Open Access Article
Open Access ArticleIntegration of transcriptome and proteome data reveals ochratoxin A biosynthesis regulated by pH in Penicillium citrinum
Lina Zhaoa,
Yaping Penga,
Xiaoyun Zhanga,
Jun Lib,
Xiangfeng Zhenga,
Qiya Yanga,
Maurice Tibiru Apaliyaa and
Hongyin Zhang *a
*a
aSchool of Food and Biological Engineering, Jiangsu University, Zhenjiang 212013, Jiangsu, People's Republic of China. E-mail: zhanghongyin126@126.com
bSchool of the Environment and Safety Engineering, Jiangsu University, Zhenjiang 212013, Jiangsu, People's Republic of China
First published on 3rd October 2017
Abstract
Ochratoxin A (OTA) has been found in a wide range of commodities and is highly toxic to both humans and animals. Therefore, a good understanding of the mechanisms of OTA production by fungi will contribute to the development of eco-friendly methods to mitigate this toxin. In this study, the results showed that Penicillium citrinum X9-4, which was isolated from infected grapes in our laboratory, produced the highest amount of OTA at pH 5 in culture media, and toxin production was restrained under an acidic environment (pH 3). Then, differentially expressed proteins of P. citrinum X9-4 cultured under these two conditions were analyzed by proteomic technology. Additionally, through the analysis of the transcriptome data of P. citrinum cultured at pH 3 and 5, the differentially expressed genes have been found to be involved in many metabolic pathways including amino acid transport and metabolism, transport and metabolism of carbohydrates, inorganic ion transport and metabolism, biosynthesis of secondary metabolites, and energy and supply metabolism, which are likely to be involved in the regulation of OTA biosynthesis. It was also revealed that the expression levels of some OTA synthesis-related enzymes, such as acetyltransferase, acyl coenzyme A oxidase, alcohol oxidase, cytochrome P450, acetyl xylan esterase, and turn ketol enzyme, and genes for the regulation of toxin synthesis pathways were reduced under acidic culture conditions.
1 Introduction
Ochratoxin A (OTA) is found in a variety of plant and animal products after infection of the produce by filamentous species belonging to some genera of Aspergillus and Penicillium. OTA is classified as a possible human carcinogen (Group 2B) by the International Agency of Research on Cancer (IARC, 1993), and it exhibits nephrotoxicity, hepatotoxicity, neurotoxicity, teratogenicity, and immunotoxicity;1–3 hence, it poses potential hazard to human and animals. OTA is a common mycotoxin contaminant of a wide range of foods and their products, which includes cereals, grapes, coffee, nuts, spices, cocoa beans, and other products processed from these substances. After cereals, grapes and its derived products are the second most contaminated products with OTA. The role of OTA in the etiology of Balkan endemic nephropathy and its association with urinary tract tumors have also been proved. OTA forms a benzoquinone electrophile following activation by cytochrome P450 enzymes and radical species triggered by enzymes with peroxidase activities.4,5 OTA is particularly toxic to the human body due to its high stability.6 Based on the strong toxicity and pathogenicity of OTA, some organizations and institutions have established official regulations and guidelines for maximum levels of OTA content in food.OTA was first reported to be caused by Penicillium verrucosum and Aspergillus ochraceus in cereals and its derived products.7 The main OTA-producing strains include Penicillium8 and Aspergillus genera, a few Petromyces genera, and Cladosporium strains.9–11 Black aspergilli (i.e., Aspergillus niger and Aspergillus carbonarius) have been found to produce OTA in commodities such as wine, grapes, and raisins in occident.12 The rate of OTA synthesis is influenced by the kind of substrate contamination, environmental conditions, and geographical regions.13 In temperate climate, OTA is mainly produced by Penicillium species, whereas in tropical and subtropical areas, it is produced by Aspergillus species.14
Although the pathway of OTA biosynthesis has not been completely elucidated in detail to date, some steps of the toxin biosynthesis pathway have been clearly elucidated.15–17 It is clear that the pathway of ochratoxin biosynthesis involves several crucial steps such as biosynthesis of the isocoumarin group through a reaction catalyzed by polyketide synthase (PKS), ligation of the isocoumarin group with amino acid phenylalanine through the carboxyl group in a reaction catalyzed by a peptide synthetase, and chlorination; however, the order of the reactions is not well defined to date.18 There is a clear evidence of the involvement of polyketide synthase in OTA production in A. ochraceus,19 Penicillium nordicum/verrucosum,20 and A. carbonarius,21 and the involvement of the non-ribosomal peptide synthetase (nrps) has been unequivocally demonstrated in A. carbonarius only.22 The pathway is still fragmentary, and some genes are missing during the virtual overlap between the hypothesized biochemical pathway and the genes found in putative OTA clusters of both Aspergillus and Penicillium.19,23 It is reported that mycotoxin biosynthetic genes are often coordinately regulated and arranged in clusters.24–26
Studies based on differential expression profiles are often performed to identify genes and proteins related to OTA biosynthesis. To identify the putative OTA biosynthetic genes in A. ochraceus, O'Callaghan et al.15 focused on two putative cytochrome P450 monooxygenase genes by comparing their expression profiles in permissive and restrictive media. Additionally, it was reported that polyketide synthase gene (pks) and nonribosomal peptide synthetase gene (nrps) were related to OTA biosynthesis in P. nordicum.27 A putative OTA biosynthetic cluster has been identified, containing biosynthetic genes encoding (i) PKS (otapksPN), (ii) nonribosomal peptide synthetase (NRPS) (otanpsPN) putatively responsible for the formation of the peptide bond between polyketide and phenylalanine, (iii) a gene (otachlPN) with some homology to chlorinating enzymes, which may be involved in the chlorination step, and (iv) a gene (otatraPN) with high homology to a transport protein hypothesized to be involved in OTA export.28 An interesting thing is that otapksPN gene is present only in P. nordicum and not in P. verrucosum; this indicates that genetic differences also exist between both OTA-producing Penicillium species.29 However, none of the necessary genes for the biosynthesis of this harmful toxin have been found to date.
The effect of pH on the growth and OTA production of P. citrinum was investigated. Proteomics and transcriptomic analysis of P. citrinum cultured at different pH values were performed to reveal the underlying mechanisms of OTA biosynthesis regulated by pH. This study will reveal the regulation mechanisms of OTA biosynthesis, enrich the gene database of OTA production genera, and assist in developing effective strategies for the protection of food and feed from OTA contamination.
2 Materials and methods
2.1 Fungal strains
P. citrinum X9-4 was isolated from the infected grape berries in our laboratory and deposited in the China Center for Type Culture Collection (CCTCC), and the collection number was CCTCC AF 2016008. The strain P. citrinum X9-4 was incubated on a potato dextrose agar medium (PDA: 200 mL of extract from 200 g boiled potatoes, 20 g dextrose, 20 g agar, and 800 mL distilled water) for 7 days at 25 °C before being used for the experiment; then, conidia were counted using a hemocytometer and adjusted to a concentration of 1 × 106 conidia per mL with sterile distilled water. OTA production was determined using Czapek Yeast Extract agar (CYA, purchased from Oxoid Ltd., Basingstoke, Hampshire, England), which contained (per liter of medium) 1 g KH2PO4, 10 mL Cza-concentrate, 1 mL trace metal solution, 5 g yeast extract, 30 g sucrose, and 15 g agar.2.2 pH assay and determination of the growth and OTA production of P. citrinum
The pH value of the medium was adjusted to 3, 5, 7, 9, and 11 by adding HCl or NaOH before autoclaving. The pH value was measured using the pH meter PHS-3B (Shanghai Lida Instrument Factory, Shanghai, China). For the acidified medium (pH 3.0), agar was sterilized separately and then added to the sterilized medium to avoid solidification. The pH value of each medium was checked after sterilization. Aliquots of 100 μL of P. citrinum spore suspension with a concentration of 1 × 106 conidia per mL were inoculated using the Petri dish-smearing method. Petri dishes were incubated at 25 °C in dark. Each assay was performed in duplicate. Then, two weeks after inoculation, mycelium and spores were obtained for protein and RNA extraction.To determine the mycelium growth of the strain P. citrinum X9-4 under different culture conditions, the colony diameter was chosen as the evaluation index. Then, 2.5 μL of 1 × 106 conidia per mL of P. citrinum X9-4 was inoculated in the single point of the culture solid medium at 25 °C (aw = 90%) for 7 days; after this, the colony diameter was measured using a Vernier caliper. Each plate was measured three times, and the experiment was repeated twice. For the determination of OTA production of the strain X9-4 under different pH culture conditions, 100 μL of 1 × 106 conidia per mL of P. citrinum X9-4 was spread by a coated rod on CYA plates (containing 30 mL CYA medium) for 2 weeks; then, a whole plate was extracted using 5 mL methanol by shaking to obtain a uniform mixture and deposited in dark for 3 hours. The extracts were filtered through a 0.22 μm filter for OTA detection. OTA production was determined by a high-performance liquid chromatograph equipped with a fluorescence detector (Agilent Technologies 1260, America). Agilent ZORBAX SB-C18 reversed-phase HPLC analytical column (250 mm × 4.6 mm, 5 μm) was used, and the column temperature was 30 °C. The excitation and emission wavelengths were set at 330 nm and 460 nm, respectively. The mobile phase comprised a mixture of acetonitrile and 1% acetic acid (60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40, v/v) at a flow rate of 1 mL min−1 in the isocratic elution mode.
40, v/v) at a flow rate of 1 mL min−1 in the isocratic elution mode.
2.3 Proteome analysis
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g, and the supernatant was transferred into a new Eppendorf tube; after this, equal volume of acetone containing 20% TCA was added. The protein was then precipitated at −20 °C for 12–16 hours. The precipitated proteins were obtained after centrifugation for 60 min at 12
000 × g, and the supernatant was transferred into a new Eppendorf tube; after this, equal volume of acetone containing 20% TCA was added. The protein was then precipitated at −20 °C for 12–16 hours. The precipitated proteins were obtained after centrifugation for 60 min at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g, and then, the precipitate was washed twice with cold acetone (stored at −20 °C more than 30 min) and centrifuged for 10 min at 12
000 × g, and then, the precipitate was washed twice with cold acetone (stored at −20 °C more than 30 min) and centrifuged for 10 min at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g. The precipitate was centrifuged instantaneously, and all visible liquid in the tube was discarded. Then, the protein extracts were air-dried for 5 min before being dissolved in 100 μL lysis buffer (2 M thiourea, 7 M urea, 4% (w/v) CHAPS, 65 mM DTT, and 0.2% (v/v) carrier ampholytes/Bio-Rad) and stored at −80 °C until further analysis.
000 × g. The precipitate was centrifuged instantaneously, and all visible liquid in the tube was discarded. Then, the protein extracts were air-dried for 5 min before being dissolved in 100 μL lysis buffer (2 M thiourea, 7 M urea, 4% (w/v) CHAPS, 65 mM DTT, and 0.2% (v/v) carrier ampholytes/Bio-Rad) and stored at −80 °C until further analysis.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 V h. After focusing, the strips were kept at 500 V (for no more than 15 h) to prevent deviation of the focused proteins until they were used directly for electrophoresis. Proteins were separated using EttanDALTsix. Electrophoresis System was carried out in SDS electrophoresis buffer (25 mM Tris, pH 8.8, 192 mM glycine, and 0.1% SDS) at 1 W per strip for 1 h and then at 15 W per strip until the BPB solvent settled at the bottom of the gels. All abovementioned instruments were purchased from GE Healthcare. The gels were stained using a modified Coomassie-protocol, washed twice with MilliQ water, and then stained for 2 h in Coomassie staining solution (45% methanol, 45% MilliQ water, 10% glacial acetic acid, and 2.5 g R250). After being stained, the gels were washed with discoloration solution (45% alcohol, 45% MilliQ water, and preserved in 10% glacial acetic acid).
000 V h. After focusing, the strips were kept at 500 V (for no more than 15 h) to prevent deviation of the focused proteins until they were used directly for electrophoresis. Proteins were separated using EttanDALTsix. Electrophoresis System was carried out in SDS electrophoresis buffer (25 mM Tris, pH 8.8, 192 mM glycine, and 0.1% SDS) at 1 W per strip for 1 h and then at 15 W per strip until the BPB solvent settled at the bottom of the gels. All abovementioned instruments were purchased from GE Healthcare. The gels were stained using a modified Coomassie-protocol, washed twice with MilliQ water, and then stained for 2 h in Coomassie staining solution (45% methanol, 45% MilliQ water, 10% glacial acetic acid, and 2.5 g R250). After being stained, the gels were washed with discoloration solution (45% alcohol, 45% MilliQ water, and preserved in 10% glacial acetic acid).MS analysis was performed as described by Zhang et al.,31 with some modifications. Selected spots were manually excised from gels and placed in Eppendorf tubes. Each gel piece of protein was washed with 350 μL of deionized water for 10 min, and the procedure was repeated twice. Afterward, the gel pieces were dehydrated with 50 μL of 10 mM (NH4)2CO3/acetonitrile (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) for 10 min. This process was repeated until the color faded. Then, the gel pieces were dehydrated with 25 mM (NH4)2CO3 and 50% acetonitrile. Next, 3 μL trypsin solution (10 ng μL−1 trypsin in 25 mM (NH4)2CO3) was added, and the mixture was kept at 4 °C for 30 min until the entire trypsin solution was absorbed by the gel pieces. The gel pieces were then covered with 15 μL of 25 mM (NH4)2CO3 and digested at 37 °C overnight for proteolysis. After this, 2% TFA was added and centrifuged immediately to terminate the reaction. Herein, 1 μL of the peptide solution was spotted per well in perforated plates and evaporated at room temperature, and this step was repeated once.
1) for 10 min. This process was repeated until the color faded. Then, the gel pieces were dehydrated with 25 mM (NH4)2CO3 and 50% acetonitrile. Next, 3 μL trypsin solution (10 ng μL−1 trypsin in 25 mM (NH4)2CO3) was added, and the mixture was kept at 4 °C for 30 min until the entire trypsin solution was absorbed by the gel pieces. The gel pieces were then covered with 15 μL of 25 mM (NH4)2CO3 and digested at 37 °C overnight for proteolysis. After this, 2% TFA was added and centrifuged immediately to terminate the reaction. Herein, 1 μL of the peptide solution was spotted per well in perforated plates and evaporated at room temperature, and this step was repeated once.
The identification of the vast majority of the proteins was performed using search engine MASCOT Peptide Mass Fingerprint of Matrix Science and compared with NCBInr Swissport databases. The parameters used for MS search included taxonomy, all series; allowed modifications, carbamidomethyl of cysteine (fixed) and oxidation of methionine (variable); and peptide tolerance, ±0.3 Da. Only the highest Mowse score was considered as the most probable identification and was significant (p < 0.05) when protein scores were greater than 88 (NCBInr) or 70 (swissport). The data were analyzed by analysis of variance (ANOVA) using the statistical program SPSS/PC version II.x, (SPSS Inc. Chicago, Illinois, USA), and the Duncan's multiple range test was used for mean separation. In addition, when two groups of data were compared, the independent-sample t test was applied for mean separation. The statistical significance was assessed at p < 0.05.
2.4 RNA extraction and transcriptome sequencing
2.5 Reverse transcription and real-time quantitative PCR
Herein, one microgram of DNase-treated RNA was reverse-transcribed using a reverse transcription kit (Thermo Scientific, Lithuania). The RT-qPCR reactions of interested genes were performed using the SYBR Premix Ex Taq™ (Tli RNaseH Plus) quantitative fluorescence kit (TaKaRa, Dalian, China) in ABI StepOnePlus Real-Time PCR Systems (Applied Biosystems, CA, USA) following the manufacture's instructions. The primers used for amplification were designed using primer 5, with the product size between 150 and 250 bp, and the primer sequences are listed in Table 3. The relative expression was calculated using the 2−ΔΔCT method, as described by Schmittgen and Livak.32 Actin binding gene was used as an internal reference. In this pH assay, the reference was a pH 3.0 medium.333 Results and discussion
3.1 Effect of pH on the growth and OTA production of the strain X9-4
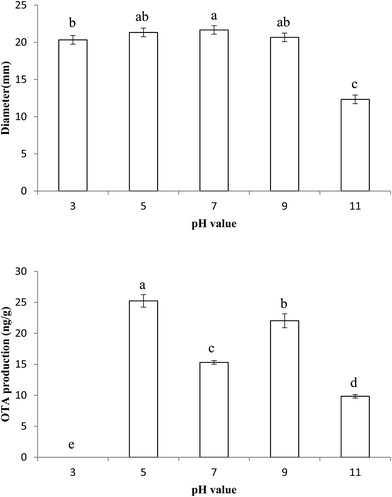
The growth of P. citrinum X9-4 is shown in Fig. 1. The strain could grow within a wide pH range, but the alkaline environment was unsuitable for its growth. When the strain was cultured at pH 11, the colony diameter was 12 mm, half of that of the strain cultured at pH 7, which was the most favorable pH for growth. In contrast, colony diameters were very close when the pH of the culture medium was between 3 and 9. With regard to the influence of pH on toxin production by the strain, toxin production was high in the medium with pH 5 and 9, and the highest concentration of the toxin was 25.6 ng g−1 at pH 5. It was also observed that when the strain was cultured at pH 3, no OTA was detected; this indicated that its biosynthesis was inhibited in an acidic environment. It was found that the OTA-producing strain A. niger not only adapted to a wider pH range for growth, but also produced OTA in a wider pH range.34 It was also reported that A. carbonarius and A. niger could grow and produce OTA well in a wider pH range,35 and the highest level of OTA production was observed at pH 5.35. Our result was well consistent with that reported by Passamani about the effect of pH on OTA production in A. carbonarius and A. niger.353.2 Relative expression of proteins involved in OTA biosynthesis in response to different pH conditions
The production of OTA by P. citrinum in an acidic environment was lower than the minimum limit of detection of an HPLC fluorescence detector, but OTA concentration reached 25.6 ng g−1 at pH 5. This means that OTA production under acidic conditions is restrained. To investigate the mechanism of how an acidic environment restricts the OTA biosynthesis, total proteins of P. citrinum cultured at pH 3 and 5 were extracted. As shown in Fig. 2, a total of 300 protein spots were detected on each gel using PDQuest, containing 90 differentially expressed proteins (ratio ≥ 2.0), and 25 of the best-resolved spots were identified by MALDI-TOF/TOF MS (Table 1).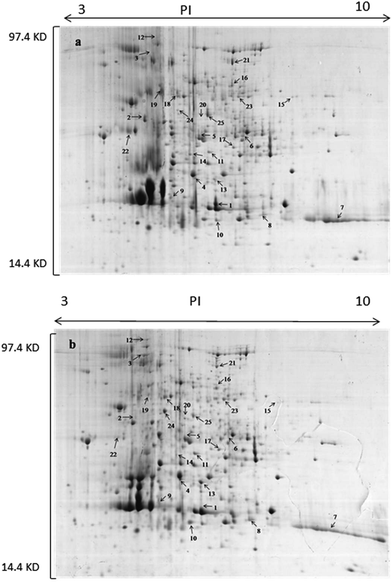 | ||
| Fig. 2 Two-dimensional pattern of total proteins of P. citrinum cultured at different pH values. (A) P. citrinum cultured at pH 3, (B) P. citrinum cultured at pH 5. | ||
| Function classification | Protein spot | NCBI accession | Protein name | Species | Mass/PI(Da) | Score | Peptide hits | Expression change |
|---|---|---|---|---|---|---|---|---|
| Metabolism and synthesis related protein | 11 | gi|582959135 | Fructose-bisphosphate aldolase | Drechslerella stenobrocha | 5.52/39![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 347.56 347.56 |
64 | 1(1) | Up-regulated |
| 4 | gi|9955865 | Fructose1,6-bisphosphate aldolase | Aspergillus oryzae | 6.02/39![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 162.33 162.33 |
63 | 1(1) | Up-regulated | |
| 14 | gi|115400267 | Transaldolase | Aspergillus terreus | 6.16/35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 057.20 057.20 |
126 | 1(1) | Down-regulated | |
| 17 | gi|425774591 | Nucleoside-diphosphate-sugar epimerase | Penicillium digitatum | 5.62/36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 935.70 935.70 |
96 | 1(1) | Up-regulated | |
| Energy metabolism | 21 | gi|121701281 | Transketolase | Aspergillus clavatus | 6.06/74![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 671.48 671.48 |
153 | 2(2) | Down-regulated |
| 10 | gi|115399504 | Riosephosphate isomerase | Aspergillus terreus | 5.42/27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 084.90 084.90 |
387 | 6(3) | Down-regulated | |
| 19 | gi|425772348 | Adenosine kinase | Penicillium digitatum | 5.21/38![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 013 013 |
106 | 2(1) | Down-regulated | |
| 24 | gi|74664773 | Enolase | Penicillium citrinum | 5.33/47![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 275.45 275.45 |
480 | 6(4) | Up-regulated | |
| 18 | gi|654941862 | Leucine/isoleucine/valine transporter ATP-binding subunit | Bacillus sp | 6.72/29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 165.91 165.91 |
61 | 1(0) | Up-regulated | |
| 25 | gi|425767748 | 6-Phosphogluconate dehydrogenase, decarboxylating | Penicillium digitatum | 5.85/53![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 908.23 908.23 |
77 | 2(0) | Down-regulated | |
| Stress protein | 23 | gi|700449320 | Aldehyde dehydrogenase | Penicillium expansum | 5.99/53![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 823.25 823.25 |
170 | 3(0) | Up-regulated |
| 3 | gi|14423733 | Heat shock 70 kDa protein70 kDa | Penicillium citrinum | 4.99/55![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 125.80 125.80 |
340 | 7(2) | Up-regulated | |
| Transporter related protein | 9 | gi|494679012 | Peptide ABC transporter substrate-binding protein | Acetobacteraceae bacterium | 6.58/59![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 424.33 424.33 |
63 | 1(1) | Down-regulated |
| 8 | gi|557724566 | Glutathione S-transferase GstA | Byssochlamys spectabilis | 6.47/29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 102.79 102.79 |
217 | 2(2) | Up-regulated | |
| Regulatory protein | 12 | gi|503068397 | Integrase domain-containing protein | Ignisphaera aggregans | 10.34/37![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 022.51 022.51 |
65 | 1(1) | Up-regulated |
| Others | 22 | gi|119483728 | Class V chitinase ChiB1 | Neosartorya fischeri | 5.10/47![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 667.27 667.27 |
131 | 3(1) | Down-regulated |
| 20 | gi|114077 | Apovitellenin, Protein component of VLDL | Aves | 9.40/9492.11 | 65 | 1(1) | Up-regulated | |
| 16 | gi|34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 069 069 |
Unnamed protein product | Homo sapiens | 5.12/39![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 218.58 218.58 |
96 | 2(0) | Down-regulated | |
| Hypothetical protein | 2 | gi|115491105 | Hypothetical protein | Aspergillus terreus | 5.25/326391.46 | 79 | 1(1) | Down-regulated |
| 13 | gi|740092337 | Hypothetical protein | Sulfitobacter sp. | 5.42/22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 273.12 273.12 |
71 | 1(1) | Up-regulated | |
| 15 | gi|531280525 | Hypothetical protein | Peptoclostridium difficile CD196 | 7.91/4376.12 | 49 | 2(0) | Down-regulated | |
| 6 | gi|358386419 | Hypothetical protein | Trichoderma virens | 6.57/42![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 545.25 545.25 |
109 | 2(0) | Down-regulated | |
| 1 | gi|734656611 | Hypothetical protein | Metarhizium album | 6.76/26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 146.62 146.62 |
84 | 1(1) | Down-regulated | |
| 7 | gi|503691373 | Hypothetical protein | Parachlamydia acanthamoebae | 9.17/50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 868.18 868.18 |
64 | 1(1) | Down-regulated | |
| 5 | gi|799157440 | Hypothetical protein | Streptomyces sp | 9.13/24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 830.99 830.99 |
66 | 1(1) | Down-regulated |
3.3 Analysis of the differentially expressed proteins
The synthesis of OTA by P. citrinum may be associated with basic metabolism (Fig. 3). When P. citrinum was cultured at pH 5, metabolism- and synthesis-related proteins, such as fructose-bisphosphate aldolase (FBA, spots 11 and 4) and nucleoside-diphosphate-sugar epimerase were up-regulated. FBA can reversibly catalyze fructose 1,6-diphosphate to two triose, dihydroxyacetone phosphate and glyceraldehyde 3-phosphate. This reaction exists in both glycolytic/gluconeogenesis pathway and pentose phosphate pathway (PP pathway), which provide energy and substrate for biological synthesis and metabolism. Transaldolase (spot 14) was down-regulated when P. citrinum was cultured at pH 5. This enzyme is involved in PP pathway and can transfer the dihydroxyacetone part of 7-phosphosedoheptose to glyceraldehyde-3-phosphate, thus the down-regulation of transaldolase may be decreased acetone necessary for OTA synthesis in P. citrinum.Energy is essential for the biosynthesis of OTA, and PP pathway is an important source to provide NADPH.36 In this study, the expression of 6-phosphogluconate dehydrogenase (6PGDH, spot 25) was decreased when the strain was cultured at pH 5. 6PGDH is generally considered to catalyze the following reactions with the generation of NADPH: 6-phosphogluconate + NADP+ → ribulose 5-phosphate + CO2 + NADPH. That is, the PP pathway was more active when the strain was cultured at pH 3. Transketolase (spot 21) is an important enzyme in the pentose phosphate cycle and reduced pentose phosphate cycle, with pyrophosphate thiamine and Mg2+ as the prosthetic group. Triose phosphate isomerase (spot 10), which catalyzes triose phosphate isomer to transform between dihydroxyacetone phosphate and type D glyceraldehyde 3-phosphate, plays an important role in the glycolysis process and energy supply. However, the expression of triose phosphate isomerase was down-regulated at pH 5. Adenylate kinase (spot 19) is an important enzyme in energy metabolism, which catalyzes adenosine triphosphate (ATP) to make adenosine monophosphate (AMP) and adenosine diphosphate (ADP) by phosphorylation. Herein, two proteins involved in energy metabolism were up-regulated when cultured at a pH value of 5: phosphopyruvate hydratase and amino acid transporter ATP-binding subunit. Phosphopyruvate hydratase (spot 24), which is involved in the process of sugar dysplasia catalytic-D-2-phosphoric acid by glycolysis glyceric acid (PGA)-enol phosphate type adverse reactions of pyruvate (PEP), transform PEP to PGA, it plays an important role in the process of energy metabolism in cells.37 Adenosine phosphate protein subunits (spot 18) are involved in the process of generation of biological energy, and dipeptide adenosine phosphate is involved in the synthesis of dTDP-4-ethyl amide group-alpha-D-trehalose (dTDP-N-acetylthomosamine) in Haemophilus parasuis; the results are presented in Uniprot. Under the acidic conditions, a part of the proteins involved in energy metabolism was up-regulated, and other was down-regulated in the strain X9-4.
6PGDH (spot 25) in the process of metabolism in the PP pathway produces a substance called NADPH, which can protect cells from oxidation. Thus, 6PGDH is involved in energy metabolism and can cause stress. Aldehyde dehydrogenase (spot 23) plays an important role in improving the resistance of organisms. Guo et al. showed that under a low-temperature stress, genetically modified (SoBADH gene) strain exhibits high aldehyde dehydrogenase activity and significant improvement in frost-resistance ability.38 There were also differentially expressed proteins, such as 70 kDa heat stress protein (spot 3), related to the immune response.
The toxin needs to be discharged by the translocator after its production. The expression of glutathione S-transferases39 (GSTs, spot 8) was up-regulated under the culture condition of pH 5. GSTs are a set of multifunctional isozymes widely distributed in various organisms, and their function involves coupling the electrophilic group of substances with the mercapto group of reduced glutathione; this makes it easy for the substances to cross the cell membranes by increasing their hydrophobicity. It was observed that there was positive correlation between up-regulated enzymes of GSTs and the efficiency of OTA production, the concentration of OTA in P. citrinum was increased when the expression of GSTs was up-regulation. ABC transporters (spot 9) is a type of protein across the membrane, which widely exists in various organisms in the range from bacteria to humans. Its main function is to transfer the substrate combined with it outside the cell using ATP. SAM (Sterile Alpha Motif) function domain (spot 12), which is up-regulated under the culture condition of pH 5, is a protein interaction between protein models, for example, SAM functional domains at the C terminal of p63 and p73 work by recruiting other protein effectors to regulate the functions of p63 and p73.
3.4 Relative expression of the genes involved in OTA biosynthesis in response to different pH conditions
Via sequencing, the large-scale transcriptome data were obtained from P. citrinum (Table 2), and there was a total of 9.91 Gb clean and high-quality sequences, which were suitable for subsequent analysis.| Samples | Clean reads | Clean data | GC content | % ≥ Q30 |
|---|---|---|---|---|
| a % ≥ Q30: the percentage of bases that clean data quality value is greater than or equal to 30. | ||||
| pH 3 | 21089411 | 5272352750 | 50.32 | 94.27 |
| pH 5 | 18568067 | 4642016750 | 50.53 | 94.6 |
| No. | Unigenes | Description | Forward primer (5′ to 3′) | Revers primer (5′ to 3′) | Relative expression |
|---|---|---|---|---|---|
| 1 | TRINITY_DN4069_c0_g1 | Acetyltransferase | GGCTCCTCAACCAACCTC | TCTCCCAATCCCTTTACC | +1.28 |
| 2 | TRINITY_DN5955_c0_g3 | Polyketide synthase phosphopantetheine-binding domain | AATCGCTGAACAACCATCCG | GGCTTCCATTCCAAACTC | +1.6 |
| 3 | TRINITY_DN1510_c0_g1 | Acyl-CoA dehydrogenase oxidase | GTAATGCCGTCGCAAGAT | GATTCGGTTCAAAGGGTT | +1.71 |
| 4 | TRINITY_DN3456_c0_g2 | Alcohol oxidase | TTCGTGTACTTGTCCCATT | TATCGTCCGCTGCCTTCT | +3.73 |
| 5 | TRINITY_DN10315_c0_g1 | NAD(P)-binding protein | CTCAGAAGCACCGATTTT | CAACATCCAACCCGTCCA | +6.29 |
| 6 | TRINITY_DN9885_c0_g1 | Acetylxylan esterase | TACATCGGTGCTTCTTTCGTC | TCGGTGCCATAGCCTTGA | +8.04 |
| 7 | TRINITY_DN526_c0_g2 | Cytochrome P450 | TATCCCAGCCCATACCATC | CACCCTCTTCCTCCAACG | +8.15 |
| 8 | TRINITY_DN4980_c0_g4 | ABC transporter, putative | TCTTTCCAGGTTCTATTCGG | CTGCTCGTTGGTTCGTCT | +1.51 |
| 9 | TRINITY_DN598_c0_g1 | Glutathione S-transferase | GAGTTTTGGATAAGTGGTTG | TTTGACTGATTCTATGGTGG | +1.40 |
| 10 | TRINITY_DN6150_c0_g1 | Transketolase C-terminal domain | GCCGACTATGTCTTTCCG | ACCGTGGCATTACTACCC | +1.02 |
| 11 | TRINITY_DN1616_c0_g1 | Alcohol dehydrogenase | TGGCTGCGATTACACCTT | CCCTTGATACCACCGAAA | −7.81 |
| 12 | TRINITY_DN2593_c0_g1 | Transaldolase | CGCTTGGCTGTCAGAATA | CTTGATAGCGATGCGTGT | −6.09 |
| 13 | TRINITY_DN6090_c0_g2 | Beta-ketoacyl synthase domain-containing protein | TAGGACGCTGGAGTGCTTCTGGT | CTCGACGGGCTGAGTTGT | −4.55 |
| 14 | TRINITY_DN6972_c0_g2 | Heat shock 70 kDa protein | AGTGTTGTGGGGGTTCAT | CTCCTGTGTGGGTGTGTT | −1.65 |
| 15 | TRINITY_DN4080_c0_g2 | Actin biding | CGAACAGGACAACGCAGC | CCACTGTCTGAAGGACGAAC | +1.019 |
There were different gene expressions at different time and space phases; external stimulus and internal environment also affected gene expression. Differentially expressed genes (DEGs) are the genes differently expressed under two different conditions (e.g., control and processing, wild type and mutant type, different time, different organizations, etc.). In this study, the difference in OTA synthesis ability may be caused by differences in gene expression; therefore, it is essential to study the differently expressed unigenes of P. citrinum cultured at pH 3 and 5. By comparing the results, we found 2200 DEGs between two samples (fold change ≥ 2 and FDR < 0.05); among them, 1024 genes were up-regulated and 1176 were down-regulated. P3_vs._P5 were used to categorize differentially expressed genes; henceforth, up-regulated genes have a higher expression level in the sample group P5 than that in the sample group P3 and vice versa.
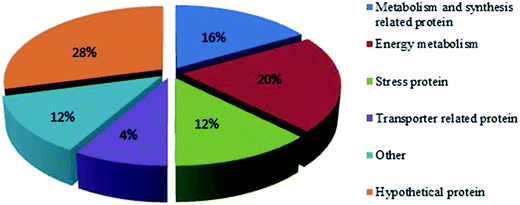
To analyze the function of these differentially expressed genes, we performed the GO and COG annotations and classification (Fig. 4). We divided 4545 unigenes into three GO function categories, which were further divided into 57 categories; many unigenes were assigned to different categories simultaneously. Then, the percent of DEG unigenes from each category was calculated. From the results of “biological process” category, the highest category of differentially expressed unigenes was “metabolism” of 1002 (12.28%); the differentially expressed unigenes in “Molecular function” category was “catalytic activity of 822 (12.61% of gaia category), and the “combination”, 623 (11.91%); the two high percentage categories of “cells” category were “cell part” (582, 10.39%) and “cell” (579, 10.42%). These results suggest that the difference in OTA production by P. citrinum may be due to the several differentially expressed genes in the categories, and differentially expressed genes in these categories accounted for a big proportion; this provides the direction for the subsequent analysis.
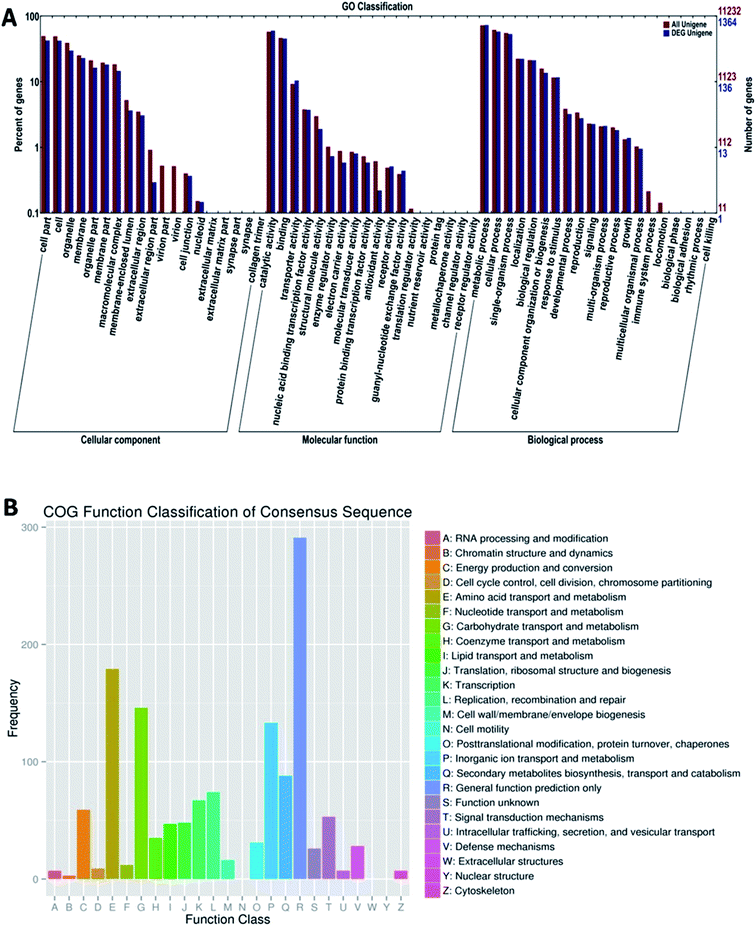 | ||
| Fig. 4 Functional classification of DEG in P. citrinum. (A) GO classification. (B) COG classification. | ||
Regarding the transcriptome data for P. citrinum cultured at different pH values, it was observed that the acidic pH inhibited the toxin synthesis due to the low expression of some metabolic pathway-related enzymes. Additionally, the expression levels of some genes involved in the regulation of the biosynthesis of toxins were inhibited in P. citrinum cultured under an acidic environment; this could also reduce the synthesis of toxins. The DEGs were classified in the most enriched categories including amino acid transport and metabolism, transport and metabolism of carbohydrates, inorganic ion transport and metabolism, biosynthesis of secondary metabolites, and energy and supply metabolism. These differentially expressed genes are likely to be involved in the synthesis and OTA regulation process.
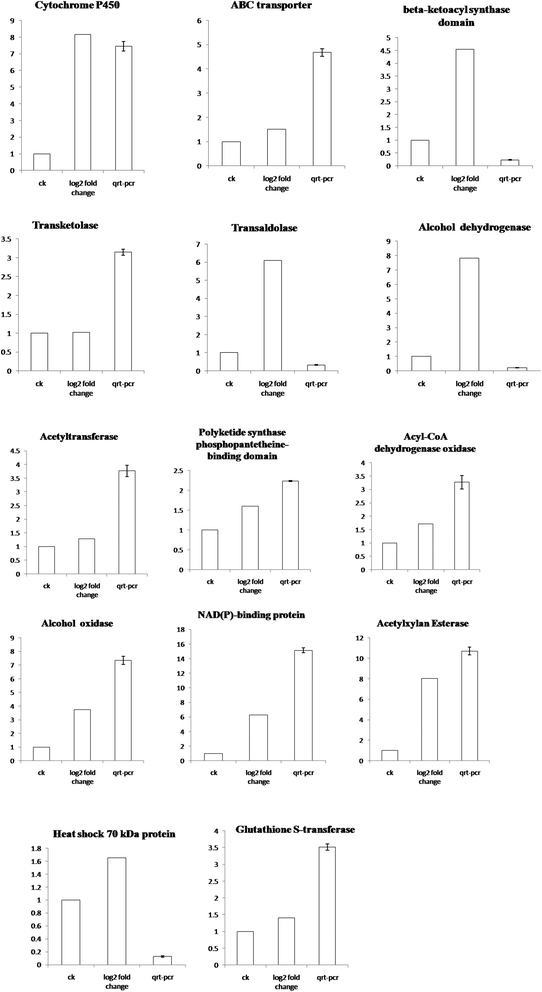
To verify the differences in these genes expressed in the OTA synthesis, the mRNA levels of 14 unigenes, which were differently expressed genes and associated with toxin production, were analyzed by RT-qPCR (Fig. 5). From the results of P. citrinum cultured at pH 5, the expression levels of these genes were obviously higher than those of the strain cultured at pH 3. These results were consistent with those of transcriptome data analysis.
In conclusion, the results indicated that low pH conditions inhibited the biosynthesis of OTA in P. citrinum. The proteomics technique employed to investigate differentially expressed proteins of P. citrinum under different pH culture conditions revealed that the expression levels of 11 proteins, accounting for 44% of the total number of proteins, were up-regulated in the medium of pH 5 as compared to those at pH 3. In contrast, the expression levels of most basic metabolism- and synthesis-related proteins, as well as of those related to energy supply, were decreased; this made it difficult for cells to maintain the life activities. Fructose-diphosphate aldolase, acetone phosphate acid hydratase, adenosine phosphate subunits, and glutathione S-transferase activities were decreased under the low pH (3) culture condition of P. citrinum. Fructose-diphosphate aldolase provides ATP and substrate for organisms, and glutathione S-transferase may be associated with toxin transport. Under the low pH (3) condition, the expression levels of fructose-diphosphate aldolase and glutathione were limited, whereas those of the stress-related proteins, such as some signal transduction and dehydrogenate-related proteins, increased. The expression level of glucose-6-phosphate dehydrogenase was increased under low pH (3) conditions; this protected the strain from stress.
Through the analysis of the transcriptome data of P. citrinum cultured under different pH conditions, the mechanism of OTA biosynthesis and metabolic regulation has been revealed. Under the two pH conditions of 3 and 5, 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 598 unigenes were identified. The results indicated that some genes played important roles in OTA synthesis by P. citrinum. These genes mainly encoded acetyltransferase, polyketone synthase phosphate generic acyl cysteamine-combined domain, acyl coenzyme A oxidase, alcohol oxidase, NAD(P)-binding protein, acetyl xylan esterase, cytochrome P450, ABC transporters, beta ketone enzyme domain protein, turn ketol enzyme, aldol enzyme, and alcohol dehydrogenase. Some proteins were consistent with the results of the two-dimensional technique; however, many important enzymes could not be identified by this technique due to technical reasons. These transcriptome findings provided the basis for the molecular level for further studies of OTA biosynthesis by P. citrinum and Penicillium species. Finally, the transcriptome data for P. citrinum not only revealed the possible mechanism of OTA synthesis and molecular regulation in P. citrinum, but also provided scientific guidance for controlling the OTA synthesis.
598 unigenes were identified. The results indicated that some genes played important roles in OTA synthesis by P. citrinum. These genes mainly encoded acetyltransferase, polyketone synthase phosphate generic acyl cysteamine-combined domain, acyl coenzyme A oxidase, alcohol oxidase, NAD(P)-binding protein, acetyl xylan esterase, cytochrome P450, ABC transporters, beta ketone enzyme domain protein, turn ketol enzyme, aldol enzyme, and alcohol dehydrogenase. Some proteins were consistent with the results of the two-dimensional technique; however, many important enzymes could not be identified by this technique due to technical reasons. These transcriptome findings provided the basis for the molecular level for further studies of OTA biosynthesis by P. citrinum and Penicillium species. Finally, the transcriptome data for P. citrinum not only revealed the possible mechanism of OTA synthesis and molecular regulation in P. citrinum, but also provided scientific guidance for controlling the OTA synthesis.
Ethical statement
This article does not contain any studies with human participants or animals performed by any of the authors.Conflicts of interest
The authors declare that they have no conflict of interest.Acknowledgements
This research was supported by the National Key Research Project (sub project) of China (2016YFD0400902-04) and the National Natural Science Foundation of China (31571899, 31701971).References
- S. Raghubeer, S. Nagiah, A. Phulukdaree and A. Chuturgoon, J. Cell. Biochem., 2015, 116, 2947 CrossRef CAS PubMed.
- L. Rutigliano, L. Valentini, N. A. Martino, F. Pizzi, A. Zanghì, M. E. Dell'Aquila and F. Minervini, Reprod. Toxicol., 2015, 57, 121–129 CrossRef CAS PubMed.
- D. Feier and M. Tofana, Bulletin of the University of Agricultural Sciences & Veterinary, 2009, 66, 1843–5386 Search PubMed.
- A. Pfohl-Leszkowicz and R. A. Manderville, Mol. Nutr. Food Res., 2007, 51, 61–99 CAS.
- R. Manderville and A. Pfohlleszkowicz, World Mycotoxin J., 2008, 1, 357–367 CrossRef CAS.
- K. A. El and A. Atoui, Toxins, 2010, 2, 461 CrossRef PubMed.
- S. C. Duarte, A. Pena and C. M. Lino, Food Microbiol., 2010, 27, 187–198 CrossRef CAS PubMed.
- H. D. Nguyen, D. R. Mcmullin, E. Ponomareva, R. Riley, K. R. Pomraning, S. E. Baker and K. A. Seifert, Fungal Biol., 2016, 120, 1041 CrossRef CAS PubMed.
- M. R. Bragulat, E. Martínez, G. Castellá and F. J. Cabañes, Int. J. Food Microbiol., 2008, 126, 43 CrossRef CAS PubMed.
- L. Covarelli, G. Beccari, A. Marini and L. Tosi, Food Control, 2010, 21, 347–356 Search PubMed.
- M. Sánchezhervás, J. V. Gil, F. Bisbal, D. Ramón and P. V. Martínezculebras, Int. J. Food Microbiol., 2008, 125, 336 CrossRef PubMed.
- M. Bau, M. R. Bragulat, M. L. Abarca, S. Minguez and F. J. Cabañes, Int. J. Food Microbiol., 2005, 98, 125–130 CrossRef CAS PubMed.
- M. Khalesi and N. Khatib, Environ. Toxicol. Pharmacol., 2011, 32, 113–121 CrossRef CAS PubMed.
- W. Masoud and C. H. Kaltoft, Int. J. Food Microbiol., 2006, 106, 229–234 CrossRef CAS PubMed.
- J. O'Callaghan, M. X. Caddick and A. D. Dobson, Microbiology, 2003, 149, 3485 CrossRef PubMed.
- A. Karolewiez and R. Geisen, Syst. Appl. Microbiol., 2005, 28, 588–595 CrossRef CAS PubMed.
- A. Gallo, G. Perrone, M. Solfrizzo, F. Epifani, A. Abbas, A. D. W. Dobson and G. Mulè, Int. J. Food Microbiol., 2009, 129, 8–15 CrossRef CAS PubMed.
- A. Gallo, K. S. Bruno, M. Solfrizzo, G. Perrone, G. Mulè, A. Visconti and S. E. Baker, Appl. Environ. Microbiol., 2012, 78 Search PubMed.
- J. O'Callaghan, P. C. Stapleton and A. D. Dobson, Fungal Genet. Biol., 2006, 43, 213 CrossRef PubMed.
- A. Karolewiez and R. Geisen, Syst. Appl. Microbiol., 2005, 28, 588–595 CrossRef CAS PubMed.
- A. Cresposempere, L. Gonzálezcandelas and P. V. Martínezculebras, Int. J. Food Microbiol., 2010, 142, 170–179 CrossRef CAS PubMed.
- A. Gallo, G. Perrone, M. Solfrizzo, F. Epifani, A. Abbas, A. D. Dobson and G. Mulè, Int. J. Food Microbiol., 2009, 129, 8–15 CrossRef CAS PubMed.
- R. Geisen, M. Schmidt-Heydt and A. Karolewiez, Mycotoxin Res., 2006, 22, 134–141 CrossRef CAS PubMed.
- H. J. Pel, J. H. D. Winde, D. B. Archer, P. S. Dyer, G. Hofmann, P. J. Schaap, G. Turner, R. P. D. Vries, R. Albang and K. Albermann, Nat. Biotechnol., 2007, 25, 221 CrossRef PubMed.
- G. S. Sidhu, Eur. J. Plant Pathol., 2002, 108, 705–711 CrossRef CAS.
- J. Yu, P. K. Chang, K. C. Ehrlich, J. W. Cary, D. Bhatnagar, T. E. Cleveland, G. A. Payne, J. E. Linz, C. P. Woloshuk and J. W. Bennett, Appl. Environ. Microbiol., 2004, 70, 1253 CrossRef CAS PubMed.
- R. Geisen, Z. Mayer, A. Karolewiez and P. Färber, Syst. Appl. Microbiol., 2004, 27, 501–507 CrossRef CAS PubMed.
- H. J. Pel, J. H. D. Winde, D. B. Archer, P. S. Dyer, G. Hofmann, P. J. Schaap, G. Turner, R. P. D. Vries, R. Albang and K. Albermann, Nat. Biotechnol., 2007, 25, 221 CrossRef PubMed.
- P. Farber and R. Geisen, Eur. J. Plant Pathol., 2004, 110, 661–669 CrossRef.
- M. M. Bradford, Anal. Biochem., 1976, 72, 248–254 CrossRef CAS PubMed.
- L. Zhang, Z. Yu, L. Jiang, J. Jiang, H. Luo and L. Fu, J. Proteomics, 2011, 74, 1135 CrossRef CAS PubMed.
- K. J. Livak and T. D. Schmittgen, Methods, 2001, 25, 402–408 CrossRef CAS PubMed.
- J. Zeng, Y. Liu, W. Liu, X. Liu, F. Liu, P. Huang, P. Zhu, J. Chen, M. Shi and F. Guo, PLoS One, 2013, 8, e53409 CAS.
- A. Esteban, M. L. Abarca, M. R. B. Cabañes and F. J. Cabañes, Int. J. Food Microbiol., 2006, 108, 188–195 CrossRef CAS PubMed.
- F. R. Passamani, T. Hernandes, N. A. Lopes, S. C. Bastos, W. D. Santiago, M. D. Cardoso and L. R. Batista, J. Food Prot., 2014, 77, 1947–1952 CrossRef PubMed.
- R. Geisen, Mol. Nutr. Food Res., 2004, 48, 532–540 CAS.
- B. Sun, Z. Wang and F. Zhu, Aquaculture, 2016, 455, 87–96 CrossRef CAS.
- B. Guo, Y. Zhang, H. Li, L. Du, Y. Li, J. Zhang, S. Chen and Z. Zhu, Acta Bot. Sin., 2000, 42, 279–283 Search PubMed.
- B. Mannervik, U. H. Danielson and B. Ketterer, Crit. Rev. Biochem. Mol. Biol., 1988, 23, 283 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2017 |