Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Effect of pRi T-DNA genes and elicitation on morphology and phytoecdysteroid biosynthesis in Ajuga bracteosa hairy roots†
Waqas Khan Kayani‡
 a,
Javier Palazònb,
Rosa M. Cusidòb and
Bushra Mirza‡
a,
Javier Palazònb,
Rosa M. Cusidòb and
Bushra Mirza‡ *a
*a
aDepartment of Biochemistry, Faculty of Biological Sciences, Quaid-i-Azam University, Islamabad 45320, Pakistan. Fax: +92-51-90644050; Tel: +92-51-90643007
bPlant Physiology Laboratory, Faculty of Pharmacy, University of Barcelona, Avda Joan XXIII s/n, 08028 Barcelona, Spain. Fax: +34 93 402 90 43; Tel: +34 93 402 0267
First published on 12th October 2017
Abstract
The medicinal plant Ajuga bracteosa is a rich source of biologically active and metabolism-enhancing phytoecdysteroids. Transgenic hairy roots from A. bracteosa were obtained by infection with Agrobacterium rhizogenes strains A4, LBA-9402 and ARqua1. The 59 selectively established and rolC-positive hairy root lines increased in size up to 6.6-fold (L42) after one month of in vitro culturing and produced a phytoecdysteroid content ranging from 69.3 to 4449 μg g−1 (L3 and A2, respectively). The clones of these transgenic roots were maintained for successive subcultures on hormone-free medium to obtain a stabilized morphology. Hairy roots displayed four different morphologies: typical hairy root (THR) (59%), callus-like (CM) (17%), thick (TK) (14%) and thin (TN) (10%). The growth rate of the transgenic hairy root lines varied according to their morphology, with CM showing the highest rate (3.93-fold per month). However, THR exhibited the highest phytoecdysteroid content (1538.5 μg g−1). Considering the hairy root morphology in relation to the pRi TR-DNA genes, 100% of the hairy root lines harbored the mas1 and ags genes and 90% of CM lines the aux1 gene. Strikingly, the clones with TK morphology were all positive for the TL- and TR-DNA genes. In contrast, 0% of TN clones harbored the aux1 and ags genes, and only 16.7% the mas1 gene. All CM and TK hairy roots were positive for the ags gene, while mas1 was present in all hairy root types except TN. As expected, all the root lines considered in this work were positive for the rol C gene. Eleven hairy root lines displaying a high growth rate and phytoecdysteroid content were elicited with methyl jasmonate (MeJ) and coronatine (Cor). MeJ doubled the phytoecdysteroid content after 14 days of elicitation (8356 μg g−1 in L2) compared with unelicited control hairy roots, and in in vitro-grown untransformed roots the increase was 5.6-fold.
Introduction
Ajuga bracteosa (Lamiaceae) is a perennial medicinal herb of the sub-tropical and temperate regions of Kashmir, Bhutan, Pakistan, Afghanistan, China and Malaysia and highly used in folk medicine.1 Studies report that A. bracteosa alleviates liver fibrosis,2 possesses promising anti-inflammatory activity,3,4 and exhibits a dose-dependent inhibition of chronic arthritis in rats5 and analgesic effects in mice.3,6 Extracts of A. bracteosa are valuable antiplasmodial agents.7,8Major compounds found in A. bracteosa are phytoecdysteroids, neo-clerodane diterpenoids, withanolides and iridoid glycosides. Phytoecdysteroids are structural analogs of the insect molting hormone ecdysone. In plants, these compounds are responsible for some physiological functions, while in mammals they have a huge variety of applications. Phytoecdysteroids increase protein-synthesizing processes,9 body mass10 and blood protein content in rats,10 improve kidney functioning,11 suppress albuminuria,12 activate human lymphocytes,13 reduce lipid peroxidation,14 and increase the copulative function and improve sperm quality.15 They are neuroprotective,16 anti-hyperglycemic,17 anti-inflammatory,18 anti-diabetic19 and also possess antifungal and antibacterial activity.20
Hairy roots are a valuable biotechnological tool for the production of plant secondary metabolites due to their high productivity and growth rate, and genetic stability.21 Agrobacterium rhizogenes infect plants and generate hairy roots in response to the transfer and integration of DNA (T-DNA) from the large root-inducing agrobacterial plasmid (pRi) into the plant genome.22,23 Agropine type strains contain two T-DNA regions on the Ri plasmid known as TL-DNA and TR-DNA, which can be independently incorporated into the plant genome.22,24 The Ri TL-DNA carrying rolA, B, C and D genes is responsible for hairy root induction,24,25 while Ri TR-DNA possesses the genes responsible for opine biosynthesis,26 as well as those involved in the additional route for the formation of indole acetic acid. The TR-region is also important in the determination of the hairy root morphology.27,28 Elicitation of hairy roots can further enhance the biosynthesis of valuable secondary metabolites.21
Phytoecdysteroid biosynthesis in A. reptans hairy roots was found to be closely related to their growth: in a root line (Ar-4) that increased in weight 230-fold, the content of 20-hydroxyecdysone (20-HE) increased 4-fold after 45 days of culture.29 The regenerated plants from these hairy roots showed altered plant morphology30 but the same production of 20-HE as in original root line.31 Hairy roots obtained from A. multiflora produced 10 times more 20-HE compared to the wild type.32 Moreover, elicitors are found to induce higher phytoecdysteroid biosynthesis in Spinacia oleracea plants.33
To the best of our knowledge, in the present study, we evaluated for the first time the potential of transformed roots of A. bracteosa to produce phytoecdysteroids and the effect of elicitation. Moreover, the phenotypic effects of the integration of TR-DNA genes from A. rhizogenes into the hairy root genome were also investigated.
Methods
Hairy roots cultures
Hairy roots of Ajuga bracteosa were obtained by the transformation of the plant genome with three agropine Agrobacterium rhizogenes strains: A4, LBA-9402 and ARqua1 as described by Kayani, et al.34 Briefly, leaves with petioles at the proximal end were surface sterilized and infected with YEB (yeast extract and beef medium)-grown agrobacteria. Explants were infected with agrobacteria adhered to the surface of a sterile surgical blade. The inoculated leaf discs were placed on Schenk–Hildebrandt (SH) medium35 with a pH of 7 and containing 3% sucrose, 0.1% myoinositol and 0.8% phytagel at 27 °C in the dark. After a 2 day co-cultivation, the explants were shifted to new SH medium supplemented with claforan (500 mg l−1). The hairy roots were retained with the mother explants for 20–30 days, and were then excised and grown on optimized hormone-free half-strength Murashige and Skoog (MS) medium36 (half-strength of salts) supplemented with 30 g l−1 sucrose and solidified with 0.8% phytagel. Fifty nine hairy root clones were used to study growth and the phytoecdysteroid profile, and eleven of these (elite root clones in growth and production) were maintained in the growth room in darkness at 27 °C and routinely subcultured every 2 weeks.Growth quotient
To screen actively growing hairy roots, 200 mg of fresh weight of root inoculum was cultured on half-strength media for one month. After the incubation period, each root line was harvested and its fresh weight measured. The growth quotient was obtained by dividing the hairy root fresh weight by the fresh weight of the inoculum. For the extraction of ecdysteroids, the harvested hairy roots were freeze-dried, weighed (dry weight) and ground to a fine powder. The powder was stored at −20 °C until further processing.Treatment with elicitors
Methyl jasmonate (MeJ) and coronatine (Cor) (Sigma-Aldrich, St. Louis, MO, USA) were added to the medium optimized for stable growth and production (half-strength MS) prior to inoculation. Both the elicitors were filter-sterilized (0.22 μm sterile PES filters, Millipore, Billerica, MA, USA) and added to the media just before pouring to make a final concentration of 100 μM MeJ and 1 μM Cor. 200 mg of fresh inoculum of each hairy root line was cultured for 14 and 21 days. Each treatment was conducted in triplicate, and to compare the effect of elicitors, control (unelicited) hairy roots were treated with 2.5 ml ethanol (MeJ control). For analysis, three plates for each root line studied were harvested after 14 and 21 days of elicitation.Molecular analysis
For the PCR analysis, genomic DNA was isolated from hairy roots and control roots (untransformed) by an optimized method37 and its quantitative and qualitative parameters were checked by NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies Wilmington, DE, USA). Integration of TL-DNA into the genome of hairy roots was confirmed by the PCR of the rolC gene. To ensure the hairy root clones were free from agrobacterial contamination, PCR was carried out to confirm the absence of the virD1 gene. TR-DNA integration into the hairy root genome was confirmed by the PCR analysis of aux1, mas1, and ags genes. The rolC and actin genes were used in the expression analysis by sq-RT-PCR. Total RNA was isolated with the TRIzol® Plus RNA Purification Kit (Life Technologies, Germany) following the manufacturer's instructions. cDNA was synthesized with SuperScript II reverse transcriptase (Invitrogen, Carlsbad CA) according to their instructions. PCR Master Mix (Life Technologies, Spain) was used for the PCR reaction in a thermocycler (Perkin-Elmer Gene Amp PCR System 9600, USA). The genes, primers, primer sequences and PCR conditions are described in Table 1. PCR conditions were fixed except for the annealing temperature. They consisted of: 5 minutes at 95 °C; 35 cycles of 35 s at 95 °C, 35 s (primer annealing temperature given in Table 1), and 1 min at 70 °C; and 10 min at 70 °C. Amplified PCR products were resolved at 1.5 percent (w/v) agarose gel electrophoresis in TBE running buffer and visualized with UV-Trans illuminator (Life Technology, USA).| Gene | Sequence | Size (bp) | Tm (°C) |
|---|---|---|---|
| a PCR conditions were fixed except for annealing temperature. They were; 5 minutes 95 °C, followed by 35 cycles of 35 seconds at 95 °C, 35 s for primers annealing (temperature given in table), extension of 1 min at 70 °C and a final extension of 10 min at 70 °C. | |||
| rolC | F:5′-TAACATGGCTGAAGACGACC-3′, R:5′-AAACTTGCACTCGCCATGCC-3′ | 534 | 60 |
| virD1 | F:5′-ATGTCGCAAGGCAGTAAGCCC-3′, R:5′-GAAGTCTTTCAGCATGGAGCA-3′ | 438 | 56 |
| ags | F:5′-GGCGTGAGCACCTCATATCCG-3′, R:5′-TTCGAAGCCTTTGCCTGCAAA-3′ | 347 | 62 |
| mas1 | F:5′-ACCTTGGTACTGCCCAGCCAC-3′, R:5′-CTTCAGTGGTCCATACCCACC-3′ | 343 | 62 |
| aux1 | F:5′-ATGTCGCAAGGCAGTAAGCCC-3′, R:5′-GAAGTCTTTCAGCATGGAGCA-3′ | 438 | 56 |
| actin (SQ-RT-PCR) | F:5′-ATCAGCAATACCAGGGAACATAGT-3′, R:5′-AGGTGCCCTGAGGTCTTGTTCC-3′ | 160 | 60 |
| rolC (SQ-RT-PCR) | F:5′-CTGTACCTCTACGTCGACT-3′, R:5′-AAACTTGCACTCGCCATGCC-3′ | 363 | 62 |
Extraction of ecdysteroids
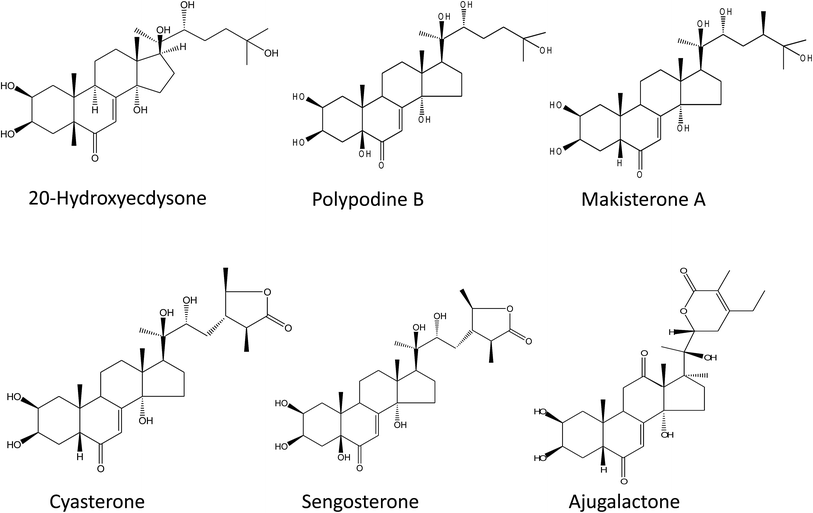
The six ecdysteroids screened in the study were 20-hydroxyecdysone (20-HE), ajugalactone (AJL), sengosterone (SG), cyasterone (CY), polypodine (PoB) and makisterone A (MKA) (Fig. 1).3,34 For the ecdysteroid extraction, a previously optimized protocol38 was followed with some modifications.34 In brief, powdered plant material (∼500 mg) was extracted first with 10 ml of methanol (MeOH) two times and then the residue was extracted again with 85% MeOH and both fractions (100% MeOH and 85% MeOH) were mixed and dried. The resulting pellet was resuspended in 85% MeOH, sonicated for 20 min and subjected to partial purification through previously activated/equilibrated column cartridges (Strata C18-E, 55 μm, 70 A, Phenomenex, USA) with 85% MeOH. The filtrate obtained was dried, and the pellet was resuspended in 5 mL of 85% MeOH and used for the HPLC injection.RP-HPLC analysis
Analytical HPLC was performed according to already optimized protocols34,39 and the mobile phases were water (A) and acetonitrile (B). The HPLC system, chromatographic column with its dimensions, injection volume, flow rate, the gradient program and the acquisition of UV spectra were same as described before.34Results and discussion
Phenotypic characterization and phytoecdysteroid content in transgenic roots
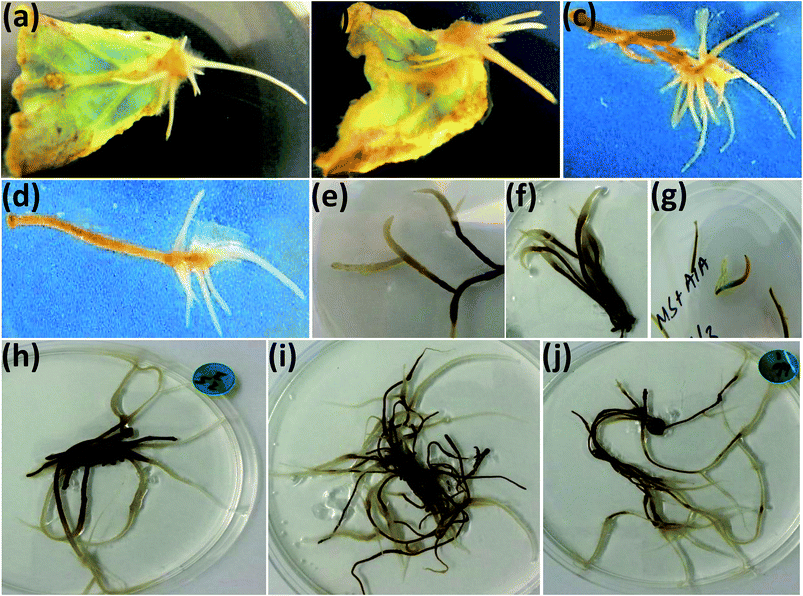
Transgenic roots were observed in the proximal end of A. bracteosa leaves infected with A. rhizogenes strains A4, LBA-9402 and ARqua1 carrying pRi within 9–12 days (Fig. 2a–d). The cultures were maintained for successive subcultures without changes in morphology. Among the selected hairy root lines, two were obtained from the A4 strain (A1 and A2), while the others were obtained from the infection with the LBA-9402 strain (L1 to L42).A variety of media (MS, B5 and SH with different combinations) was used for the optimization of hairy root induction, stabilization and steady growth (ESI Table 1†). Among 1596 explants infected, LBA infected explants were 56.3%. SH medium supported maximum hairy roots induction as 289 hairy roots (90% ramified) were produced from the only 60 infected explants. Moreover, unlike to that of MS or B5 media, the transgenic hairy roots were induced within a week over SH medium (ESI Table 2†). Initially, half strength MS medium with IBA was found proliferating the hairy roots (ESI Table 3†) but for the stable growth of well ramified hairy roots IBA was alleviated (ESI Table 4†). Besides the media and hormones, the ex vitro source plants were found as the best explant source for hairy roots induction (ESI Table 5†).
Only one hairy root line with moderate growing capacity was obtained from ARqua1 strain and named as AR1. The established hairy root cultures grew actively on hormone-free half-strength medium, and showed four different morphologies (Fig. 2e–j). Most of the hairy roots (59%) exhibited typical hairy root morphology (THR), as described previously.40 Among the rest of the hairy root lines established, those with callus-like (CM), thick (TK) and thin (TN) morphology accounted for 17%, 14% and 10%, respectively of the total (Table 2). All hairy root lines obtained were fast-growing, ramified and plagiotropic except those with CM, which were less ramified. The CM lines exhibited the capacity to dedifferentiate and produce callus tissue in hormone-free culture medium.
| Root morphology | Incidence | Growth rate | PE content (μg g−1 DW) | virD1 | rolC | aux1 | mas1 | ags |
|---|---|---|---|---|---|---|---|---|
| a Growth capacity of transgenic clones of different root lines expressed as (harvested FW/inoculum FW) after 28 days of culture in hormone-free half strength MS medium. Each value is the average of 3 replicates. THR: typical hairy root, CM: callus like morphology, TK: hairy root with thick morphology, TN: hairy root with thin morphology, PE: phytoecdysteroid content. | ||||||||
| THR | 58.62% | 3.277 ± 0.32 | 1538 ± 48.2 | 0% | 100% | 70.6% | 100% | 73.5% |
| CM | 17.24% | 3.93 ± 0.3 | 1509.56 ± 37.3 | 0% | 100% | 90% | 100% | 100% |
| TK | 13.79% | 2.675 ± 0.23 | 1217 ± 17.9 | 0% | 100% | 100% | 100% | 100% |
| TN | 10.34% | 2.35 ± 0.52 | 1202.3 ± 63 | 0% | 100% | 0.0% | 16.7% | 0.0% |
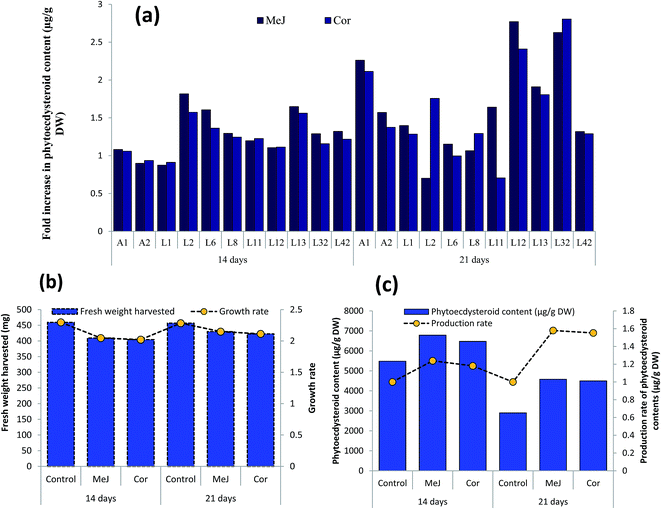
All the hairy root lines were subjected to HPLC analysis (Fig. 3) and growth rate measurements (Fig. 4). The growth rate of hairy roots (fresh weight harvested/fresh weight of inoculum) was found to be significantly high in 11 transgenic hairy root lines (Fig. 4a, Table 3). These fast-growing hairy root lines also possessed significantly high phytoecdysteroid contents, e.g. 4449 and 4122 μg g−1 DW accumulated by A2 and L1 hairy root lines, respectively (Fig. 4b). The growth rate of the transgenic hairy root lines varied according to their morphology (Fig. 4c). After 30 days of culture, on average, CM hairy roots showed a higher growth rate (3.93 times) by producing 786 mg of fresh weight in one month of culture than the other hairy root lines (Fig. 4c). However, THR showed more phytoecdysteroid content (1538.48 μg g−1 DW) than CM lines (1509.56 μg g−1 DW). The lowest phytoecdysteroid levels were found in TN and TK hairy roots (Table 2). A possible reason for the decrease in phytoecdysteroids in CM roots is the loss of organized tissue. Similarly, Mallol and coworkers found that ginsenoside production in Panax ginseng transgenic hairy roots with CM and THR morphology (levels almost the same) was significantly higher than in roots with thin morphology.27
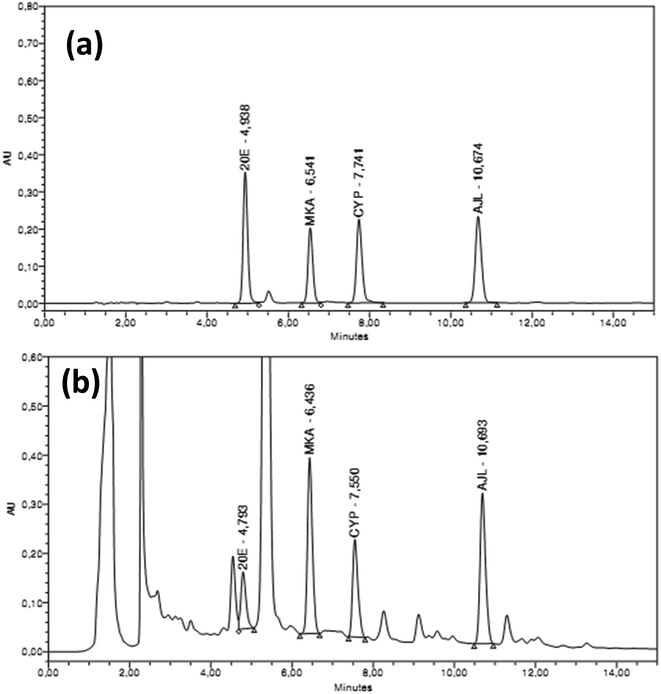 | ||
| Fig. 3 HPLC chromatographs showing the elution of phytoecdysteroids. (a) Standard ecdysteroids (100 mg ml−1 of each standard), (b) phytoecdysteroid elution pattern in one of the hairy root extract. | ||
| Source | DF | SS | MS | F-Value | P value |
|---|---|---|---|---|---|
| a Coefficient of variation: 6.71%. ANOVA of transgenic hairy root lines was done against 59 established root clones. DF, degree of freedom; SS, sum of squares; MS, means square. *** means that the transgenic lines, phytoecdysteroids and their interaction is significant at P < 0.001. | |||||
| Transgenic hairy roots | 58 | 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 012 012![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 543.866 543.866 |
379![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 526.618 526.618 |
590.6268 | *** |
| Ecdysteroids | 3 | 39![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 953 953![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 845.301 845.301 |
13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 317 317![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 948.434 948.434 |
20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 725.6527 725.6527 |
*** |
| Transgenic hairy roots × phytoecdysteroids | 174 | 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 722 722![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 812.986 812.986 |
107![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 602.373 602.373 |
167.4529 | *** |
| Error | 472 | 303![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 299.093 299.093 |
642.583 | ||
| TOTAL | 707 | 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 992 992![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 501.246 501.246 |
|||
TL- and TR-DNA integration into the genome of transgenic hairy root lines
We related the presence of T-DNA genes with the morphology and growth rate of transgenic hairy roots, and their phytoecdysteroid content (ESI Table 6†). Three TR-DNA genes of A. rhizogenes, aux1, ags and mas1, were screened along with the rolC gene located in the TL-DNA, which is a key gene responsible for hairy root development. The ags gene is involved in agropine biosynthesis, while the aux1 gene, as indicated previously, is considered to play an additional role in the formation of indole-3-acetic acid in transgenic material.41 The mas1 gene is involved in manopine biosynthesis.42When comparing the different hairy root morphologies in relation with the genes of the pRi T-DNA, it was observed that 100% of the CM root lines harbored rolC, mas1 and ags genes, and 90% of the total hairy root lines established were positive for aux1 (Fig. 5), which was not found in any of the TN roots (Table 2). A semi quantitative PCR analysis of the high phytoecdysteroid yielding transgenic hairy root lines has shown high rolC expression (Fig. 6). The role of the aux1 gene (to provide the transgenic cells with an additional source of auxins) is reflected in previous studies.43 Our findings are in accordance with previous work44 in which the provision of synthetic auxins (2,4-D) enhanced callus biomass and reduced alkaloid production. It is argued that the overproduction of auxins can drive the disorganization in the transgenic hairy root lines.45 Robins reported a complete loss of nicotine in the root clones of Nicotiana rustica treated with synthetic auxins.46
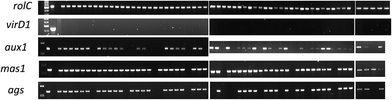 | ||
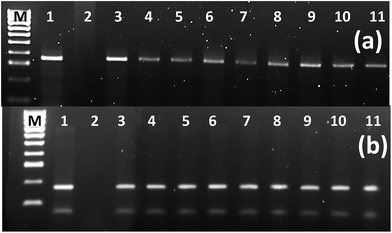
| Fig. 5 PCR analysis of the selected genes to confirm their integration into the transgenic hairy roots' genome. First well contains ladder DNA, second and third are positive and negative controls respectively while the rest of the samples are in accordance with the information provided in ESI Table 6.† | ||
Strikingly, the roots with TK morphology were 100% positive for the TL- and TR-DNA genes. In contrast, the TN roots showed 0% presence of the aux1 and ags genes, and only 16.7% of them were positive for mas1 integration into their genome. The ags gene was present in all CM and TK hairy roots, while mas1 was present in all hairy root types except TN (Table 2 and ESI Table1†). In another study, 100% of CM hairy roots from tobacco, Duboisia hybrid and Datura metel Solanaceae plants exhibited insertion of the aux1 gene.28 Less incorporation of TR-DNA into transgenic hairy roots can be due to incomplete integration events.47 In a previous study, a constitutive A. tumefaciens strain carrying pRiA4TR (aux genes deleted) produced transgenic hairy roots with exactly the same morphology (no callogenesis) and alkaloid production as the transgenic roots obtained with the strain A4.28
Effect of elicitors on phytoecdysteroid production in hairy root clones
The hairy root lines (11 independent clones, Fig. 4a and b) exhibiting the highest growth rate and phytoecdysteroid content were selected to study the effect of elicitation with MeJ (100 μM) and Cor (1 μM). Elicitation with MeJ was found to induce a maximum phytoecdysteroid content of 8356 μg g−1 DW after 14 days in the L2 line, followed by L32, which produced 8066 μg g−1 (Fig. 7a). In our study, elicitation with MeJ was more effective than with Cor. Moreover, higher phytoecdysteroid levels were obtained after 14 days of elicitation than 21 days.Phytoecdysteroids are biosynthesized through the mevalonate pathway. In a previous study, 2–3.5% radiolabeled mevalonate (14C-MVA) was incorporated into the 20-HE produced in spinach cultures after 24 hours.48 The key enzyme, 3-hydroxy-3-methylglutaryl coenzyme A reductase (HMGR-CoA reductase), irreversibly converts HMG-CoA to MVA and is considered to be the rate-limiting factor in this pathway.49 The higher phytoecdysteroid content found in our elicited hairy root clones could be a result of a high expression of HMGR. Recently, an elicitor-responsive gene involved in 20-HE production, encoding 3-hydroxy-3-methylglutaryl coenzyme A reductase (HMGR), was cloned in Cyanotis arachnoidea. The resulting enhanced biosynthesis of 20-HE in MeJ-treated cells and a correspondingly high expression of HMGR suggest that 20-HE biosynthesis may be the result of the up-regulation of CaHMGR expression.50 HMGR provides mevalonate for the biosynthesis of 20-HE and other secondary metabolites. In another study, elicitation of Achyranthes bidentata cells with 0.6 mM MeJ for 6 days produced 2.6-fold more 20-HE.51
The hairy root lines elicited for 21 days had a higher growth rate/biomass than those treated for 14 days in comparison with their respective controls. The growth rate was determined after culturing the hairy root clones for one month on hormone-free medium. However, despite producing more biomass, 21 day-elicited roots had lower phytoecdysteroid levels and the treatment induced detrimental effects, since the hairy roots turned from yellow to brown and then dark brown, and ultimately died (Fig. 7b). The plasticity or tolerance of plants towards different chemicals varies according to species. For example, the treatment of spinach with MeJ induced 78% and 61% more 20-HE concentration in roots and shoots, respectively, without affecting plant growth.33
On average, MeJ-treated hairy roots produced more phytoecdysteroids after 14 days of elicitation (6789 μg g−1 DW), with a 1.24-fold higher biomass production (Fig. 7c). In contrast, 21 days of elicitation with MeJ and Cor resulted in the average production of 4577 and 4498 μg g−1 DW of phytoecdysteroids, respectively, which was nevertheless 1.58- and 1.55-fold higher compared to the control. This reduction of phytoecdysteroid content after a longer elicitation period is in accordance with the experiments performed by Mangas and coworkers. The production of sterols in Centella asiatica, and Galphimia glauca increased after two weeks of MeJ treatment and then gradually declined during the third and fourth weeks.52 Decrease in phytoecdysteroid production could also be caused by a higher concentration of elicitors. In another Ajuga species, A. turkestanica, the 20-HE content increased 3-fold when the cell suspension culture was treated with 125 μM MeJ and it dramatically decreased when supplemented with 250 μM MeJ compared to the control/untreated cultures.53
We report for the first time the biotechnologically enhanced production of phytoecdysteroids in A. bracteosa both via transformation and elicitation. pRi TR-DNA genes were found to play a significant role in the determination of the morphology of hairy roots. While those with callus-like morphology produced more biomass, typical transgenic hairy roots produced more phytoecdysteroids. Phytoecdysteroid levels were further enhanced by 14 days of MeJ elicitation. Based on these findings, it can be suggested that typical transgenic hairy roots treated with 14 days of MeJ elicitation constitute a useful tool to achieve high phytoecdysteroid production in A. bracteosa.
Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
Ecdysteroids standards were kindly provided by Prof. Josep Coll-Toledano, Department of Biological Chemistry and Molecular Modeling, Spanish National Research Council (SNRC), Barcelona, Spain. This project was financed by the Higher Education Commission of Pakistan. Part of this work was financially supported by the Spanish MEC (BIO2014-51861-R) and the Generalitat de Catalunya (2014SGR215).References
- W. K. K. Barkatullah, M. Ibrar, A. Rauf, T. B. Hadda, M. S. Mubarak and S. Patel, J. Ethnopharmacol., 2015, 169, 335–346 CrossRef PubMed.
- W. T. Hsieh, Y. T. Liu and W. C. Lin, J. Ethnopharmacol., 2011, 135, 116–125 CrossRef PubMed.
- W. K. Kayani, E. Dilshad, T. Ahmed, H. Ismail and B. Mirza, BMC Complementary Altern. Med., 2016, 16, 375 CrossRef PubMed.
- R. Gautam, S. M. Jachak and A. Saklani, J. Ethnopharmacol., 2011, 133, 928–930 CrossRef PubMed.
- G. Kaithwas, R. Gautam, S. M. Jachak and A. Saklani, Asian Pac. J. Trop. Biomed., 2012, 2, 185–188 CrossRef CAS PubMed.
- A. Pal and R. S. Pawar, Int. J. Curr. Biol. Med. Sci., 2011, 1, 12–14 Search PubMed.
- S. Chandel and U. Bagai, Parasitol. Res., 2011, 108, 801–805 CrossRef PubMed.
- G. Fekete, L. A. Polgár, M. Báthori, J. Coll and B. Darvas, Pest Manage. Sci., 2004, 60, 1099–1104 CrossRef CAS PubMed.
- T. Otaka, M. Uchiyama, T. Takemoto and H. Hikino, Chem. Pharm. Bull., 1969, 17, 1352–1355 CrossRef CAS PubMed.
- V. N. Syrov, S. S. Nasyrova and Z. A. Khushbaktova, Eksp. Klin. Farmakol., 1996, 60, 41–44 Search PubMed.
- Z. Saatov, D. A. Agzamkhodzhaeva and V. N. Syrov, Chem. Nat. Compd., 1999, 35, 186–191 CrossRef CAS.
- V. N. Syrov and Z. A. Khushbaktova, Eksp. Klin. Farmakol., 2000, 64, 56–58 Search PubMed.
- D. S. Trenin and V. V. Volodin, Arch. Insect Biochem. Physiol., 1999, 41, 156–161 CrossRef CAS PubMed.
- A. I. Kuzmenko, E. Niki and N. Noguchi, J. Oleo Sci., 2001, 50, 497–506 CrossRef CAS.
- I. Mirzaev, V. N. Syrov, S. A. Khrushev and S. D. Iskanderova, Eksp. Klin. Farmakol., 1999, 63, 35–37 Search PubMed.
- W. Wang, T. Wang, W. Y. Feng, Z. Y. Wang, M. S. Cheng and Y. J. Wang, Neurosci. Res., 2014, 81, 21–29 CrossRef PubMed.
- Q. Chen, Y. Xia and Z. Qiu, Life Sci., 2006, 78, 1108–1113 CrossRef CAS PubMed.
- A. G. Kurmukov and V. N. Syrov, Med. Zh. Uzb., 1988, 10, 68–70 Search PubMed.
- D. K. Najmutdinova and Z. Saatov, Arch. Insect Biochem. Physiol., 1999, 41, 144–147 CrossRef CAS PubMed.
- V. U. Ahmad, S. M. Khaliq-Uz-Zaman, M. Ali, S. Perveen and W. Ahmed, Fitoterapia, 1996, 67, 88–91 CAS.
- L. Pistelli, A. Giovannini, B. Ruffoni, A. Bertoli and L. Pistelli, in Bio-Farms for Nutraceuticals, Springer, 2010, pp. 167–184 Search PubMed.
- M. D. Chilton, D. A. Tepfer, A. Petit, C. David, F. Cassedelbart and J. Tempe, Nature, 1982, 295, 432–434 CrossRef CAS.
- M.-D. Chilton, M. H. Drummond, D. J. Merlo, D. Sciaky, A. L. Montoya, M. P. Gordon and E. W. Nester, Cell, 1977, 11, 263–271 CrossRef CAS PubMed.
- F. Vilaine and F. Casse Delbart, Mol. Gen. Genet., 1987, 206, 17–23 CrossRef CAS.
- M. Cardarelli, D. Mariotti, M. Pomponi, L. Spano, I. Capone and P. Costantino, Mol. Gen. Genet., 1987, 209, 475–480 CrossRef CAS PubMed.
- A. D. Paolis, M. L. Mauro, M. Pomponi, M. Cardarelli, L. Spano and P. Costantino, Plasmid, 1985, 13, 1–7 CrossRef PubMed.
- A. Mallol, R. M. Cusidò, J. Palazòn, M. Bonfill, C. Morales and M. T. Piñol, Phytochemistry, 2001, 57, 365–371 CrossRef CAS PubMed.
- E. Moyano, S. Fornalé, J. Palazón, R. M. Cusidó, M. Bonfill, C. Morales and M. T. Piñol, Phytochemistry, 1999, 52, 1287–1292 CrossRef CAS.
- T. Matsumoto and N. Tanaka, Agric. Biol. Chem., 1991, 55, 1019–1025 CAS.
- N. Tanaka and T. Matsumoto, Plant Tissue Cult. Lett., 1993, 10, 78–83 CrossRef CAS.
- N. Tanaka and T. Matsumoto, Plant Cell Rep., 1993, 13, 87–90 CrossRef CAS PubMed.
- O. T. Kim, M. Manickavasagm, Y. J. Kim, M. R. Jin, K. S. Kim, N. S. Seong and B. Hwang, J. Plant Biol., 2005, 48, 258–262 CrossRef CAS.
- I. R. Soriano, I. T. Riley, M. J. Potter and W. S. Bowers, J. Chem. Ecol., 2004, 30, 1885–1899 CrossRef CAS PubMed.
- W. K. Kayani, J. Palazòn, R. M. Cusidò and B. Mirza, RSC Adv., 2016, 6, 22700–22708 RSC.
- R. U. Schenk and A. Hildebrandt, Can. J. Bot., 1972, 50, 199–204 CrossRef CAS.
- T. Murashige and F. Skoog, Physiol. Plant., 1962, 15, 473–497 CrossRef CAS.
- J. J. Doyle and J. L. Doyle, Focus, 1990, 12, 13–15 Search PubMed.
- A. Castro, J. Coll, Y. A. Tandrón, A. K. Pant and C. S. Mathela, J. Nat. Prod., 2008, 71, 1294–1296 CrossRef CAS PubMed.
- J. J. Wu, K. W. Cheng, H. Wang, W. C. Ye, E. T. Li and M. Wang, Phytochem. Anal., 2009, 20, 58–63 CrossRef CAS PubMed.
- C. David, M.-D. Chilton and J. Tempé, Nat. Biotechnol., 1984, 2, 73–76 CrossRef CAS.
- O. Nilsson and O. Olsson, Physiol. Plant., 1997, 100, 463–473 CrossRef CAS.
- D. Bouchez and J. Tourneur, Plasmid, 1991, 25, 27–39 CrossRef CAS PubMed.
- R. O. Morris, Annu. Rev. Plant Physiol., 1986, 37, 509–538 CrossRef CAS.
- J. Palazón, T. Altabella, R. M. Cusidó, M. Ribó and M. T. Piñol, Biol. Plant., 1995, 37, 161–168 CrossRef.
- K. H. Jung, S. S. Kwak, C. Y. Choi and J. R. Liu, Plant Cell Rep., 1995, 15, 51–54 CrossRef CAS PubMed.
- R. J. Robins, Nat. Prod. Rep., 1998, 15, 549–570 RSC.
- V. Gaudin, T. Vrain and L. Jouanin, Plant Physiol. Biochem., 1994, 32, 11–29 CAS.
- A. Bakrim, A. Maria, F. Sayah, R. Lafont and N. Takvorian, Plant Physiol. Biochem., 2008, 46, 844–854 CrossRef CAS PubMed.
- J. Chappell, Plant Physiol., 1995, 107, 1–6 CrossRef CAS PubMed.
- Q. J. Wang, L. P. Zheng, P. F. Zhao, Y. L. Zhao and J. W. Wang, Plant Physiol. Biochem., 2014, 84, 1–9 CrossRef CAS PubMed.
- Q. J. Wang, L. P. Zheng, Y. H. Sima, H. Y. Yuan and J. W. Wang, Plant Omics, 2013, 6, 116–120 CAS.
- S. Mangas, M. Bonfill, L. Osuna, E. Moyano, J. Tortoriello, R. M. Cusidò, M. T. Piñol and J. Palazón, Phytochemistry, 2006, 67, 2041–2049 CrossRef CAS PubMed.
- D. M. Cheng, G. G. Yousef, M. H. Grace, R. B. Rogers, J. Gorelick-Feldman, I. Raskin and M. A. Lila, Plant Cell, Tissue Organ Cult., 2008, 93, 73–83 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra06399g |
| ‡ Present address: Department of Plant Breeding, Swedish University of Agricultural Sciences, Växtskyddsvägen 1, SE-230 53 Alnarp, Sweden, E-mail: bushramirza@qau.edu.pk. |
| This journal is © The Royal Society of Chemistry 2017 |