Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Utilization of chromic polydiacetylene assemblies as a platform to probe specific binding between drug and RNA†
Anothai Kamphanab,
Changjun Gongc,
Krishnagopal Maitic,
Souvik Surc,
Rakchart Traiphol*abd and
Dev P. Arya *c
*c
aDepartment of Chemistry, Faculty of Science, Naresuan University, Phitsanulok 65000, Thailand
bLaboratory of Advanced Polymers and Nanomaterials, School of Materials Science and Engineering, Center of Excellence for Innovation in Chemistry, Faculty of Science, Mahidol University at Salaya, Phuttamonthon 4 Road, Salaya, Nakhon Pathom 73170, Thailand
cLaboratories of Medicinal Chemistry, Department of Chemistry, Clemson University, Clemson, South Carolina 29634, USA. E-mail: dparya@clemson.edu
dNANOTEC-MU Excellence Center on Intelligent Materials and Systems, Faculty of Science, Mahidol University, Rama 6 Road, Ratchathewi, Bangkok 10400, Thailand. E-mail: rakchart.tra@mahidol.ac.th
First published on 24th August 2017
Abstract
Recognition of nucleic acids remains an important endeavor in biology. Nucleic acids adopt shapes ranging from A-form (RNA and GC rich DNA) to B-form (AT rich DNA). We show, in this contribution, shape-specific recognition of A–U rich RNA duplex by a neomycin (Neo)–polydiacetylene (PDA) complex. PDA assemblies are fabricated by using a well-known diacetylene (DA) monomer, 10,12-pentacosadiynoic acid (PCDA). The response of poly(PCDA) assemblies is generated by mixing with a modified neomycin–PCDA monomer (Neo–PCDA). The functionalization by neomycin moiety provides specific binding with homopolyribonucleotide poly(rA)–poly(rU) stimulus. Various types of alcohols are utilized as additives to enhance the sensitivity of poly(PCDA)/Neo–PCDA assemblies. A change of absorption spectra is clearly observed when a relatively low concentration of poly(rA)–poly(rU) is added into the system. Furthermore, poly(PCDA)/Neo–PCDA shows a clear specificity for poly(rA)–poly(rU) over the corresponding DNA duplex. The variation of linker between neomycin moiety and conjugated PDA backbone is found to significantly affect its sensitivity. We also investigate other parameters including the concentration of Neo–PCDA and the DA monomer structure. Our results provide here preliminary data for an alternative approach to improve the sensitivity of PDA utilized in biosensing and diagnostic applications.
1. Introduction
A number of antibiotics that stop the growth of foreign pathogens, such as bacteria and viruses, are known to bind nucleic acids such as DNA and RNA. One such class of antibiotics are aminoglycosides, well known to bind bacterial ribosomal RNA.1 Neomycin is one such antibiotic, belonging to aminoglycosides containing the deoxystreptamine core. The binding of neomycin, and other aminoglycosides drugs, to bacterial RNA affects the production of proteins through miscoding in translation, ultimately leading to bacterial death.2–5 In addition to binding to ribosomal RNA, aminoglycosides have been shown to bind numerous other RNA,6–10 DNA,11–17 hybrid duplex,18,19 triplex20–23 and quadruplex24–26 structures of therapeutic interest.27Conventional methods for the study of neomycin drug–RNA interactions utilize established analytical tools. For example, circular dichroism (CD) spectroscopy is used to study conformational changes of RNA during ligand–RNA binding.11,28 Isothermal titration calorimetry (ITC), differential scanning calorimetry (DSC) and UV-vis spectroscopy are used to elucidate the thermodynamic parameters of ligand–RNA interactions.11,28 Fluorescence intercalator displacement (FID) assays can be used for the study of selectivity, affinity, stoichiometry and binding site size of RNA and ligand.11,28 New and simple approaches that can help us study small molecule's interaction with biopolymers would be welcome addition to these established rigorous methods of ligand–nucleic acids interactions.
Polydiacetylene (PDA) is a conjugated polymer, which has received tremendous attention as a chromatic sensor. PDA can be prepared via 1,4-addition of highly organized assemblies of diacetylene monomers, normally initiated by UV light irradiation.29,30 PDA possesses color transition, typically from blue to red, upon the exposure to various external stimuli such as heat,31–42 acids and bases,40,43–46 chemicals47–56 and biomolecules.57–66 The perturbation by these stimuli generally weakens segmental interactions within the PDA assemblies. This results in the reorientation of conjugated backbone and alkyl chain. The decrease of conjugation length of PDA provides the shift of absorption spectrum to lower energy region and hence induces color transition.30,67–70 Many prior reports demonstrate the ability of PDA-based biosensors for the detection of viral RNA with quick and reliable testing without requiring advanced instruments. Amano et al. reviewed the construction of PDA with desirable physical, optical and electrical properties that responds to the influenza virus's RNA. The exposure of influenza virus to modified-PDA solution causes blue shift in the absorption spectra, corresponding to blue-to-red color transition of PDA.71 Park et al. demonstrates the detection of RNA by using PDA conjugated with DNA complementary to RNA of pathogenic bacteria. The hybridization of a specific DNA to RNA enhances fluorescent intensity of PDA-based sensor, leading to the application as a biochip in the detection of pathogenic microorganisms.72 In this report, we demonstrate the use of PDA in a modified approach for the investigation of RNA–ligand interactions.
Here, PCDA modified neomycin head groups (Neo–PCDA) is synthesized to generate a specific binding site for RNA. Previous calorimetry and FID studies have shown that the binding of poly(rA)–poly(rU), a homopolymeric dsRNA, with neomycin has high Ka values (∼107 M−1), and low AC50 (0.02 ± 0.01 μM) respectively, representing high affinities.11,14 These studies are in concert with other data showing nanomolar binding of neomycin to RNA targets. Therefore, poly(rA)–poly(rU) was used here as a model RNA for investigating the neomycin–RNA interaction. We also demonstrate here that the sensitivity of poly(PCDA)/Neo–PCDA assemblies can be significantly improved by using alcohol additives. An addition of alcohols weakens the strength of interactions within the poly(PCDA)/Neo–PCDA assemblies resulting in the increase of colorimetric response compared to its pure system.73 The developments of specific and rapid color transition of PDA to biomolecules could open wide-range of possibilities for biosensor, drug detection, and diagnostic applications.
2. Experiment
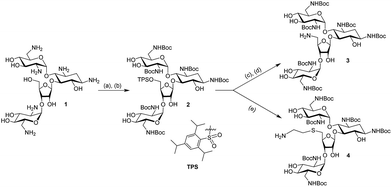
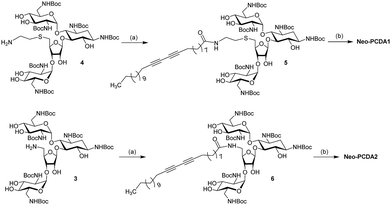
2.1 The synthesis of Neo–PCDA1 and Neo–PCDA2
The chemical structures of neomycin Neo–PCDA1 and Neo–PCDA2 are shown in Fig. 1. Neo–PCDA1 and Neo–PCDA2 were synthesized as shown in Schemes 1 and 2. Boc-protected neomycin amine derivatives 3 and 4 were prepared from commercially available neomycin B (1) according to published schemes (Scheme 1).11,14,22,74–76 All the compounds were characterized and spectral data was matched with the reported values from the literatures. Amine functionalized neomycin derivatives 3 and 4 were coupled with 10,12-pentacosadiynoic acid (PCDA) monomer (Aldrich) in the presence of coupling reagent O-(Benzotriazol-1-yl)-N,N,N′,N′-tetramethyluronium tetrafluoroborate (TBTU) in DMF solution, followed by deprotection with trifluoroacetic acid in dichloromethane, leading to neomycin-PCDA conjugates, Neo–PCDA1 and Neo–PCDA2 (Scheme 2). All the four new compounds 5, 6, Neo–PCDA1 and Neo–PCDA2 were characterized by nuclear magnetic resonance spectroscopy (NMR, Bruker Advance-500 and 300) and MALDI-TOF mass spectrometry (Bruker Microflex) and the spectra are provided in the ESI.† One of the α-anomeric protons of two pyranoside units in 5, appeared as a singlet at 5.38 ppm whereas the β-anomeric proton of furanoside unit resonates at 5.15 ppm as a singlet, in 1H NMR spectrum of 5 (Fig. S2†). The peak at 99.09 and 100.52 ppm corresponded to α-anomeric carbon of two pyranoside moieties whereas β-anomeric carbon of furanoside moiety appeared at 111.08 in 13C NMR spectrum of 5 (Fig. S3†). Further, the formation of the compound 5 was confirmed by the appearance of molecular ion peak at 1653.268 [M + Na]+, as the base peak in MALDI-TOF mass spectrum of 5 (Fig. S1†). In 1H NMR spectrum of 6 (Fig. S8†), β-anomeric proton of the furanoside moiety appeared as a singlet at 5.08 ppm whereas the corresponding anomeric carbon resonated at 111.48 ppm, in 13C NMR spectrum of 6 (Fig. S9†). The anomeric peaks of compound 6 were assigned by the help of HMQC spectrum (Fig. S10†). The formation of compound 6 was further confirmed by MALDI-TOF mass spectrum (Fig. S7†), where the molecular ion peak appeared as the base peak at 1593.065 [M + Na]+.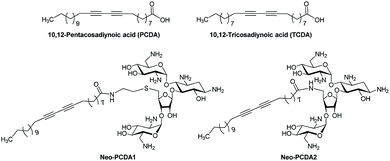 | ||
| Fig. 1 Chemical structures of diacetylene monomer, 10,12-pentacosadiynoic acid (PCDA) and 10,12-tricosadiynoic acid (TCDA); and synthesized neomycin–PCDA conjugates, Neo–PCDA1 and Neo–PCDA2. | ||
The details of the synthetic procedures for compounds 5, 6, Neo–PCDA1 and Neo–PCDA2 are given below.
2.2 The preparation of PDA/Neo–PCDA assemblies
The assemblies of PDA/Neo–PCDA were prepared by using two DA monomers, including 10,12-pentacosadiynoic acid (PCDA) and 10,12-tricosadiynoic acid (TCDA). DA monomer and Neo–PCDA powder were dissolved in ethanol and then filtered by using a 0.45 μm nylon filter to remove polymerized impurities. The ratios of Neo–PCDA were 0, 5, 10, 15 and 20 mol%. The DA/Neo–PCDA solutions in ethanol were slowly dried at 60 °C in a water bath. Phosphate-buffered saline solutions (PBS buffer) at various pH values (6, 7, and 8) were added into the DA/Neo–PCDA films to provide 0.25 mM aqueous suspension. The pH of PBS buffer was adjusted by using HCl and NaOH solutions. The samples were sonicated at 70 °C for 90 min to disperse DA/Neo–PCDA films into aqueous medium. The suspensions were allowed to equilibrate at room temperature and then stored at 4 °C overnight to induce self-assembly process. The cloudy suspensions of DA/Neo–PCDA were irradiated under UV light (10 W, λ ∼ 254 nm) for 5 min, resulting in blue PDA/Neo–PCDA vesicles. Effect of Neo–PCDA structure was studied by using Neo–PCDA1 and Neo–PCDA2 prepared by the same method.2.3 The study of colorimetric response of PDA/Neo–PCDA to poly(rA)–poly(rU)
Polyadenylic–uridylic acid (Poly(rA)–poly(rU)) was purchased from Midland Certified Company. Tuning colorimetric response of PDA/Neo–PCDA to poly(rA)–poly(rU) was induced by using two alcohols, butanol and hexanol. The alcohols were added into PDA/Neo–PCDA solution at various concentrations by using a micropipette. After the addition of alcohols, the solution was shaken by using vortex for a few seconds. Poly(rA)–poly(rU) solution in PBS buffer (pH 8) was added into PDA/Neo–PCDA solution at various concentrations by using a micropipette. The mixture was shaken for a few seconds prior to each addition. The color transition of PDA/Neo–PCDA assemblies was followed by measuring the absorption spectra (UV-vis spectrophotometer, Agilent technologies, Cary series). The concentration of PDA/Neo–PCDA assemblies in each experiment was controlled to be the same by adjusting the absorbance value. To quantify the extent of blue-to-red color transition of PDA/Neo–PCDA assemblies, the colorimetric response (% CR) was defined and calculated as follows: % CR = [(PB0 − PB)/PB0] × 100. The PB is the percent blue calculated from A640/(A540 + A640), where A540 and A640 are the absorbance of red (λ = 540) and blue (λ = 640) phases of vesicles, respectively. The initial PB0 value was determined before the addition of poly(rA)–poly(rU). Morphology of these solutions was also studied by using transmission electron microscope (TEM, Hitachi H7600). The samples were prepared by dropping diluted PDA/Neo–PCDA solutions onto copper grids, followed by air-drying at room temperature.The specificity of PDA/Neo–PCDA was study by testing with a control DNA sequence containing only adenine and thymine bases. The poly(PCDA)/Neo–PCDA1 containing 0.5% v/v hexanol was used. The solution of control DNA in PBS buffer (pH 8) was added into poly(PCDA)/Neo–PCDA1 solution by using the same testing process as poly(rA)–poly(rU) system. The colorimetric response of poly(PCDA)/Neo–PCDA1 vesicles was followed by measuring the absorption spectra.
3. Results and discussion
3.1 Morphologies of poly(PCDA)/Neo–PCDA assemblies and the optimization
Pure poly(PCDA), poly(PCDA)/Neo–PCDA1 and poly(PCDA)/Neo–PCDA2 solutions show a blue color, indicating proper packing of PCDA monomers required for the topopolymerization process. Morphologies of resultant poly(PCDA)/Neo–PCDA assemblies revealed by TEM are shown in Fig. 2 (bottom). A model of the Neo–PCDA compounds bound to RNA is also included in Fig. 2 (top). It has been observed that pure poly(PCDA) shows spherical shape with featureless internal pattern.21 The TEM image of poly(PCDA)/Neo–PCDA1 also shows spherical particles with diameter of about 100 nm (see Fig. 2a). In contrast, poly(PCDA)/Neo–PCDA2 assemblies exhibit relatively large rod-liked and sheet-liked shapes as shown in Fig. 2b. This is attributed to the difference of chemical linker between PCDA and neomycin head group (see Fig. 1). The Neo–PCDA2 constitutes much shorter linker compared to that of the Neo–PCDA1, which in turn significantly affects their packing structure. Neo–PCDA2 has a shorter linker compared to Neo–PCDA1 (see Fig. 1), resulting in a larger head group repulsion. This provides the increase of head group area and the decrease in the packing parameter, leading to the differences in molecular organization and ultimately different shape and size of poly(PCDA)/Neo–PCDA assembly.39,77,78 Jung,66,79 Cheng80 and Ye81 also demonstrate that the variation of head group structure provides different shape and size of PDA assemblies. Flower-like morphology, rectangular shape, fiber, ribbon with various size in the range of nanometer to micrometer are observed. It is important to note that the differences of shape and size of PDA significantly affect its colorimetric response as well. This will be discussed in the following section.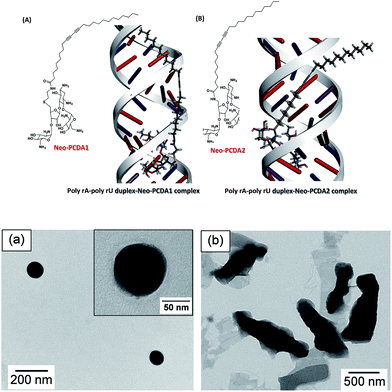 | ||
| Fig. 2 In silico models (above) and TEM images (below) of poly(PCDA) prepared by mixing with 10 mol% of (a) Neo–PCDA1 and (b) Neo–PCDA2. | ||
To achieve the highest response of poly(PCDA)/Neo–PCDA assemblies to poly(rA)–poly(rU), concentration of Neo–PCDA and pH of PBS buffer were optimized. We observed that the poly(PCDA)/Neo–PCDA1 assemblies containing 20 mol% of Neo–PCDA1 at pH 8 provided the highest response to poly(rA)–poly(rU) (see Fig. S14 in ESI†). RNA-binding affinity of an aminoglycoside is known to vary with the change in pH, as the protonated states of the amino groups are affected.82 However, this system exhibits large precipitation, which is clearly undesirable for sensing applications. Therefore, the highest possible concentration of Neo–PCDA, 10 mol%, providing clear blue solution and no precipitation of the assemblies, was used for the entire study.
3.2 Colorimetric response of poly(PCDA)/Neo–PCDA1 to poly(rA)–poly(rU)
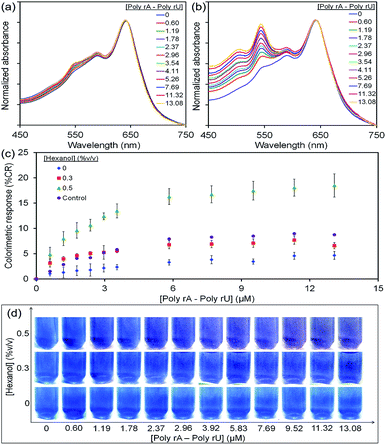
Poly(PCDA)/Neo–PCDA1 assemblies were prepared by mixing of PCDA and 10 mol% Neo–PCDA1 in PBS buffer at pH 8. Initial absorption spectrum of poly(PCDA)/Neo–PCDA1 exhibits λmax at ∼640 nm. The spectrum hardly changes after the addition of poly(rA)–poly(rU) up to ∼13 μM (see Fig. 3a). Interestingly, the colorimetric response of poly(PCDA)/Neo–PCDA1 drastically increases when a small amount of hexanol, 0.5% v/v, is added into the system. Absorption peak at ∼540 nm grows significantly upon the addition of poly(rA)–poly(rU) (see Fig. 3b), indicating the formation of red poly(PCDA)/Neo–PCDA1. A blue-to-purple color transition of poly(PCDA)/Neo–PCDA1 can be observed as shown in Fig. 3d. According to our previous studies, the added hexanol can penetrate into the poly(PCDA)/Neo–PCDA1 layers which in turn weakens inter-/intrachain interactions within the assemblies.51,52,73 This perturbation enhances colorimetric response of poly(PCDA)/Neo–PCDA1 to poly(rA)–poly(rU).The study of its morphology by TEM supports our hypothesis. The morphology of poly(PCDA)/Neo–PCDA1 changes from spherical shape to sheet-like structure after the addition of 0.5% v/v hexanol as shown in Fig. 4a. This observation suggests that the presence of hexanol induces segmental reorganization within the assemblies, enhancing the colorimetric response to poly(rA)–poly(rU). We also observe that the perturbation by poly(rA)–poly(rU) further induces the agglomeration of poly(PCDA)/Neo–PCDA1 (see Fig. 4b). It is worthwhile to note that the morphologies revealed by TEM could be different from those of PDA assemblies in the solution state due to the sample preparation process. However, the change of morphology in dried state still reflects the properties of PDA assemblies in solution.
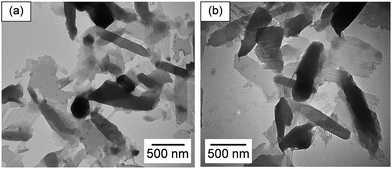 | ||
| Fig. 4 TEM images of poly(PCDA)/Neo–PCDA1 (a) with the addition of 0.5% v/v hexanol and then (b) was disturbed by the addition of 9.52 μM poly(rA)–poly(rU). | ||
The plots of % CR clearly show that the increase of hexanol concentration systematically enhances poly(PCDA)/Neo–PCDA1 response to poly(rA)–poly(rU) (see Fig. 3c). The higher amounts of hexanol causes increased perturbation of poly(PCDA)/Neo–PCDA1 assemblies,51,52,73 resulting in more blue-to-purple color transition as shown in Fig. 3d. However, the further increase of hexanol concentration results in a cloudy suspension arising from its limited solubility in water.83 A control set of poly(PCDA)/Neo–PCDA1 assemblies containing 0.5% v/v hexanol was also investigated. The absorption spectra of this solution were measured at the same time interval with the measurement of other solutions without the addition of poly(rA)–poly(rU) solution. This system shows a slight increase of CR value to ∼8% while the addition of poly(rA)–poly(rU) provides the CR value of ∼20% as shown in Fig. 3c. The difference of these two sets indicates the color transition of poly(PCDA)/Neo–PCDA1 induced by interaction between Neo–PCDA1 and poly(rA)–poly(rU).
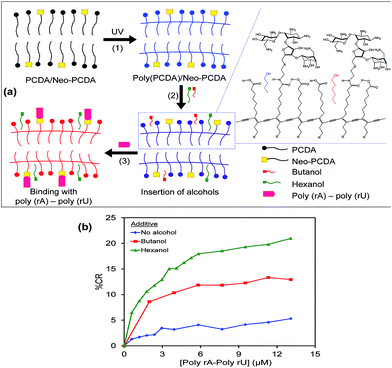
Schematic illustration in Fig. 5a concludes our study in this first part. The self-assembling of PCDA and Neo–PCDA monomers takes place in a solution state. A blue phase of poly(PCDA)/Neo–PCDA assemblies is obtained via photopolymerization process. The insertion of alcohols into poly(PCDA)/Neo–PCDA layers weakens the interactions within the system, which in turn enhances the sensitivity. The specific binding between neomycin moiety and poly(rA)–poly(rU) induces the color transition. In this study, butanol was also used as an additive in order to investigate the effect of alcohol alkyl tail on the colorimetric response of poly(PCDA)/Neo–PCDA1. The highest concentration of butanol, 5% v/v, was added into poly(PCDA)/Neo–PCDA1 while the suspension still remained blue. We observe that the addition of butanol provides lower response of poly(PCDA)/Neo–PCDA1 compared to the use of hexanol (see Fig. 5b). Although the added concentration of butanol is 10 times higher than that of hexanol, the % CR reaches plateau at about 12% upon increasing concentration of poly(rA)–poly(rU) to ∼13 μM. Our results suggests that hexanol with relatively long chain can penetrate into deeper region of poly(PCDA) layer. This induces larger extent of perturbation (see model in Fig. 5a), providing higher colorimetric response.51,73 It is possible to use other types of alcohols as additives to enhance the sensitivity of PDA. The effects of alcohol structures on colorimetric response of PDAs are previously reported by our group.51,73 It is worthwhile to note that a phospholipid (dimyristoylphosphatidylcholine, DMPC) is normally used for improving sensitivity of PDA assemblies to biomolecules in previous studies.53,60,61 We demonstrate in this study that alcohol is an interesting additive for enhancing the sensitivity of PDA to biomolecules. The alcohol is also much cheaper compared to the DMPC.
3.3 The specificity of poly(PCDA)/Neo–PCDA1 to poly(rA)–poly(rU)
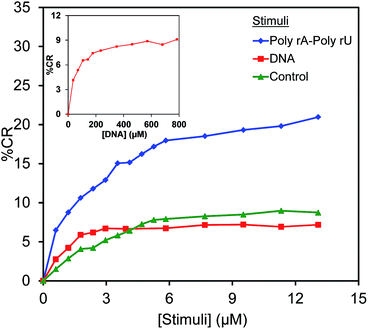
High specificity of PDA to its target biomolecules is necessary for the utilization as a biosensor. Here, we explore the specificity of poly(PCDA)/Neo–PCDA1 to poly(rA)–poly(rU) by comparing the colorimetric response to an AT rich DNA (5′-ATATATATATATATAT-3′). The results are illustrated in Fig. 6. As discussed earlier, the addition of 13.06 μM of poly(rA)–poly(rU) induces the increase of % CR to ∼22%. In contrast, the addition of DNA hardly induces any colorimetric response. The plot of % CR is similar to that of the control set. No significantly change is observed in this system. The further increase of DNA concentration to 800 μM still shows similar behavior. This result indicates that the poly(PCDA)/Neo–PCDA1 has high specificity to poly(rA)–poly(rU) attributed to the specific binding with neomycin moiety. This corresponds to low AC50 for the binding of neomycin moiety and poly(rA)–poly(rU).11,283.4 The effect of Neo–PCDA structure on colorimetric response
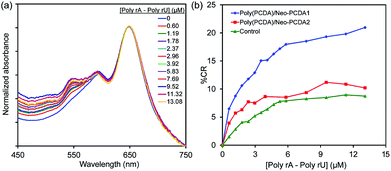
Specific binding site of Neo–PCDA is an important key for the recognition of poly(rA)–poly(rU). Here, we investigate the effect of Neo–PCDA structure on the binding ability of poly(PCDA)/Neo–PCDA to poly(rA)–poly(rU). Our result shows that the change of linker between neomycin moiety and PCDA significantly influences the colorimetric response of their assemblies. The Neo–PCDA2 constitutes a shorter linker compared to that of the Neo–PCDA1 (see Fig. 1). Fig. 7a shows that the addition of poly(rA)–poly(rU) into poly(PCDA)/Neo–PCDA2 assemblies causes slight growth of absorption peak at ∼540 nm, corresponding to the formation of some red phase. The plot of % CR clearly shows that the color response of poly(PCDA)/Neo–PCDA2 is much lower than that of poly(PCDA)/Neo–PCDA1 (see Fig. 7b). The CR value of poly(PCDA)/Neo–PCDA2 system is slightly higher than that of the control set of poly(PCDA)/Neo–PCDA1 system. The study of their morphologies by TEM (see Fig. 2) reveals that the poly(PCDA)/Neo–PCDA1 and poly(PCDA)/Neo–PCDA2 assemblies exhibit a different shape and size. The poly(PCDA)/Neo–PCDA2 assemblies exhibit much larger size compared to the system of poly(PCDA)/Neo–PCDA1. It has been known that the smaller PDA has higher response to external stimuli.84 Therefore, the larger size of poly(PCDA)/Neo–PCDA2 assemblies likely causes the lower response to poly(rA)–poly(rU).We further investigate the color-transition behaviors of poly(PCDA)/Neo–PCDA1 and poly(PCDA)/Neo–PCDA2 upon increasing temperature (see ESI Fig. S15†). Significant drop of absorbance in the system of poly(PCDA)/Neo–PCDA2 is observed upon increasing temperature attributed to the formation of large aggregates and then precipitation. In addition, this system also shows lower % CR compared to pure poly(PCDA) and poly(PCDA)/Neo–PCDA1, corresponding to higher stability. This result is consistent with the lower response of poly(PCDA)/Neo–PCDA2 to poly(rA)–poly(rU).
3.5 The effect of DA monomer structure on colorimetric response
The variation of DA monomer structure is an approach for enhancing colorimetric response of PDA to its stimuli. In this part, the PDA/Neo–PCDA1 assemblies were fabricated by using TCDA monomer which has slightly shorter alkyl tail than PCDA monomer. Fig. 8a shows the absorption spectra of poly(TCDA)/Neo–PCDA1 containing 0.5% v/v hexanol upon the addition of poly(rA)–poly(rU) at various concentrations. We observe significant growth of absorption peak at ∼540 nm. The plot of % CR clearly shows that poly(TCDA)/Neo–PCDA1 has higher CR values compared to the system of poly(PCDA)/Neo–PCDA1 (see Fig. 8b) attributed to the shorter alkyl tail of poly(TCDA). The decrease of alkyl chain length results in weaker dispersion interaction along the PDA layers.51,52,73 Therefore, we observe higher response of poly(TCDA)/Neo–PCDA1 to poly(rA)–poly(rU) compared to poly(PCDA)/Neo–PCDA1.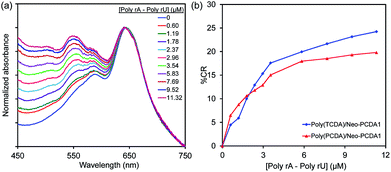 | ||
| Fig. 8 Absorption spectra of (a) poly(TCDA)/Neo–PCDA1 and (b) the plot of % CR shows the effect of DA monomer structure on the response to poly(rA)–poly(rU). | ||
4. Conclusions
Sensitive and specific PDA-based biosensor for the detection of homopolyribonucleotide poly(rA)–poly(rU) has been developed in this study. The poly(PCDA)/Neo–PCDA1 assemblies provide blue-to-purple color transition while pure poly(PCDA) vesicles show no change after the addition of poly(rA)–poly(rU). The addition of control DNA to poly(PCDA)/Neo–PCDA1 assemblies does not induce the color transition, indicating high specificity of poly(PCDA)/Neo–PCDA1 to poly(rA)–poly(rU). Sensitivity of poly(PCDA)/Neo–PCDA1 to poly(rA)–poly(rU) is enhanced by using alcohols as additives and the variation of DA monomer structure. The addition of small amount of hexanol results in significant increase of poly(PCDA)/Neo–PCDA1 response to poly(rA)–poly(rU). The penetration of alcohol molecules weakens interactions within the poly(PCDA)/Neo–PCDA1 assemblies, leading to the increase of colorimetric response. The variation of Neo–PCDA structure also affects its sensitivity. The use of Neo–PCDA1 with longer linker provides a higher response compared to that of the Neo–PCDA2. The PDA/Neo–PCDA assemblies prepared by using TCDA monomer exhibit a larger change compared to that of PCDA system. The development of sensitive and specific PDA to target biomolecules expands our toolkit for the utilization of PDA in biosensing, drug detection, and diagnostic applications.Conflicts of interest
There are no conflicts to declare.List of abbreviations
| AC50 | Concentration at 50% of maximum activity |
| CD | Circular dichroism spectroscopy |
| DA | Diacetylene (monomer) |
| DMF | N,N-Dimethylformamide |
| DMPC | Dimyristoylphosphatidylcholine |
| DNA | Deoxyribonucleic acid |
| FID | Fluorescence intercalator displacement assays |
| ITC | Isothermal titration calorimetry |
| MALDI | Matrix assisted laser desorption/ionization-time of flight mass spectrometry |
| Neo | Neomycin |
| Neo-PCDA | Conjugate of neomycin and PCDA |
| NMR | Nuclear magnetic resonance spectrophotometer |
| PBS buffer | Phosphate-buffered saline solutions |
| PCDA | 10,12-Pentacosadiynoic acid |
| PDA | Polydiacetylene |
| Poly(rA)–poly(rU): | Polyadenylic–uridylic acid |
| RNA | Ribonucleic acid |
| TBTU | O-(Benzotriazol-1-yl)-N,N,N′,N′-tetramethyluronium tetrafluoroborate |
| TCDA | 10,12-Tricosadiynoic acid |
| TEM | Transmission electron microscope |
| TFA | Trifluoroacetic acid |
| TPS | 2,4,6-Triisopropylbenzenesulfonyl chloride |
| UV | Ultraviolet |
Acknowledgements
We thank National Institutes for Health for financial support (grants R42GM097917, AI114114) to DPA. The research was also supported by the Thailand Research Fund (Grant BRG6080008). A. Kamphan thanks the Royal Golden Jubilee Ph.D. Program for supporting her Ph.D. scholarship. This work has partially been supported by the Nanotechnology Center (NANOTEC), Ministry of Science and Technology, Thailand, through its program of Center of Excellence Network.Notes and references
- D. P. Arya, Aminoglycoside antibiotics: from chemical biology to drug discovery, John Wiley & Sons, 2007 Search PubMed.
- M. A. Nowak, S. Bonhoeffer, A. M. Hill, R. Boehme, H. C. Thomas and H. McDade, Proc. Natl. Acad. Sci. U. S. A., 1996, 93, 4398–4402 CrossRef CAS.
- J. L. Seyec, P. Chouteau, I. Cannie, C. Guguen-Guillouzo and P. Gripon, J. Virol., 1999, 73, 2052–2057 Search PubMed.
- J. Zhong, P. Gastaminza, G. Cheng, S. Kapadia, T. Kato, D. R. Burton, S. F. Wieland, S. L. Uprichard, T. Wakita and F. V. Chisari, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 9294–9299, DOI:0503596102 [pii].
- S. A. Waksman, H. A. Lechevalier and D. A. Harris, J. Clin. Invest., 1949, 28, 934–939, DOI:10.1172/jcI102182 [doi].
- S. Kumar, P. Kellish, W. E. Robinson Jr, D. Wang, D. H. Appella and D. P. Arya, Biochemistry, 2012, 51, 2331–2347 CrossRef CAS PubMed.
- S. Kumar and D. P. Arya, Bioorg. Med. Chem. Lett., 2011, 21, 4788–4792 CrossRef CAS PubMed.
- N. Ranjan, S. Kumar, D. Watkins, D. Wang, D. H. Appella and D. P. Arya, Bioorg. Med. Chem. Lett., 2013, 23, 5689–5693 CrossRef CAS PubMed.
- S. Nahar, N. Ranjan, A. Ray, D. P. Arya and S. Maiti, Chem. Sci., 2015, 6, 5837–5846 RSC.
- S. Kumar, N. Ranjan, P. Kellish, C. Gong, D. Watkins and D. P. Arya, Org. Biomol. Chem., 2016, 14, 2052–2056 CAS.
- S. Kumar, L. Xue and D. P. Arya, J. Am. Chem. Soc., 2011, 133, 7361–7375 CrossRef CAS PubMed.
- B. Willis and D. P. Arya, Bioorg. Med. Chem. Lett., 2009, 19, 4974–4979 CrossRef CAS PubMed.
- B. Willis and D. P. Arya, Biochemistry, 2009, 49, 452–469 CrossRef PubMed.
- D. P. Arya, L. Xue and B. Willis, J. Am. Chem. Soc., 2003, 125, 10148–10149 CrossRef CAS PubMed.
- B. Willis and D. P. Arya, Adv. Carbohydr. Chem. Biochem., 2006, 60, 251–302 CrossRef CAS PubMed.
- D. P. Arya and B. Willis, J. Am. Chem. Soc., 2003, 125, 12398–12399 CrossRef CAS PubMed.
- B. Willis and D. P. Arya, Biochemistry, 2006, 45, 10217–10232 CrossRef CAS PubMed.
- N. N. Shaw, H. Xi and D. P. Arya, Bioorg. Med. Chem. Lett., 2008, 18, 4142–4145 CrossRef CAS PubMed.
- I. Charles, E. Davis and D. P. Arya, Biochemistry, 2012, 51, 5496–5505 CrossRef CAS PubMed.
- H. Xi, S. Kumar, L. Dosen-Micovic and D. P. Arya, Biochimie, 2010, 92, 514–529 CrossRef CAS PubMed.
- D. P. Arya, Acc. Chem. Res., 2010, 44, 134–146 CrossRef PubMed.
- L. Xue, I. Charles and D. P. Arya, Chem. Commun., 2002, 70–71 RSC.
- L. Xue, N. Ranjan and D. P. Arya, Biochemistry, 2011, 50, 2838–2849 CrossRef CAS PubMed.
- N. Ranjan, K. F. Andreasen, S. Kumar, D. Hyde-Volpe and D. P. Arya, Biochemistry, 2010, 49, 9891–9903 CrossRef CAS PubMed.
- D. Watkins, N. Ranjan, S. Kumar, C. Gong and D. P. Arya, Bioorg. Med. Chem. Lett., 2013, 23, 6695–6699 CrossRef CAS PubMed.
- N. Ranjan and D. P. Arya, Molecules, 2013, 18, 14228–14240 CrossRef PubMed.
- D. P. Arya, DNA Binders and Related Subjects, 2005, pp. 149–178 Search PubMed.
- H. Xi, E. Davis, N. Ranjan, L. Xue, D. Hyde-Volpe and D. P. Arya, Biochemistry, 2011, 50, 9088–9113 CrossRef CAS PubMed.
- S. Okada, S. Peng, W. Spevak and D. Charych, Acc. Chem. Res., 1998, 31, 229–239 CrossRef CAS.
- R. W. Carpick, D. Y. Sasaki, M. S. Marcus, M. Eriksson and A. R. Burns, J. Phys.: Condens. Matter, 2004, 16, R679 CrossRef CAS.
- B. Yoon, S. Lee and J. Kim, Chem. Soc. Rev., 2009, 38, 1958–1968 RSC.
- M. van den Heuvel, D. W. Löwik and J. C. van Hest, Biomacromolecules, 2008, 9, 2727–2734 CrossRef CAS PubMed.
- L. Rougeau, D. Picq, M. Rastello and Y. Frantz, Tetrahedron, 2008, 64, 9430–9436 CrossRef CAS.
- I. S. Park, H. J. Park and J. Kim, ACS Appl. Mater. Interfaces, 2013, 5, 8805–8812 CAS.
- S. Wacharasindhu, S. Montha, J. Boonyiseng, A. Potisatityuenyong, C. Phollookin, G. Tumcharern and M. Sukwattanasinitt, Macromolecules, 2010, 43, 716–724 CrossRef CAS.
- M. Gou, G. Guo, J. Zhang, K. Men, J. Song, F. Luo, X. Zhao, Z. Qian and Y. Wei, Sens. Actuators, B, 2010, 150, 406–411 CrossRef CAS.
- J. Kim, J. Lee, H. Choi, D. Sohn and D. J. Ahn, Macromolecules, 2005, 38, 9366–9376 CrossRef CAS.
- J. Pang, L. Yang, B. F. McCaughey, H. Peng, H. S. Ashbaugh, C. J. Brinker and Y. Lu, J. Phys. Chem. B, 2006, 110, 7221–7225 CrossRef CAS PubMed.
- T. Pattanatornchai, N. Charoenthai and R. Traiphol, J. Colloid Interface Sci., 2014, 432, 176–181 CrossRef CAS PubMed.
- Q. Cheng and R. C. Stevens, Langmuir, 1998, 14, 1974–1976 CrossRef CAS.
- A. Kamphan, C. Khanantong, N. Traiphol and R. Traiphol, J. Ind. Eng. Chem., 2017, 46, 130–138 CrossRef CAS.
- N. Traiphol, A. Chanakul, A. Kamphan and R. Traiphol, Thin Solid Films, 2017, 622, 122–129 CrossRef CAS.
- Z. Yuan and T. W. Hanks, Polymer, 2008, 49, 5023–5026 CrossRef CAS.
- S. J. Kew and E. A. Hall, Anal. Chem., 2006, 78, 2231–2238 CrossRef CAS PubMed.
- H. Jeon, J. Lee, M. H. Kim and J. Yoon, Macromol. Rapid Commun., 2012, 33, 972–976 CrossRef CAS PubMed.
- D. Seo and J. Kim, Adv. Funct. Mater., 2010, 20, 1397–1403 CrossRef CAS.
- J. Lee and J. Kim, Chem. Mater., 2012, 24, 2817–2822 CrossRef CAS.
- S. Lu, F. Luo, X. Duan, C. Jia, Y. Han and H. Huang, J. Appl. Polym. Sci., 2014, 131, 40634 CrossRef.
- S. Lu, C. Jia, X. Duan, X. Zhang, F. Luo, Y. Han and H. Huang, Colloids Surf., A, 2014, 443, 488–491 CrossRef CAS.
- F. Delbecq and T. Kawai, Colloids Surf., A, 2013, 430, 85–90 CrossRef CAS.
- T. Pattanatornchai, N. Charoenthai, S. Wacharasindhu, M. Sukwattanasinitt and R. Traiphol, J. Colloid Interface Sci., 2013, 391, 45–53 CrossRef CAS PubMed.
- N. Charoenthai, T. Pattanatornchai, S. Wacharasindhu, M. Sukwattanasinitt and R. Traiphol, J. Colloid Interface Sci., 2011, 360, 565–573 CrossRef CAS PubMed.
- K. Parambath Kootery, H. Jiang, S. Kolusheva, T. Vinod, M. Ritenberg, L. Zeiri, R. Volinsky, D. Malferrari, P. Galletti and E. Tagliavini, ACS Appl. Mater. Interfaces, 2014, 6, 8613–8620 CAS.
- S. Scoville and W. Shirley, J. Appl. Polym. Sci., 2011, 120, 2809–2820 CrossRef CAS.
- Y. Su, React. Funct. Polym., 2006, 66, 967–973 CrossRef CAS.
- Y. Su, J. Li and L. Jiang, Colloids Surf., B, 2004, 39, 113–118 CrossRef CAS PubMed.
- R. Jelinek, Drug Dev. Res., 2000, 50, 497–501 CrossRef CAS.
- Y. Zhang, B. Ma, Y. Li and J. Li, Colloids Surf., B, 2004, 35, 41–44 CrossRef CAS PubMed.
- G. Ma and Q. Cheng, Talanta, 2005, 67, 514–519 CrossRef CAS PubMed.
- C. Wang, Z. Ma and Z. Su, Sens. Actuators, B, 2006, 113, 510–515 CrossRef CAS.
- K. Kim, H. Choi, G. S. Lee, D. J. Ahn and M. Oh, Colloids Surf., B, 2008, 66, 213–217 CrossRef CAS PubMed.
- M. van den Heuvel, D. W. Löwik and J. C. van Hest, Biomacromolecules, 2010, 11, 1676–1683 CrossRef CAS PubMed.
- Y. K. Jung, T. W. Kim, J. Kim, J. Kim and H. G. Park, Adv. Funct. Mater., 2008, 18, 701–708 CrossRef CAS.
- Y. Xia, J. Deng and L. Jiang, Sens. Actuators, B, 2010, 145, 713–719 CrossRef CAS.
- D. H. Kang, H. Jung, J. Lee, S. Seo, J. Kim, K. Kim and K. Suh, Langmuir, 2012, 28, 7551–7556 CrossRef CAS PubMed.
- Y. K. Jung and H. G. Park, Biosens. Bioelectron., 2015, 72, 127–132 CrossRef CAS PubMed.
- D. Lee, S. K. Sahoo, A. L. Cholli and D. J. Sandman, Macromolecules, 2002, 35, 4347–4355 CrossRef CAS.
- Y. Lifshitz, Y. Golan, O. Konovalov and A. Berman, Langmuir, 2009, 25, 4469–4477 CrossRef CAS PubMed.
- X. Huang, S. Jiang and M. Liu, J. Phys. Chem. B, 2005, 109, 114–119 CrossRef CAS PubMed.
- X. Wang, J. E. Whitten and D. J. Sandman, J. Chem. Phys., 2007, 126, 184905 CrossRef PubMed.
- Y. Amano and Q. Cheng, Anal. Bioanal. Chem., 2005, 381, 156–164 CrossRef CAS PubMed.
- M. Park, K. Kim, D. J. Ahn and M. Oh, Biosens. Bioelectron., 2012, 35, 44–49 CrossRef CAS PubMed.
- A. Kamphan, N. Charoenthai and R. Traiphol, Colloids Surf., A, 2016, 489, 103–112 CrossRef CAS.
- D. P. Arya, L. Xue and P. Tennant, J. Am. Chem. Soc., 2003, 125, 8070–8071 CrossRef CAS PubMed.
- I. Charles, L. Xue and D. P. Arya, Bioorg. Med. Chem. Lett., 2002, 12, 1259–1262 CrossRef CAS PubMed.
- K. Michael, H. Wang and Y. Tor, Bioorg. Med. Chem., 1999, 7, 1361–1371 CrossRef CAS PubMed.
- H. Y. Erbil, Surface chemistry of solid and liquid interfaces, Wiley Online Library, 2006 Search PubMed.
- R. Nagarajan, Langmuir, 2002, 18, 31–38 CrossRef CAS.
- Y. K. Jung, H. G. Park and J. Kim, Biosens. Bioelectron., 2006, 21, 1536–1544 CrossRef CAS PubMed.
- Q. Cheng, M. Yamamoto and R. C. Stevens, Langmuir, 2000, 16, 5333–5342 CrossRef CAS.
- Q. Ye, G. Zou, X. You, X. Yu and Q. Zhang, Mater. Lett., 2008, 62, 4025–4027 CrossRef CAS.
- F. Zhao, Q. Zhao, K. F. Blount, Q. Han, Y. Tor and T. Hermann, Angew. Chem., Int. Ed., 2005, 44, 5329–5334 CrossRef CAS PubMed.
- W. M. Haynes and D. R. Lide, Handbook of Chemistry and Physics; National Institute for Standards and Technology, CRC Press, New York, NY, 2010 Search PubMed.
- C. X. Guo, P. Boullanger, T. Liu and L. Jiang, J. Phys. Chem. B, 2005, 109, 18765–18771 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra07178g |
| This journal is © The Royal Society of Chemistry 2017 |