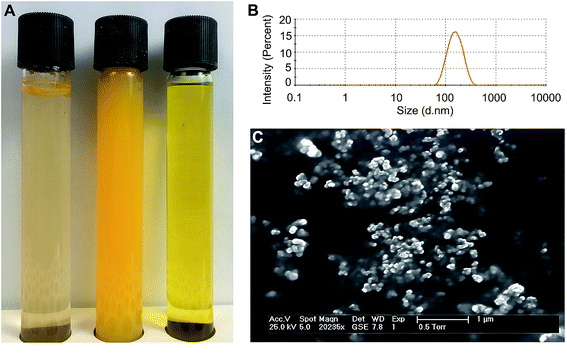
Both the CNP and the CCD formed stable aqueous dispersions with curcumin concentrations up to 1.2 mM. At the same curcumin concentration, the two formulations differed in their appearance (Fig. 2A). CNP dispersions were less transparent and had higher visual color intensity, presumably due to significant light scattering. Measurements by dynamic light scattering indicated that while dispersed in aqueous medium, CNP's had a hydrodynamic diameter of 146 ± 46 nm and narrow monomodal particle size distributions (PDI < 0.1) (Fig. 2B). Study by Doppler velocimetry showed that the particles had a positive ζ-potential of +42.6 ± 4.6 mV. Imaging by electron microscopy (Fig. 2C) revealed nanoparticles of roughly spherical shape with rather uniform sizes, though aggregated in course of freeze-drying.3.2. Cellular uptake of curcumin
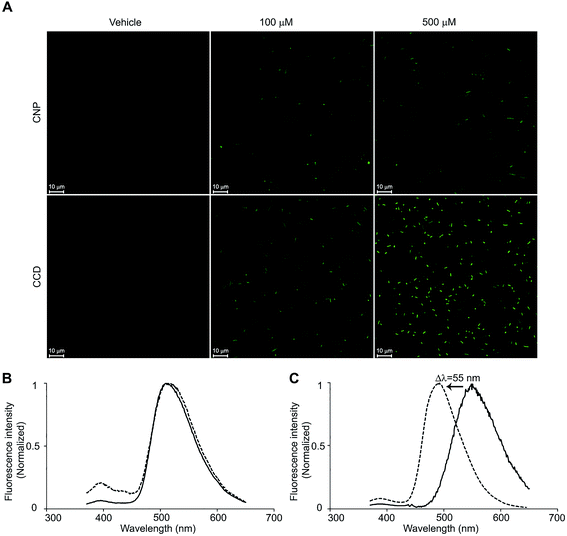
Curcumin fluorescence has been previously utilised for the registration of its uptake by human cells.20 Haukvik et al. used the fluorescence microscopy to investigate the curcumin delivery to bacterial cells but, due to resolution limitations, were unable to discern between the curcumin internalisation and adherence to the cell surfaces.21 We applied the confocal microscopic method to compare between the delivery efficiencies of CCD and CNP (Fig. 3A). The bacterial cells treated with CNP exhibited only weak fluorescent signals evident following prolonged exposure (above 8 hours), suggesting a rather slow delivery of curcumin from the nanoparticles to bacteria. This can be explained by slow, sustained release of curcumin from the nanoparticles that was observed in vitro (Fig. S2†), in a good agreement with previous reports.13 Peculiarly, the fluorescent signal of the bacteria treated with CNP remained unchanged with increasing CNP concentration. Though only limited quantitative information can be obtained from the microscopic experiments, this feature implies that curcumin released by the nanoparticles saturates its cellular binding sites and its cellular concentration reaches a plateau at low CNP concentrations. On the contrary, the bacteria treated with CCD showed significantly higher fluorescence detectable within minutes after the treatment. Furthermore, the fluorescent signal was progressively enhanced with increase in curcumin–MBCD concentration. Apparently, curcumin delivery capacity of CCD inclusion complex superseded that of the nanoparticles and possibly even followed a different path.In order to further investigate the difference in the delivery mechanisms between CCD and CNP we studied the cellular fate of both curcumin formulations. Small bacterial cell size and curcumin photobleaching preclude high-resolution microscopic determination of sub-cellular localization of curcumin. Thus, we exploited the environment-sensitive fluorescence of curcumin to address the question of subcellular localisation by means of fluorescence spectroscopy. The fluorescent emission wavelength of curcumin is known to be dependent on the polarity of the environment and to be shifting towards longer wavelengths with increasing solvent polarity.11 The fluorescence spectrum of bacterial cells treated with curcumin–CD closely coincided with the spectrum of the complex itself with an emission maximum at 511 nm (Fig. 3B) reflecting the curcumin situated in the hydrophobic inner core of cyclodextrin molecule and supporting the suggestion that CCD is taken-up by bacterial cells as a whole complex. In the case of curcumin nanoparticles, on the other hand, the fluorescence spectrum underwent a significant change with emission maximum shifting from 550 nm to 495 nm (Fig. 3C), close to that of curcumin in toluene (ε = 2.38) which argues for the curcumin partition from the nanoparticles into the more hydrophobic environment, probably the lipid bilayer, in line with recent reports showing that curcumin molecules in water-based dispersions undergo adsorption and partition to lipid bilayers.22 Therefore, it is reasonable to suggest that while curcumin upon the release from the CNP's undergoes partition into the bacterial membranes, CCD penetrates across the membranes and enters the cell.
The observed facilitation of curcumin delivery to the bacterial cells by cyclodextrin is in good agreement with previous reports showing that complexation with cyclodextrin facilitates cellular permeability and activity of numerous antimicrobial agents.9,23,24 Two possibly coexisting mechanisms, explaining this effect, have been proposed. The first mechanism is based on porins and other outer membrane proteins promoting the transport of drug–cyclodextrin complex through the membrane. First such system was discovered in Klebsiella oxytoca where CymA, a membrane-bound c-type cytochrome was demonstrated to function as a porin specific for cyclodextrins facilitating their uptake as intact entities.25,26 Such systems appear to be widespread among bacteria. Analogues to this system have been identified in Bacillus subtilis27 and in several other bacterial species including Escherichia coli.28,29 The outer-membrane proteins bearing structural similarity to CymA may enable penetration of cyclodextrin–drug complexes into bacterial cells. The second mechanism relies on the destabilisation and thus permeabilization of the outer membrane by cyclodextrin. Notably, it was established that cyclodextrin–drug complexes are capable of altering lipid packing density thus increasing the fluidity and permeability of membranes and the diffusion rate of permeating compounds.30–32 Both mechanisms are expected to contribute to an increased permeability rate of the complexed compounds, leading effectively to a more efficient delivery to their intracellular target sites and hence increasing their antimicrobial capacity. The observed results seem to suggest the existence of different delivery mechanisms for nanoparticles and molecular inclusion complexes: while molecular inclusion complexes efficiently deliver the curcumin to its molecular targets inside the bacterial cell, the nanoparticles act via releasing the curcumin into surrounding medium followed by its slow penetration inside the cell. This result emphasizes the importance of delivery systems capable of effective delivery of curcumin inside the bacterial cells for successful realization of its antimicrobial capacity.
3.3. Curcumin–CD complex is bactericidal while curcumin nanoparticles are bacteriostatic
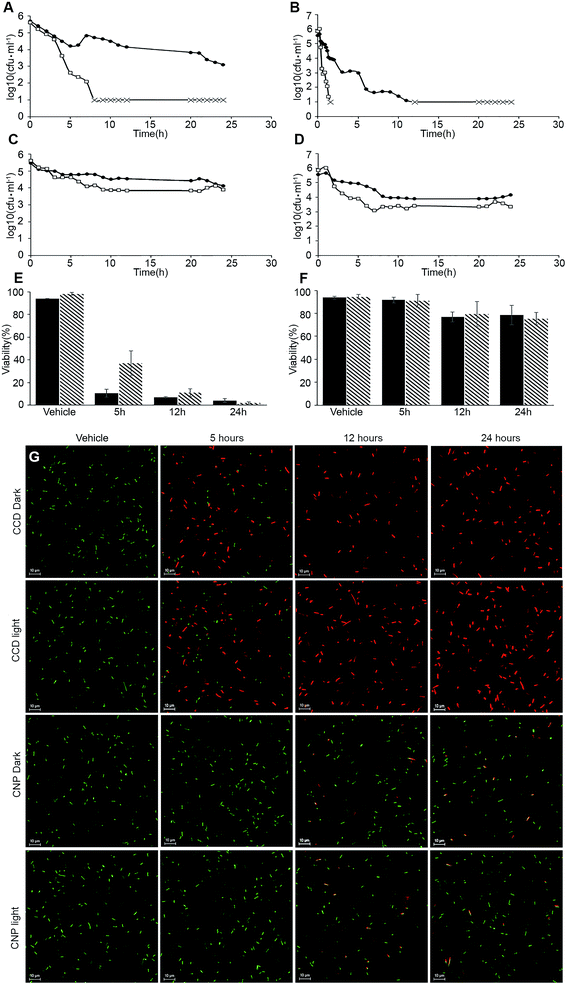
The changes in the viability of E. coli cells exposed to both curcumin formulations were assessed by two complementary methods: plating and live/dead staining (Fig. 4). Fig. 4A–D represents viable counts of E. coli exposed to CCD and CNP in MIC and >MIC curcumin doses, either in the dark or under the light. The MIC concentration of CCD in the dark (500 μM curcumin) reduced the viable CFU count by 2.6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log after 24 h treatment. Upon increasing the exposure to CCD at concentration approx. 1.3× MIC (625 μM) in the dark, the number of viable CFU was reduced by 3
log after 24 h treatment. Upon increasing the exposure to CCD at concentration approx. 1.3× MIC (625 μM) in the dark, the number of viable CFU was reduced by 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log (T99.9% 5 h) and proceeded to drop below the limit of detection after 8 h (Fig. 4A), with a very sharp transition from inhibitory to bactericidal concentration (MBC to MIC ratio of 1.25), suggesting a very potent bactericidal effect.33 In the good agreement with our previous results,10 illumination lowered MIC from 500 μM to 90 μM. Under the light, the MIC concentration appeared to be bactericidal (MIC to MBC ratio of 1) with T99.9% of 6 hours (Fig. 4B). The viable count dropped below the limit of detection after 12 hours upon the illumination (Fig. 4B). Moreover, the increase of CCD curcumin concentration from 90 μM to 120 μM (1.33× MIC) reduced the T99.9% from 6 h to just 1 h (Fig. 4B). The exposure to CNP, on the other hand, inhibited the proliferation of bacterial cells with only minor decrease in viability, suggesting the bacteriostatic rather than bactericidal action of the nanoparticles (Fig. 4C). The illumination had only limited effect on CNP toxicity towards bacteria (Fig. 4D). This pattern did not change significantly even at the highest concentration tested (2× MIC, 800 μM). Noteworthy, while the results of live/dead staining were in a good agreement with the culturability-based CFU counts for CCD (Fig. 4E and G), in the case of curcumin nanoparticles LIVE/DEAD assays proved to be significantly more conservative indicators of bacterial viability: while CFU-based method showed 1.7
log (T99.9% 5 h) and proceeded to drop below the limit of detection after 8 h (Fig. 4A), with a very sharp transition from inhibitory to bactericidal concentration (MBC to MIC ratio of 1.25), suggesting a very potent bactericidal effect.33 In the good agreement with our previous results,10 illumination lowered MIC from 500 μM to 90 μM. Under the light, the MIC concentration appeared to be bactericidal (MIC to MBC ratio of 1) with T99.9% of 6 hours (Fig. 4B). The viable count dropped below the limit of detection after 12 hours upon the illumination (Fig. 4B). Moreover, the increase of CCD curcumin concentration from 90 μM to 120 μM (1.33× MIC) reduced the T99.9% from 6 h to just 1 h (Fig. 4B). The exposure to CNP, on the other hand, inhibited the proliferation of bacterial cells with only minor decrease in viability, suggesting the bacteriostatic rather than bactericidal action of the nanoparticles (Fig. 4C). The illumination had only limited effect on CNP toxicity towards bacteria (Fig. 4D). This pattern did not change significantly even at the highest concentration tested (2× MIC, 800 μM). Noteworthy, while the results of live/dead staining were in a good agreement with the culturability-based CFU counts for CCD (Fig. 4E and G), in the case of curcumin nanoparticles LIVE/DEAD assays proved to be significantly more conservative indicators of bacterial viability: while CFU-based method showed 1.7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log (98%) reduction in the dark and 2.5
log (98%) reduction in the dark and 2.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log (99.7%) reduction upon illumination, the results of LIVE/DEAD assays showed only modest decrease in bacterial viability (21.5 ± 8.5% and 24.6 ± 5.4% in the dark and upon illumination, respectively) (Fig. 4F and G). These results suggest the presence of viable but non-culturable (VBNC) E. coli cells as a result of CNP treatment. The phenomenon of VBNC state is regarded as a survival strategy of bacteria in the face of adverse environmental conditions34 and is known to be triggered by a broad range of stimuli such as temperature shift, changes in nutrients, or pH, irradiation, desiccation, or exposure to antimicrobial substances.35 Recently, induction of VBNC state was demonstrated in human pathogen Pseudomonas aeruginosa, following exposure to silver nanoparticles.36 Further studies involving more indicators of the VBNC state are required to test the validity of this hypothesis. This observation emphasises the importance of using complementary viability evaluation techniques in the assessment of antimicrobial efficacy of nanomaterials.
log (99.7%) reduction upon illumination, the results of LIVE/DEAD assays showed only modest decrease in bacterial viability (21.5 ± 8.5% and 24.6 ± 5.4% in the dark and upon illumination, respectively) (Fig. 4F and G). These results suggest the presence of viable but non-culturable (VBNC) E. coli cells as a result of CNP treatment. The phenomenon of VBNC state is regarded as a survival strategy of bacteria in the face of adverse environmental conditions34 and is known to be triggered by a broad range of stimuli such as temperature shift, changes in nutrients, or pH, irradiation, desiccation, or exposure to antimicrobial substances.35 Recently, induction of VBNC state was demonstrated in human pathogen Pseudomonas aeruginosa, following exposure to silver nanoparticles.36 Further studies involving more indicators of the VBNC state are required to test the validity of this hypothesis. This observation emphasises the importance of using complementary viability evaluation techniques in the assessment of antimicrobial efficacy of nanomaterials.
It should be noted that one should not automatically assume the superiority of bactericidal formulation over a bacteriostatic one. At least in some cases, rapid bacterial killing by bactericidal agents may lead to some devastating consequences. Treatment of cultures of Pseudomonas aeruginosa with bactericidal antibiotics has been shown to increase the release of lipopolysaccharide, increasing the lethality in mice.37 Another notable example is endotoxin surge following the treatment with bactericidal antibiotics in infants with Gram-negative bacterial meningitis.38 The differences in antibacterial activity of CCD and CNP led us to hypothesise that these two curcumin formulations act through different toxicity mechanisms.
3.4. Two curcumin formulations exhibit different toxicity profiles
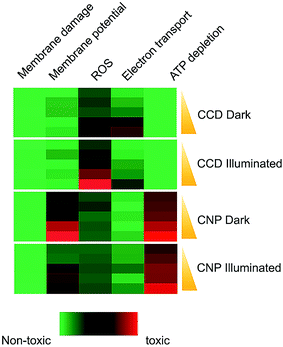
Studying the effects of CCD and CNP on five toxicity indicators: membrane damage, membrane depolarization, ROS generation, electron transport activity and ATP content (see the ESI and Methods for a detailed description†) allowed us comparing the antimicrobial mechanisms of the curcumin formulations. The outcome of this experiment is shown in the heat map in Fig. 5. Neither of formulations had an effect on membrane permeability at the concentrations studied herein. Although curcumin is known to cause membrane disruption,39 this mechanism takes place only at relatively high concentrations.40 The toxicity of CCD had shown a strong correlation (rdark = −0.95, p-value < 0.05; rlight = −0.96, p-value < 0.05) with the decrease in electron transport activity. The RedoxSensor™ Green reagent, employed in this assay, serves as an indicator of bacterial reductase activity which in turn depends upon NADH reducing equivalents. Therefore, this result is in agreement with recent evidence that E. coli cells attenuate curcumin toxicity by decreasing the NADH biosynthesis.10 Importantly, although certain increase in the level of oxidative stress (expressed in ROS generation) was observed in E. coli treated with CNPs both under dark and illuminated conditions it was not statistically significant (p-value > 0.1). By contrast, treatment with CCD led to oxidative stress which strongly correlated with the ROS production (r = 0.97, p-value < 0.01) in illuminated cells and with only statistically borderline correlation (r = 0.8, p-value > 0.1) in the dark. The data suggests that treatment with CCD followed by illumination with blue light initiates a particularly strong ROS response, even at low concentrations, and is consistent with prior reports that this formulation lead to greater cellular oxidative stress in E. coli.10 Weak ROS generation in the course of nanoparticle antimicrobial action is in a good agreement with previous works showing the lack of phototoxicity and ROS generation in slow-release curcumin nanoparticle formulations.41 This result is of importance for applications of curcumin-based nanomaterials in which ROS generation is undesirable, such as antimicrobial skin health products.Fig. 5 shows that the antimicrobial activity of CNP did not just represent a weakened CCD effect but was based on a peculiar mode of action. The two characteristic features of CNP toxicity were the severe disruption of membrane potential and depletion of cellular ATP pool. Under both light and dark regimes, the membrane depolarization correlated significantly with curcumin toxicity (rdark = 0.98, p-value < 0.01; rlight = 0.93, p-value < 0.05). The depletion of the ATP pool, also correlated significantly with CNP toxicity (rdark = 0.94, p-value < 0.05; rlight = 0.92, p-value < 0.05). Our finding that the membrane potential disruption and ATP biosynthesis, both membrane-related phenomena, are the most prominent predictors of CNP toxicity, suggests that membrane perturbation plays a central role in antibacterial mechanism of curcumin nanoparticles. Furthermore, both the membrane depolarization and ATP depletion might be nanoscale-related phenomena, rather than a result of direct curcumin action. Nanoparticles were shown to collapse the membrane potential by virtue of their interaction with a cell membrane.42 Moreover, the positive ζ-potential of the nanoparticles may play a role in the observed biological response. A considerable evidence indicates that bacterial viability and metabolic activity are affected by the interaction with a charged surfaces.43,44 The link between the surface charge and ATP level was established by Hong and Brown.45 These authors also proposed that when the bacterial cell interacts with a positively charged surface, this results in an increase in local pH at the cell surface and a corresponding drop in proton motive force and hence the reduced ATP levels. These two models are not mutually exclusive and it is possible that elements of both could be involved in the antimicrobial action of CNP's. Overall, the results of toxicity profiling highlight significant mechanistic differences in the pathways of toxicity between CNPs and CCDs and suggest the involvement of different antimicrobial mechanisms.
3.5. Prolonged exposure to CNPs triggers filamentation independently of SOS response
E. coli cells exposed for long time intervals (>12 h) to sub-MIC levels of the CNP both under dark and illuminated conditions formed long filaments, tens of microns in length and with considerable heterogeneity in size (Fig. 6B and D). Staining with DAPI followed by imagining with fluorescent confocal microscopy revealed that filaments contain numerous nucleoids, uniformly spread along the cell, a feature typical to filamentous phenotype in bacteria (Fig. 6E). Filamentous phenotype in bacteria46 appears when cytokinesis is not accompanying cell growth. Cytokinesis in most bacteria including E. coli is a carefully regulated process which begins with the assembly of the bacterial tubulin homolog FtsZ into a structure called the Z-ring which serves as a scaffold on top of which the cytokinesis apparatus is organized.47 A variety of stresses triggers a filamentous phenotype in bacteria. Most notable examples comprise exposure to antimicrobial compounds, damage to DNA and osmotic stress. In some instances, the septation is blocked by a direct poisoning of the cell division machinery or its components.48 Previously, curcumin was shown to bind FtsZ and inhibit its function, inducing filamentation in Bacillus subtilis.49 We note, however, that this is unlikely to be the case in the present study since the filamentation was observed only in the cells exposed to the CNP while the exposure to the same inhibitory concentration of CCD had no such effect (Fig. 6A and C).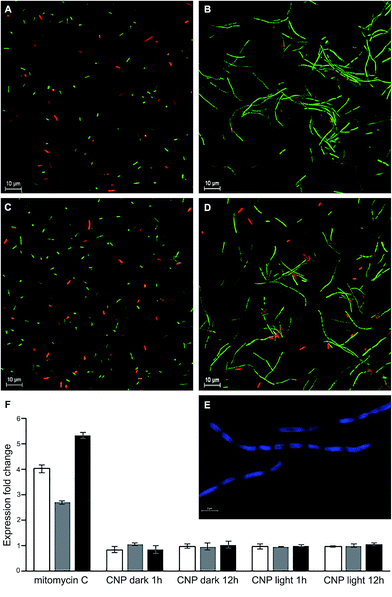 | ||
| Fig. 6 The SOS response-independent filamentation of E. coli triggered by CNP. (A–D) Representative fluorescence micrographs of E. coli cells after 12 h incubation with curcumin formulations in IC50 concentrations, stained with LIVE/DEAD assay. Treatments: (A) CCD, dark; (B) CNP, dark; (C) CCD, illuminated; (D) CNP, illuminated. This test was performed three times, and the results of a typical trial are shown. (E) Representative fluorescence micrograph of E. coli filaments stained with DAPI showing unsegregated nucleoids. (F) Expression of recA (white bars), lexA (grey bars) and SulA (black bars) determined by RT-qPCR in E. coli cells treated with CNP in the dark and upon illumination and with mitomycin C (a SOS response inducer used as a positive control). The figure presents only selected time points of the assay, after 1 and 12 h of incubation. For the full time-course study results see Fig. S2.† The error bars represent standard errors of three independent biological repetitions. | ||
The most prominent and well-studied mechanism of filamentation involves the inhibition of cytokinesis mediated by the SOS response system.18 This system is triggered by DNA damage (for example, due to ROS). The DNA damage results in the activation of RecA recombinase, which in turn activates the autoproteolysis of the LexA repressor, resulting in derepression of the SOS regulon. It is of paramount importance that while the SOS genes involved in DNA repair perform their task, the cytokinesis is put on hold, which prevents the transmission of damaged DNA to daughter cells. Thus, cell division inhibitor SulA (regulated by LexA) specifically inhibits polymerization of key division protein FtsZ leading to the formation of bacterial filaments. To assess the involvement of the abovementioned mechanism and in particular SulA in observed filamentous phenotype, we performed time-course qPCR expression studies of the recA, lexA and sulA expression in bacterial cells treated with CNPs. Mitomycin C, a known inducer of SOS response induced the expression of recA, lexA and sulA by 4.1, 2.7 and 5.3-fold respectively (Fig. 6F). The transcript levels of all three genes studied remained unchanged through the course of CNP treatment (Fig. 6F, S3a and b†) arguing against the involvement of SOS response in filamentation and suggesting that it was a result of some other mechanism. An alternative explanation might be that the filamentous phenotype was due to the envelope stress response induced by the CNP. Since membrane depolarization was the distinctive attribute of CNP toxicity towards the bacteria, it is tempting to speculate that this treatment caused further damage to envelope integrity thus generating an envelope stress response which is known to produce the filamentous phenotype.50 However, the complexity of this stress response involving multiple signalling pathways51 justifies a separate study.
4. Conclusions
The study reported here provides a strong evidence that mode of biological activity of curcumin depends on the properties of the delivery system. The curcumin–MBCD complexes not only showed higher antimicrobial potency than surface-stabilized monolithic curcumin nanoparticles but exhibited essentially different mechanisms of toxicity. While the increase in ROS generation and a decrease in electron transport activity were the best predictors of CCD antimicrobial effect, treatment with CNP depolarised the membranes and reduced cellular ATP concentrations. Importantly, we found that while curcumin–MBCD complexes exerted a bactericidal effect on E. coli, the curcumin nanoparticles had bacteriostatic effect only. Use of culture-independent methods to assess the bacterial viability allowed us to find the evidence of induction of the VBNC state in a nonpathogenic strain of E. coli ATCC 25922 by curcumin nanoparticles but not by the inclusion complex. To the best of our knowledge, this is the first study showing that the same active principle can exert its antimicrobial effect though completely different mechanisms depending on delivery system used. In the future studies, it would be interesting to expand the investigation to biological activities of other curcumin nanoformulations differing in particle characteristics, structure and composition. The results of this study should be considered in the development of antibacterial nanomaterials and evaluation of their efficacy and will facilitate the rational selection of delivery systems designed to enhance the antimicrobial properties of new materials.Conflicts of interest
There are no conflicts of interests to declare.Acknowledgements
This research was supported by the US-Israel Binational Agricultural Research and Development (BARD) Grant US-4680-13C. Contribution from the Agricultural Research Organization, the Volcani Center, Rishon LeZion, Israel, No. 779/17. The authors wish to express their gratitude to Eduard Belausov (Volcani Center Microscopy unit) for the assistance with confocal microscopy.References
- J. Lutomski, B. Kedzia and W. Debska, Planta Med., 1974, 26, 9–19 CrossRef CAS PubMed.
- R. E. W. Hancock, Trends Microbiol., 1997, 5, 37–42 CrossRef CAS PubMed.
- H. Nikaido, Microbiol. Mol. Biol. Rev., 2003, 67, 593–656 CrossRef CAS PubMed.
- A. B. Hegge, E. Bruzell, S. Kristensen and H. H. Tønnesen, Eur. J. Pharm. Sci., 2012, 47, 65–74 CrossRef CAS PubMed.
- H. H. Tønnesen, H. de Vries, J. Karlsen and G. B. Van Henegouwen, J. Pharm. Sci., 1987, 76, 371–373 CrossRef.
- T. A. Dahl, W. M. Mcgowan, M. A. Shand and V. S. Srinivasan, Arch. Microbiol., 1989, 151, 183–185 CrossRef CAS PubMed.
- M. M. Yallapu, P. K. B. Nagesh, M. Jaggi and S. C. Chauhan, AAPS J., 2015, 17, 1341–1356 CrossRef CAS PubMed.
- T. Loftsson and D. Duchêne, Int. J. Pharm., 2007, 329, 1–11 CrossRef CAS PubMed.
- G. Athanassiou, S. Michaleas, E. Lada-Chitiroglou, T. Tsitsa and E. Antoniadou-Vyza, J. Pharm. Pharmacol., 2003, 55, 291–300 CrossRef CAS PubMed.
- I. Shlar, S. Droby and V. Rodov, J. Proteomics, 2017, 160, 8–20 CrossRef CAS PubMed.
- N. Dogra, R. Choudhary, P. Kohli, J. D. Haddock, S. Makwana, B. Horev, Y. Vinokur, S. Droby and V. Rodov, J. Agric. Food Chem., 2015, 63, 2557–2565 CrossRef CAS PubMed.
- M. M. Yallapu, M. Jaggi and S. C. Chauhan, Curr. Pharm. Des., 2013, 19, 1994–2010 CAS.
- Z. Zheng, X. Zhang, D. Carbo, C. Clark, C. A. Nathan and Y. Lvov, Langmuir, 2010, 26, 7679–7681 CrossRef CAS PubMed.
- I. Shlar, E. Poverenov, Y. Vinokur, B. Horev, S. Droby and V. Rodov, Nano-Micro Lett., 2015, 7, 68–79 CrossRef CAS.
- L. Zou, B. Zheng, R. Zhang, Z. Zhang, W. Liu, C. Liu, H. Xiao and D. J. McClements, RSC Adv., 2016, 6, 3126–3136 RSC.
- R. J. Hunter, in Zeta Potential in Colloid Science, 1981, pp. 11–58 Search PubMed.
- C. Kaweeteerawat, C. H. Chang, K. R. Roy, R. Liu, R. Li, D. Toso, H. Fischer, O. A. Ivask, Z. Ji, J. I. Zink, Z. H. Zhou, Â. G. F. Chanfreau, D. Telesca, O. Y. Cohen, P. A. Holden, A. E. Nel and H. A. Godwin, ACS Nano, 2015, 9, 7215–7225 CrossRef CAS PubMed.
- É. Guerin, G. Cambray, N. Sanchez-Alberola, S. Campoy, I. Erill, S. Da Re, B. Gonzalez-Zorn, J. Barbé, M.-C. Ploy and D. Mazel, Science, 2009, 324, 1034 CrossRef PubMed.
- M. W. Pfaffl, Nucleic Acids Res., 2001, 29, e45 CrossRef CAS PubMed.
- M. M. Yallapu, M. Jaggi and S. C. Chauhan, Colloids Surf., B, 2010, 79, 113–125 CrossRef CAS PubMed.
- T. Haukvik, E. Bruzell, S. Kristensen and H. H. Tønnesen, Pharmazie, 2009, 64, 666–673 CAS.
- H. I. Ingólfsson, P. Thakur, K. F. Herold, E. A. Hobart, N. B. Ramsey, X. Periole, D. H. De Jong, M. Zwama, D. Yilmaz, K. Hall, T. Maretzky, H. C. Hemmings, C. Blobel, S. J. Marrink, A. Koçer, J. T. Sack and O. S. Andersen, ACS Chem. Biol., 2014, 9, 1788–1798 CrossRef PubMed.
- A. Zuorro, M. Fidaleo and R. Lavecchia, Bull. Korean Chem. Soc., 2010, 31, 3460–3462 CrossRef CAS.
- S. Bhargava and G. P. Agrawal, Curr. Drug Delivery, 2008, 5, 1–6 CrossRef CAS.
- M. Pajatsch, M. Gerhart, R. Peist, R. Horlacher, W. Boos and A. Böck, J. Bacteriol., 1998, 180, 2630–2635 CAS.
- M. Pajatsch, C. Andersen, A. Mathes, A. Böck, R. Benz and H. Engelhardt, J. Biol. Chem., 1999, 274, 25159–25166 CrossRef CAS PubMed.
- A. Kamionka and M. K. Dahl, FEMS Microbiol. Lett., 2001, 204, 55–60 CrossRef CAS PubMed.
- C. Baldermann, A. Lupas and H. E. J. Lubienecki, J. Bacteriol., 1998, 180, 3741–3749 CAS.
- D. A. Fajardo, J. Cheung, C. Ito, E. Sugawara, H. Nikaido and R. Misra, J. Bacteriol., 1998, 180, 4452–4459 CAS.
- F. Casterlli, G. Puglisi, R. Pignatello and S. Gurrieri, Int. J. Pharm., 1989, 52, 115–121 CrossRef.
- T. Mavromoustakos, A. Papadopoulos, E. Theodoropoulou, C. Dimitriou and E. Antoniadou-Vyza, Life Sci., 1998, 62, 1901–1910 CrossRef CAS PubMed.
- E. Antoniadou-Vyza, P. Tsitsa, E. Theodoropoulou and T. Mavromoustakos, J. Inclusion Phenom. Mol. Recognit. Chem., 1996, 25, 161–164 CrossRef CAS.
- G. A. Pankey and L. D. Sabath, Clin. Infect. Dis., 2004, 38, 864–870 CrossRef CAS PubMed.
- L. Li, N. Mendis, H. Trigui, J. D. Oliver and S. P. Faucher, Front. Microbiol., 2014, 5, 1 Search PubMed.
- J. D. Oliver, J. Microbiol., 2005, 43, 93–100 Search PubMed.
- A. M. Königs, H. C. Flemming and J. Wingender, Front. Microbiol., 2015, 6, 1–11 Search PubMed.
- J. J. Jackson and H. Kropp, J. Infect. Dis., 1992, 165, 1033–1041 CrossRef CAS PubMed.
- M. M. Mustafa, J. Mertsola, O. Ramilo, X. Sáez-Llorens, R. C. Risser and G. H. McCracken, J. Infect. Dis., 1989, 160, 891–895 CrossRef CAS PubMed.
- P. Tyagi, M. Singh, H. Kumari, A. Kumari and K. Mukhopadhyay, PLoS One, 2015, 10, 1–15 Search PubMed.
- D. G. Yun and D. G. Lee, Appl. Microbiol. Biotechnol., 2016, 100, 5505–5514 CrossRef CAS PubMed.
- D. Chopra, L. Ray, A. Dwivedi, S. K. Tiwari, J. Singh, K. P. Singh, H. N. Kushwaha, S. Jahan, A. Pandey, S. K. Gupta, R. K. Chaturvedi, A. B. Pant, R. S. Ray and K. C. Gupta, Biomaterials, 2016, 84, 25–41 CrossRef CAS PubMed.
- Y. Cui, Y. Zhao, Y. Tian, W. Zhang, X. Lü and X. Jiang, Biomaterials, 2012, 33, 2327–2333 CrossRef CAS PubMed.
- B. Gottenbos, D. W. Grijpma, H. C. van der Mei, J. Feijen and H. J. Busscher, J. Antimicrob. Chemother., 2001, 48, 7–13 CrossRef CAS PubMed.
- A. Terada, A. Yuasa, T. Kushimoto, S. Tsuneda, A. Katakai and M. Tamada, Microbiology, 2006, 152, 3575–3583 CrossRef CAS PubMed.
- Y. Hong and D. G. Brown, Appl. Environ. Microbiol., 2009, 75, 2346–2353 CrossRef CAS PubMed.
- S. S. Justice, D. A. Hunstad, L. Cegelski and S. J. Hultgren, Nat. Rev. Microbiol., 2008, 6, 162–168 CrossRef CAS PubMed.
- D. W. Adams and J. Errington, Nat. Rev. Microbiol., 2009, 7, 642–653 CrossRef CAS PubMed.
- J. M. Boberek, J. Stach and L. Good, PLoS One, 2010, 5, 1–9 Search PubMed.
- D. Rai, J. K. Singh, N. Roy and D. Panda, Biochem. J., 2008, 410, 147–155 CrossRef CAS PubMed.
- S. S. Yadavalli, J. N. Carey, R. S. Leibman, A. I. Chen, A. M. Stern, M. Roggiani, A. M. Lippa and M. Goulian, Nat. Commun., 2016, 7, 12340 CrossRef CAS PubMed.
- G. Rowley, M. Spector, J. Kormanec and M. Roberts, Nat. Rev. Microbiol., 2006, 4, 383–394 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra07303h |
| This journal is © The Royal Society of Chemistry 2017 |