Open Access Article
Open Access ArticlePerimidine based selective colorimetric and fluorescent turn-off chemosensor of aqueous Cu2+: studies on its antioxidant property along with its interaction with calf thymus-DNA†
Debashis Roya,
Arijit Chakraborty *b and
Rina Ghosh
*b and
Rina Ghosh *a
*a
aDepartment of Chemistry, Jadavpur University, Kolkata 700032, India. E-mail: roy.debashis89@gmail.com; ghoshrina@yahoo.com
bDepartment of Chemistry, Acharya B. N. Seal College, Cooch Behar, West Bengal 736101, India. E-mail: arijit_chak2002@yahoo.com
First published on 21st August 2017
Abstract
We have developed a perimidine based simple and easily synthesised chemosensor, 1,3-bis(2,3-dihydro-1H-perimidin-2-yl)benzene (1), which exhibits selective colorimetric and fluorescence “turn off” response toward nano molar Cu2+. This method allows a sensitive readout of chemosensor 1 within a wide linear range of (5–2771) × 10−8 M at λmax 346 nm, and with limit of detection (LOD) 6.19 × 10−8 M. Moreover, 1 exhibits free radical scavenging ability, six times better than that of L-ascorbic acid. The binding interaction studies of 1 with calf-thymus DNA (CT-DNA) indicate a groove binding mode.
1. Introduction
Cu2+ ions play a vital role in many biological processes such as haemopoiesis, various enzyme catalyzed biochemical reactions e.g. transformation of melanin for pigmentation of the skin and assistance of the formation of crosslinks in collagen and elastin1,2 and assistance in iron absorption. Unregulated overloading of Cu2+ can lead to lethargy, vomiting, increased blood pressure and respiratory rates, liver damage, acute haemolytic anaemia, neurotoxicity, and neurodegenerative diseases including Alzheimer's, Parkinson's and prion diseases.3–5 Furthermore, Cu2+ can entangle natural ecosystems owing to its adverse effects on micro-organisms.6 According to U. S. Environmental Protection Agency (EPA) the consumption limit of Cu2+ in drinking water should not exceed 1.3 ppm (∼20 μM).7,8 For the last 10–20 decades, Cu2+ pollution has underscored major problems all over the world due to the enormous use of Cu2+ based complexes in agriculture and industry.9 Therefore, the development of selective, sensitive, efficient and believable detection methods of aqueous Cu2+ ions have drawn a significant interest to the scientific and environmental communities.Previously, the quantification of the Cu2+ was done via a number of analytical methods including atomic absorption/emission spectroscopy (AAS/AES),10 electrochemical methods,11 inductively-coupled plasma mass spectrometry (ICPMS) and X-ray fluorescence spectroscopy (XRF).12 Nevertheless, these techniques of quantification of the Cu2+ involve the use of sophisticated instrumentation, time consuming sample pre-treatment and separation methods, thus making these inconvenient for the fast detection of aqueous Cu2+ ions.
In current period, all these techniques have been mitigated by new colorimetric as well as spectro-photometric methods. The techniques depend upon UV-vis absorption and fluorescence measurements of fluorophore moieties13–15 which can act as high performance chemo-sensors for metal ion detection. These have drawn much attention for several advantages such as high sensitivity, selectivity and comfortable operation in identifying the metal ions.
Free-radicals play an important role in the oxidative damage of organisms.16,17 An antioxidant is a chemical species that prevents the oxidation of the free-radicals. The oxidative stress induced by reactive oxygen species (ROS) is portrayed as a dynamic imbalance between the levels of free radicals generated in the body and quantity of antioxidants to scavenge them and protect against their harmful effects.18 Increasing amounts of ROS become harmful because these can initiate bio-molecular oxidation which causes cell injury and cell death; oxidative stress leads to countless diseases and disorders such as cataracts, aging, cirrhosis, cancer and atherosclerosis.19 Therefore, an evaluation of antioxidant properties of pure organic compounds is also very important because of their uses in various commodities20 and chemical reactions. DPPH assay is well-known assessment of free radical scavenging by an antioxidant and believed as one of the standard and easy colorimetric methods for the estimation of antioxidant properties of pure organic compounds.21 Herein, we are reporting our recent studies on the selective colorimetric and fluorimetric sensing of a perimidine based compound for detection of nanomolar aqueous Cu2+ solution; the free radical scavenging and the binding of the chemo-sensor with CT-DNA will also be demonstrated.
2. Experimental
2.1. General information
All the solvents and reagents used were of analytical grade and spectroscopic grade. The salts used as metal ions source were AgNO3, Ba(NO3)2, Ca(NO3)2·4H2O, Cd(NO3)2, Co(NO3)2·6H2O, Cr(NO3)3, Cu(NO3)2·3H2O, (NH4)2Fe(SO4)2·6H2O, HgNO3, MnSO4·H2O, NaNO3, Ni(NO3)2·6H2O, Pb(NO3)2 and ZnSO4·7H2O. 1H NMR and 13C NMR measurements were performed on a Bruker DPX-300 (300 and 75 MHz, respectively) spectrometer, and chemical shifts were recorded in ppm. FT-IR data were taken from PerkinElmer Spectrum Version 10.03.09. A high-resolution mass spectrum was obtained on a Waters Micromass Q-tof Micro mass spectrometer using electrospray ionization method. Melting points were determined on a LabX India digital melting point apparatus. UV-vis spectra were recorded at room temperature using a PerkinElmer UV Lambda 25 spectrometer, and fluorescence data were collected from PerkinElmer fluorescence Spectrometer LS 55. Time resolved fluorescence decay measurements were recorded in Horiba-Jobin Yvon FluoroCube fluorescence lifetime system using NanoLED at 330 nm (IBH, UK) as the excitation source and TBX photon detection module as the detector. Circular dichroism (CD) spectral measurements were run on a PC-driven JASCO J815 spectropolarimeter (Jasco International Co., Hachioji, Japan) with an attached temperature controller using a rectangular quartz cuvette of 1 cm path length.2.2. Synthesis of 1,3-bis(2,3-dihydro-1H-perimidin-2-yl) benzene (1)22
Isophthalaldehyde (1 g, 7.46 mmol) and 1,8-diaminonaphthalene (2.3587 g, 14.9 mmol) were added to 30 mL of ethanol, and the reaction mixture was refluxed for 4 hours. A rosy brown solid product was separated out from the reaction medium, and the product was filtered and washed with ethanol, distilled water and dichloro methane, successively for 2 times to get pure product. Yield: 2.9374 g (95%). Mp = 224 °C.IR spectrum (cm−1, KBr pellet): 3370 (NH str.), 3044 (CH str.), 1597 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C str.), 1482, 1425, 1414, 1252, 815, 760.1H NMR (CDCl3, 300 MHz): δ 7.96 (1H, s, H-Ar), 7.72 (2H, d, J = 7.4, H-Ar), 7.52 (1H, t, J = 7.6, H-Ar), 6.54–6.56 (4H, m, H-Ar), 5.54 (2H, s, CH), 4.54 (2H, br, s, NH), 1.63 (2H, br, s, NH). 13C NMR (75 MHz, CDCl3): δ 141.9, 141.0, 134.9, 129.4, 129.0, 127.3, 126.9, 118.1, 113.5, 106.0, 68.1. HRMS: found [M + H]+ 415.1914; ‘molecular formula C28H22N4’ requires [M + H]+415.1923.
C str.), 1482, 1425, 1414, 1252, 815, 760.1H NMR (CDCl3, 300 MHz): δ 7.96 (1H, s, H-Ar), 7.72 (2H, d, J = 7.4, H-Ar), 7.52 (1H, t, J = 7.6, H-Ar), 6.54–6.56 (4H, m, H-Ar), 5.54 (2H, s, CH), 4.54 (2H, br, s, NH), 1.63 (2H, br, s, NH). 13C NMR (75 MHz, CDCl3): δ 141.9, 141.0, 134.9, 129.4, 129.0, 127.3, 126.9, 118.1, 113.5, 106.0, 68.1. HRMS: found [M + H]+ 415.1914; ‘molecular formula C28H22N4’ requires [M + H]+415.1923.
2.3. UV-vis titration of 1 with Cu2+ ion23,24
6.26 mM solution of 1 in acetonitrile and 2.5 mM solution of Cu(NO3)2·3H2O in water were prepared as stock solutions. 2 μL of 1 (6.26 mM) was diluted to 2 mL acetonitrile in a cuvette, and the UV-vis spectrum was run. Then 2 μL of Cu2+ solution (2.5 mM) was added to the same cuvette; after mixing these homogeneously, UV-vis absorption spectrum was run at room temperature. This process of addition of Cu2+ solution (2.5 mM) was repeated for several times upto saturation.2.4. Fluorescence spectroscopic titration of 1 with Cu2+ ion25,26
5 μL of the 1 (0.25 mM) was diluted to 2 mL acetonitrile, and 1 μL of the aqueous Cu2+ solution (0.25 mM) was added each time to the diluted 1 solution. After mixing homogeneously, fluorescence spectra were recorded at room temperature by exciting 1 at 346 nm. This process was run for several times upto saturation.2.5. Job plot measurements of 1 with Cu2+ ion23
A solution (5 mM) of 1 in acetonitrile and a Cu2+ (copper nitrate trihydrate) solution (5 mM) in water were prepared as stock solutions. Then 1, 2, 3, 4, 5, 6, 7, 8 and 9 μL from the stock solution of 1 were taken and transferred to different vials. 9, 8, 7, 6, 5, 4, 3, 2 and 1 μL of the Cu2+ solution (5 mM) were added respectively, to the above mentioned vials so that the mole fraction of 1 in each vial became 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8 and 0.9, respectively. Each vial was diluted with acetonitrile to make a total volume of 2 mL. After shaking the vials for a few seconds to form 1-Cu2+ complex, UV-vis absorption spectra were recorded at 25 °C. Similar process was run for blank test with solution of 1 only.2.6. pH dependency of 127
A series of buffer solutions with pH values ranging from 1 to 10 were prepared. 2 μL of a solution (2 mM) of 1, prepared in acetonitrile, was added to 2 mL buffer of each pH. After mixing it homogeneously, fluorescence spectra (excited at 346 nm) were recorded for all pH ranging from 1 to 10 at room temperature.2.7. Evaluation of antioxidant activity by DPPH assay21
Step 1: 0.01 (M) methanolic solution of each of 1, DPPH (1,1-diphenyl-2-picrylhydrazyl) and L-ascorbic acid were prepared by accurate weighing. Then, the solution of 1 was serially diluted to 1/2, 1/4, 1/8, 1/16 and 1/32.Step 2: to seven sample vials 10 μL of 0.01 (M) DPPH solution was added. Among these, to the six sample vials 10 μL of serially diluted solutions of 1 prepared in step 1, was added. To the rest sample vial 10 μL of methanol was added. This seventh vial served as the blank. Then the content of each vial was diluted to 2 mL. After mixing homogenously, these seven solutions were allowed to react for 40 minutes at room temperature under dark. UV-vis absorption spectra of the seven solutions were run at room temperature, and the corresponding absorbance values were read at 517 nm.
Step 3: similar procedure of step 2 was also followed for L-ascorbic acid as standard.
2.8. UV-vis titration of 1 with CT-DNA28
5 mM solution of 1 in acetonitrile and 1 mM solution of CT-DNA in citrate-phosphate buffer of pH 7 were prepared as stock solutions. 5 μL of the stock solution of 1 was diluted to 2 mL citrate-phosphate buffer (pH 7) in cuvette, and the UV-vis spectrum was taken. Then 5 μL of CT-DNA (1 mM) solution was added to the same cuvette, after mixing these homogeneously, UV-vis absorption spectrum was run at room temperature. This process of addition of CT-DNA solution (1 mM) was repeated for several times upto saturation.2.9. Fluorescence spectroscopic titration of 1 with CT-DNA29
2 μL of the 1 (5 mM) was diluted to 2 mL citrate-phosphate buffer of pH 7, and 2 μL of the CT-DNA (1 mM) solution was added each time to the diluted solution of 1. After mixing homogeneously, fluorescence spectra were recorded at room temperature by exciting 1 at 346 nm. This process was run for several times upto saturation.3. Results and discussion
3.1. Synthesis and characterisation
The chemo-sensor 1 was synthesised by coupling of isophthalaldehyde with 1,8-diaminonaphthalene in absolute ethanol at refluxing condition following literature procedure22 with some modification (Scheme 1); a better yield of 1 was obtained by this modification. Compound 1 was characterised by 1H NMR, 13C NMR, FT-IR and high resolution mass spectrometry (Fig. S1–S4, ESI†).3.2. Visual and spectral changes of chemosensor 1 towards cations
Due to the poor solubility of compound 1 in water, the colorimetric sensing abilities of 1 were investigated in acetonitrile medium (40 μM) upon the treatment with various kinds of aqueous cations such as Ag+, Ba2+, Ca2+, Cd2+, Co2+, Cr3+, Cu2+, Fe2+, Hg+, Mn2+, Na+, Ni2+, Pb2+ and Zn2+. To examine the spectral changes of 1 toward cations, a solution of 1 was treated with above mentioned cations (Fig. 1).Upon the addition of aqueous Cu2+ to the solution of 1 (6.25 μM, λmax = 346 nm, molar extinction co-efficient (ε) = 2.72 × 104 M−1 cm−1 in acetonitrile), a change of colourless to chocolate colour was observed; other cations did not produce any visual change. Subsequently a change in the UV-vis absorption peak with a bathochromic shift at 475 nm and isosbestic points at 239 nm, 301 nm and 357 nm indicate the formation of a stable complex with Cu2+ ion (Fig. 1). Thus compound 1 is no-doubt a potential candidate for “naked-eye” chemo-sensor of Cu2+ ion.
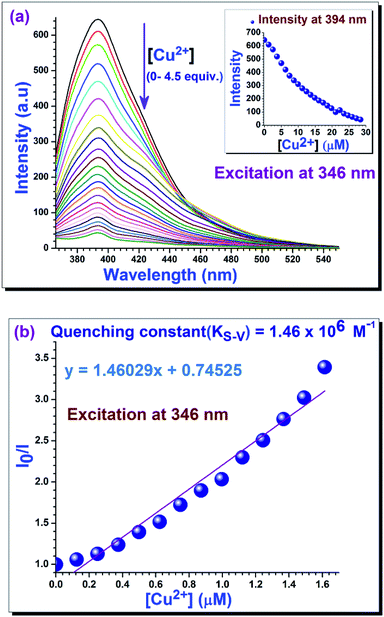
To establish fluorescent properties of 1 toward aqueous Cu2+, the emission changes were investigated. Upon gradual addition of aqueous Cu2+ to the solution of 1 in acetonitrile medium, an amazing fluorescent quenching was observed with quenching efficiency = 93% (Fig. 2).30
For collisional quenching the dramatic decrease in intensity was described by the well-known Stern–Volmer equation (eqn (1)):31
 | (1) |
3.3. Determination of binding constant
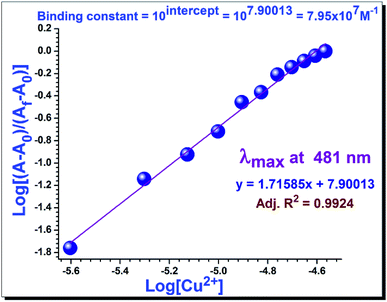
The extent of the binding of 1 toward aqueous Cu2+ ion is calculated from an experimental plot of the absorption data using the Benesi–Hildebrand relation (eqn (2)).32 According to this relation
 | (2) |
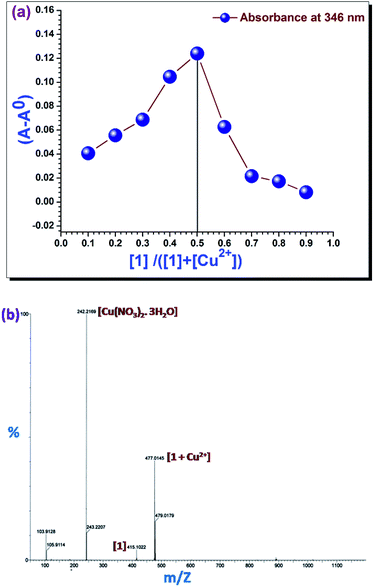
3.4. Job plot analysis
The Job plot analysis (Fig. 4a) disclosed a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometric ratio between 1 and Cu2+ ion when difference in absorption maxima were plotted against mole fraction of 1.33 This was further corroborated by the mass spectrum (Fig. 4b) of a mixture of 1 and Cu2+ in acetonitrile (1
1 stoichiometric ratio between 1 and Cu2+ ion when difference in absorption maxima were plotted against mole fraction of 1.33 This was further corroborated by the mass spectrum (Fig. 4b) of a mixture of 1 and Cu2+ in acetonitrile (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1).
1).
3.5. Determination of detection limits34
Using UV-vis titration experiment of 1 in acetonitrile solution with aqueous Cu2+ (Fig. S6, ESI†), the detection limit was calculated using the equation: detection limit = 3σ/m, where σ is the standard deviation of the blank solution (6.25 μM solution of 1 in acetonitrile only, O.D. at 346 nm), and m is the slope of the intensity versus [Cu2+] calibration curve. The detection limits of 1 toward Cu2+ ion is 6.19 × 10−8 M.3.6. Life time measurement
We measured the consequence of aqueous Cu2+ on the fluorescence decay behaviour of 1 (Fig. 5). The mean fluorescence lifetime of 1 in acetonitrile was 0.912 ns whereas, a shorter mean fluorescence lifetime 0.639 ns was observed in the presence of Cu2+ ion. We have also calculated the radiative rate constant (kr) and the total non-radiative rate constant (knr) of 1 and (1 + Cu2+) using the following equations:35| τ−1 = kr + knr | (3) |
| kr = Φ/τ | (4) |
 | ||
| Fig. 5 Time-resolved fluorescence decay curves (logarithm of normalized intensity vs. time in ns) of 1 in the absence and presence of aqueous Cu2+ ion (3 equiv.). | ||
3.7. Quantum yield measurements36,37
Quantum yield of 1 was measured by a secondary method using fluorescence spectra of 1 in the absence and in the presence of aqueous Cu2+ in acetonitrile medium using eqn (5), where ODs and ODref are the absorbances, and As and Aref are integrated emission areas of 1 and standard, respectively. A quantum yield 0.54, for quinine sulfate dihydrate in 0.1 N H2SO4, has been used as a standard. The quantum yield of 1 and 1 + Cu2+ are 0.1 and 0.0058, respectively.
 | (5) |
On absorbing photon the fluorophore 1 initiates radiating from the new excited singlet electronic state. During this time, the fluorophore 1 undergoes conformational changes and is also subject to interactions with its molecular environment. The energy of the excited state is partially dissipated, yielding a relaxed singlet excited state from which fluorescence emission originates. In case of 1, most of the absorbed energy may be engaged in conformational changes and also for the interaction with environment. Hence, lower amount of energy is emitted giving lower value of quantum yield for 1 only. The decrease in fluorescence intensity may be due to interaction of excited state of the fluorophore 1 with Cu2+ ion.
3.8. pH dependency of 1
For many biological applications, it is very important that the chemo-sensor shows no change in the physiological pH range.Therefore, we investigated the effect of pH ranging from 1 to 10 (Fig. 6).27 A sharp intensity change was observed between pH 1 to 2, but there was no appreciable change in the intensity beyond pH 2, except a minute increase at pH 8; the reason for this increase is not clear. To get the intensity changes of 1, it was excited at 346 nm for all pH values.
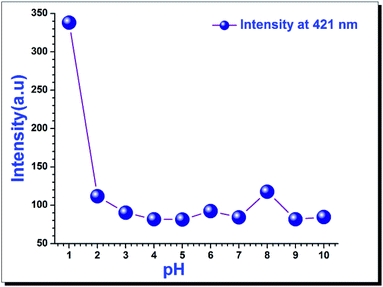 | ||
| Fig. 6 Intensity of 1 (2 μM) at 421 nm being excited at 346 nm in buffer solutions of various pH ranges (1 to 10). | ||
3.9. Solvatochromism38,39
UV-vis absorption spectra and fluorescence emission spectra of 1 in a series of solvents of varying polarity are shown in Fig. 7. In aprotic solvents, the absorption maximum was progressively red shifted with increase in the polarity of the solvent, whereas for protic solvents it became blue shifted (Fig. 7a). On the other hand no considerable changes in the fluorescence emission maxima were observed in solvents of varying polarities. Although an increase in the fluorescence intensity was observed with increasing polarity of aprotic solvents, however there is a sharp decrease in emission intensity in case of the protic solvents (Fig. 7b). The fluorescence quenching in methanol and ethanol indicates the presence of an efficient non-radiative decay process from the emitting excited state of compound 1 in protic solvent. The increased probability of non-radiative decay may be attributed to the hydrogen bonding of N–H group with the protic solvents.40 | ||
| Fig. 7 (a) Absorption changes of 1 (7.2 μM) in various solvents at room temperature. (b) Intensity changes of 1 (0.5 μM) by exciting at 346 nm in various solvents at room temperature. | ||
3.10. Antioxidant activity
This experiment is based on the extent of the scavenging capacity of antioxidant 1 towards 1,1-diphenyl-2-picrylhydrazyl (DPPH). The odd electron of N in DPPH is reduced by getting hydrogen atom from antioxidant 1 to the corresponding hydrazine.41 While DPPH can accept hydrogen radical or an electron for getting stability, 1 is irreversibly oxidised. Due to having an odd electron, DPPH shows absorption maxima at 517 nm, and its solution appears deep violet, but the absorption vanishes when the electron pairs off. The methanolic solution of 50 μM DPPH is intensely coloured, and at this concentration, the Lambert–Beer's law is obeyed over the useful range of absorbance. Equation of free radical scavenging ratio:
 | (6) |
3.11. Interaction of 1 with CT-DNA28,29
Upon the addition of aqueous CT-DNA to the solution of 1 in citrate-phosphate buffer medium of pH 7, there occurs a slight absorption changes with an isosbestic point at 292 nm (Fig. 9a). The presence of the isosbestic point gives a hint that there is equilibrium between the free 1 and the CT-DNA bound 1 in the ground state. A poor binding constant with value 4.6 × 102 M−1 at 357 nm (Fig. 9b) suggests groove binding of the 1 with CT-DNA. Furthermore, the fluorescence quenching spectra disclose the interaction of 1 with CT-DNA more precisely, and a low quenching efficiency (26%) also indicates the groove binding because intercalation of small molecules into the DNA base stack usually shows a huge change of the absorbance value as well as of fluorescence intensity.We have then measured the circular dichroism (CD) spectra to ensure the mode of binding of 1 with CT-DNA. The secondary structure of DNA is recognized to be perturbed effectively by the intercalation with small molecules by giving the changes in the intrinsic CD spectrum of DNA, whereas groove binding of the small molecules has inconsiderable impact on the CD profile diagram of DNA. The CD spectra of CT-DNA prominently display that successive addition of 1 to the CT-DNA solution does not present any meaningful change in the CD spectrum of CT-DNA implying that the secondary structure of the CT-DNA remains unaffected on binding with 1.
This investigation eliminates the chance of intercalation of 1 in the DNA helix, and hence establishes the groove binding mode.
4. Conclusion
In summary, we have developed a perimidine based new colorimetric sensor for the detection of aqueous Cu2+ ion, as the third most abundant transition metal ion in human bodies. The chemo-sensor 1 can also be used as a highly selective and sensitive, naked-eye and fluorescence turn-off sensor for aqueous Cu2+ ion; it enables analysis of aqueous Cu2+ ion with a detection limit of 6.19 × 10−8 M, which is below the WHO acceptable limit (31.5 × 10−3 M).23 Moreover, the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 interaction of 1 with Cu2+ was chemically irreversible in the presence of EDTA and CN− which suggests strong binding between 1 and Cu2+. Furthermore, 1 acts as a better antioxidant (six times) than L-ascorbic acid. We have also investigated the interaction of this chemo-sensor with calf-thymus DNA in the ground state as well as in the higher excited state and established the binding mode of 1 in the DNA helix. Overall, the present work is a novel impute to the field of chemo-sensing.
1 interaction of 1 with Cu2+ was chemically irreversible in the presence of EDTA and CN− which suggests strong binding between 1 and Cu2+. Furthermore, 1 acts as a better antioxidant (six times) than L-ascorbic acid. We have also investigated the interaction of this chemo-sensor with calf-thymus DNA in the ground state as well as in the higher excited state and established the binding mode of 1 in the DNA helix. Overall, the present work is a novel impute to the field of chemo-sensing.
Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
Financial supports from SERB, DST, India (SR/FT/CS-116/2010) and CSIR, India (02/0186/14/EMR-II) are acknowledged, respectively by AC and RG. The authors are thankful to the Department of Chemistry, Maulana Azad College, Kolkata for supporting with some instrumental facilities and to FIST-DST and UGC-CAS, India for providing with the instrumental facilities in the Department of Chemistry, Jadavpur University.References
- D. Udhayakumari, S. Velmathi, Y. M. Sung and S. P. Wu, Sens. Actuators, B, 2014, 198, 285–293 CrossRef CAS
.
- P. L. Malvankar and V. M. Shinde, Analyst, 1991, 116, 1081–1084 RSC
.
- H. Xu, X. Wang, C. Zhang, Y. Wu and Z. Liu, Inorg. Chem. Commun., 2013, 34, 8–11 CrossRef CAS
.
- C. Vulpe, B. Levinson, S. Whitney, S. Packman and J. Gitschier, Nat. Genet., 1993, 3, 7–13 CrossRef CAS PubMed
.
- P. C. Bull, G. R. Thomas, J. M. Rommens, J. R. Forbes and D. W. Cox, Nat. Genet., 1993, 5, 327–337 CrossRef CAS PubMed
.
- R. Kramer, Angew. Chem., Int. Ed., 1998, 37, 772–773 CrossRef CAS
.
- W. T. Tak and S. C. Yoon, KSN, 2001, 20, 863–865.
- J. W. Liu and Y. Lu, J. Am. Chem. Soc., 2007, 129, 9838–9839 CrossRef CAS PubMed
.
- L. Kiaune and N. Singhasemanon, Rev. Environ. Contam. Toxicol., 2011, 213, 1–26 CAS
.
- A. Tong, Y. Akama and S. Tanaka, Analyst, 1990, 115, 947–949 RSC
.
- T. Poursaberi, L. H. Babaei, M. Yousefi, S. Rouhani, M. Shamsipur, M. K. Razi, A. Moghimi, H. Aghabozorg and M. R. Ganjali, Electroanalysis, 2001, 13, 1513–1517 CrossRef CAS
.
- G. P. C. Rao, K. Seshaiah, Y. K. Rao and M. C. Wang, J. Agric. Food Chem., 2006, 54, 2868–2872 CrossRef CAS PubMed
.
- M. Royzen, Z. H. Dai and J. W. Canary, J. Am. Chem. Soc., 2005, 127, 1612–1613 CrossRef CAS PubMed
.
- Y. Zhao, X. B. Zhang and Z. X. Han, Anal. Chem., 2009, 81, 7022–7030 CrossRef CAS PubMed
.
- S. J. Lee, J. E. Lee and J. Seo, Adv. Funct. Mater., 2007, 17, 3441–3446 CrossRef CAS
.
- J. M. McCord, Am. J. Med., 2000, 108, 652–659 CrossRef CAS PubMed
.
- A. Scalbert, C. Manach, C. Morand and C. Remsy, Crit. Rev. Food Sci. Nutr., 2005, 45, 287–306 CrossRef CAS PubMed
.
- A. Shirwaikar, A. Shirwaikar, R. Kuppusamy and S. R. Isaac, Biol. Pharm. Bull., 2006, 29, 1906–1910 CAS
.
- B. Halliwell and J. M. C. Gutteridge, Free radicals in biology and medicine, Oxford University Press, Oxford, 2000 Search PubMed
.
- F. Anwar, M. Ali, A. I. Hussain and M. Shahid, Flavour Fragrance J., 2009, 24, 170–176 CrossRef CAS
.
- J. Deng, W. Cheng and G. Yang, Food Chem., 2011, 125, 1430–1435 CrossRef CAS
.
- I. G. Jung, S. U. Son, K. H. Park, K. C. Chung, J. W. Lee and Y. K. Chung, Organometallics, 2003, 22, 4715–4720 CrossRef CAS
.
- H. Y. Jo, G. J. Park, Y. J. Na, Y. W. Choi, G. R. You and C. Kim, Dyes Pigm., 2014, 109, 127–134 CrossRef CAS
.
- R. L. Liu, H. Y. Lu, M. Li, S. Z. Hu and C. F. Chen, RSC Adv., 2012, 2, 4415–4420 RSC
.
- H. Xu, X. Wang, C. Zhang, Y. Wu and Z. Liu, Inorg. Chem. Commun., 2013, 34, 8–11 CrossRef CAS
.
- G. K. Li, Z. X. Xu, C. F. Chen and Z. T. Huang, Chem. Commun., 2008, 1774–1776 RSC
.
- J. Reijenga, A. V. Hoof, A. V. Loon and B. Teunissen, Anal. Chem. Insights, 2013, 8, 53–71 CrossRef CAS PubMed
.
- P. Kundu, S. Ghosh and N. Chattopadhyay, Phys. Chem. Chem. Phys., 2015, 17, 17699–17709 RSC
.
- D. Sarkar, P. Das, S. Basak and N. Chattopadhyay, J. Phys. Chem. B, 2008, 112, 9243–9249 CrossRef CAS PubMed
.
- R. M. Duke, E. B. Veale, F. M. Pfeffer, P. E. Kruger and T. Gunnlaugsson, Chem. Soc. Rev., 2010, 39, 3936–3953 RSC
.
- A. Misra, M. Shahid, P. Dwivedi, P. Srivastava, R. Ali and S. S. Razi, ARKIVOC, 2013, ii, 133–145 Search PubMed
.
- M. Dong, T. H. Ma, A. J. Zhang, Y. M. Dong, Y. W. Wang and Y. Peng, Dyes Pigm., 2010, 87, 164–172 CrossRef CAS
.
- J. Li, H. Lin, Z. Cai and H. Lin, J. Lumin., 2009, 129, 501–505 CrossRef CAS
.
- F. U. Rahman, A. Ali, S. K. Khalil, R. Guo, P. Zhang, H. Wang, Z. T. Li and D. W. Zhang, Talanta, 2017, 164, 307–313 CrossRef CAS PubMed
.
- A. B. Pradhan, S. K. Mandal, S. Banerjee, A. Mukherjee, S. Das, A. R. K. Bukhsh and A. Saha, Polyhedron, 2015, 94, 75–82 CrossRef CAS
.
- W. H. Melhuish, J. Phys. Chem., 1961, 65, 229–235 CrossRef CAS
.
- D. F. Eaton, Pure Appl. Chem., 1988, 60, 1107–1114 CrossRef CAS
.
- J. Jayabharathi, V. Thanikachalam, M. V. Perumal and K. Jayamoorthy, J. Fluoresc., 2012, 22, 213–221 CrossRef CAS PubMed
.
- S. Cha, M. G. Choi, H. R. Jeon and S. K. Chang, Sens. Actuators, B, 2011, 157, 14–18 CrossRef CAS
.
- J. D. Cheon, T. Mutai and K. Araki, Org. Biomol. Chem., 2007, 5, 2762–2766 CAS
.
- S. B. Kedare and R. P. Singh, J. Food Sci. Technol., 2011, 48, 412–422 CrossRef CAS PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra06687b |
| This journal is © The Royal Society of Chemistry 2017 |