Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Magnetic bead-based mimic enzyme-chromogenic substrate and silica nanoparticles signal amplification system for avian influenza A (H7N9) optical immunoassay†
Dan Sua,
Hanyun Lia,
Jinlin Lib,
Yali Liua,
Mi Penga,
Bingwei Fenga,
Pengfei Xua and
Yonggui Song *a
*a
aNational Pharmaceutical Engineering Center for Solid Preparation in Chinese Herbal Medicine, Jiangxi University of Traditional Chinese Medicine, 56 Yangming Road, Nanchang 330006, China. E-mail: songyonggui1999@163.com; Fax: +86 791 87119632; Tel: +86 791 87119632
bNanchang Institute for Food and Drug Control, Nanchang 330038, China
First published on 30th August 2017
Abstract
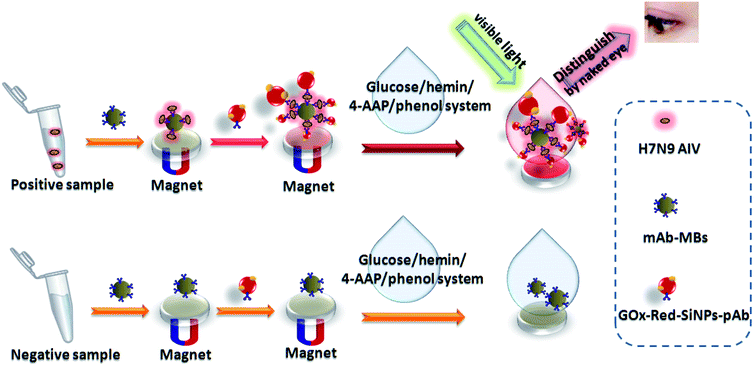
This work reports on a simple, feasible and optical color-enhanced colorimetric immunoassay with hemin (a horseradish peroxidase mimic enzyme) catalyzed color reaction and multifunctional colored silica nanoparticles for qualitative and quantitative determination of avian influenza A (H7N9) virus (H7N9 AIV) at an ultralow concentration by using the magnetic nanobeads (MBs)-mimic enzyme-multifunctional silica nanoparticles color-enhanced chromogenic substrate system. In this work, MBs were fabricated by co-immobilizing target recognition molecules (mAb–MB), which were used as both separation and enrichment carriers. The multifunctional colored silica nanoparticles were synthesized by doping Cy2 (red organic dye) into silica nanoparticles using an inverse microemulsion method, and then made a covalent modification of pAb and glucose oxidase (GOx). In the presence of H7N9 AIV, GOx catalyzed glucose to gluconic acid and hydrogen peroxide (H2O2). The latter can oxidize 4-aminoantipyrine (4-AAP) to red products and the reaction is catalyzed by hemin. The chromogenic compound and the red-silica nanoparticles (Red-SiNPs) together cause the color of the solution to change from colorless to red. Through this color-enhanced immunoassay, we can detect H7N9 AIV with the naked eye and UV detector. And the immunosensor provides a simple and reliable platform with high sensitivity and selectivity which shows great potential in early diagnosis of diseases and in public health security.
1. Introduction
Since human infections with novel H7N9 AIV were first documented in Shanghai City and Jiangsu Province in 2013,1 human cases continued to be reported in China in 2014 and 2016,2,3 which raise significant concerns that H7N9 AIV is a likely candidate for the next pandemic strains. Meanwhile, fast and sensitive virus detection techniques are urgently needed to provide immediate and appropriate clinical strategies to control the spread of the virus. Up to now, traditional virus detection methods have been employed for influenza virus detection, including virus isolation, polymerase chain reaction (PCR)4 and enzyme-linked immunosorbent assay (ELISA).5 Among these methods, ELISA based on a specific antigen–antibody interaction is a very powerful technique for the determination of clinically important analytes in a variety of biological matrixes.6,7 Usually, high sensitivity for ELISA is achieved by using an indicator system that results in the amplification of the product to be measured, e.g., enzyme labels or nanolabels.8,9 Undoubtedly, enzyme labels are utilized more widely than other labeling strategies, since a single enzymatic molecule, e.g., horseradish peroxidase (HRP), can cause the conversion of 107 substrate molecules per minute.10 Meanwhile using H2O2 as substrate, HRP can catalyze the oxidation of numerous hydrogen donors (DH2).11 Despite some advances in this field, there are some limitations, such as use of an expensive protease like HRP, harsh reaction conditions, short period of stability and lack of easy operation methods to combine visual qualitative identification and quantitative analysis methods. Moreover, for optical detection, too light a color is the important reason for the unsatisfactory detection limit.To solve the above problems, a magnetic nanobeads (MBs)-mimic enzyme chromogenic substrate-multifunctional silica nanoparticles color-enhanced system-based colorimetric immunoassay (MB–MEMSCI) has been designed. In this work, MBs were used as separation and enrichment carriers, and hemin was used in place of HRP to catalyze hydrogen peroxide and simultaneously to produce a color reaction. Moreover, a multifunctional bright red silica nanoparticle modified with antibody and GOx was prepared. The experimental results show that the catalytic activity of hemin can match that of HRP, and it also has the advantages of low cost, good stability of physical and chemical properties, easy preservation and mild reaction conditions. To model the MB–MEMSCI in terms of immunoreactions, we employed H7N9 AIV as our target analyte, with specific monoclonal antibody (mAb) and rabbit-derived polyclonal antibody (pAb) as the capture and detection antibodies, respectively. Herein, the assay is carried out on a magnetic immunosensing probe by using polyclonal anti-H7N9 AIV/GOx-labeled Red-SiNPs as the signal-transduction tag with a sandwich-type immunoassay format. In the presence of target H7N9 AIV, the carried GOx accompanying the sandwiched immunocomplex initially catalyzes glucose to gluconic acid and hydrogen peroxide, and then the regenerated hydrogen peroxide is catalyzed by hemin to produce the colored product. By monitoring the obvious color change together caused by the red product and the multifunctional Red-SiNPs with the naked eye and UV absorption, we can not only qualitatively but also quantitatively determine ultralow concentrations of target H7N9 AIV in samples with wide linear range, low detection limit, high selectivity and long-term stability. Meanwhile, the handy operational MB–MEMSCI has good accuracy to detect H7N9 AIV in real samples.
2. Materials and methods
2.1 Materials and reagents
Inactivated H7N9 AIV, H5N1 AIV, pseudo rabies virus (PRV), Newcastle disease virus (NDV) and murine origin H7N9 hemagglutinin (HA) specific mAb and rabbit-derived pAb were obtained from Wuhan Institute of Virology, Chinese Academy of Sciences. The influenza A H9N2 virus and enterovirus 71 viruses (EV71) were obtained from Academy of Military Medical Sciences, Beijing. 3-Aminopropyltriethoxysilane (APTES, 98 wt%), hemin and glucose oxidase (GOx) were obtained from Sigma-Aldrich (Shanghai, China). Glucose was purchased from Alfa Aesar (Shanghai, China). 3-[2-(2-Aminoethylamino)ethylamino]propyltrimethoxysilane (APTMS), tetraethyl orthosilicate (TEOS), N-hydroxysuccinimide (NHS), 1-ethyl-3,3-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDC) and 2-(N-morpholino)ethanesulfonic acid (MES) were obtained from Aladdin Industrial Inc. (Shanghai, China). Bovine serum albumin (BSA) was obtained from Sinopharm Chemical Reagent Co. Ltd (Shanghai, China). 4-Aminoantipyrine (4-AAP), phenol, glutaraldehyde (25 wt%), Triton X-100, cyclohexane, hexanol, and ammonia (25–28 wt%) were obtained from Fuchen Chemicals (Tianjin, China). Colloidal gold (AuNP) of 15 nm in diameter was synthesized according to our previous report.12 MB (particle size ∼100 nm) in an aqueous suspension with a concentration of 25 mg mL−1 were obtained from Chemical GmbH (Berlin, Germany). Cy2 was supplied by Zhejiang Shunlong Chemical Co. Ltd (Zhejiang, China). All reagents used for all experiments were of analytical grade and were used without further purification. Ultrapure water (18.2 MΩ cm) was purified by a Millipore-Q system. In the preparation of a carbonate buffer solution of pH 9.6, Na2CO3 (1.59 g), NaHCO3 (2.93 g), and NaN3 (0.2 g) were dissolved in 1000 mL of double-distilled water. Phosphate-buffered saline (PBS) solutions with various pH values were prepared by using Na2HPO4 and KH2PO4, and 0.1 M NaCl was used as the supporting electrolyte. Clinical serum samples were made available by Jiangxi Provincial Hospital of Traditional Chinese Medicine, China. And all experiments were in accordance with the guidelines of the National Institute of Food and Drug, Nanchang, China, and approved by the institutional ethical committee (IEC) of Jiangxi University of Traditional Chinese Medicine.2.2 Preparation of mAb–MB conjugates
Before conjugation with mAb, MBs were initially separated using an external magnet and then dried in a vacuum at 80 °C for 1 h. Following that, 50 mg of MB was added into 1 mL of anhydrous ethanol and the resulting mixture was sonicated for 10 min at room temperature (RT) to obtain a homogeneous suspension. Afterward, 30 μL of APTES (98 wt%) was injected in the mixture and continuously stirred for 6 h at RT. During this process, aminated MB was formed based on the reaction between –OCH3 and –OH on the MB. The functional MB was separated and redispersed into 1 mL of PBS (pH 7.4) containing 300 μL of glutaraldehyde (25 wt%). The suspension was stirred vigorously for 6 h at RT. After magnetic separation, the precipitate was dissolved into 1 mL of carbonate buffer (pH 9.6) solution containing 100 μg of mAb antibody and shaken on a shaker (MS, IKA GmbH, Staufen, Germany) overnight at 4 °C. The excess mAb antibody was removed by magnetic separation. Subsequently, the mAb–MB conjugates were treated with 10 wt% BSA–PBS (1.0 mL, pH 7.4) at 4 °C for 2 h to block the unreacted and nonspecific sites. Finally, 100 μL of sodium cyanoborohydride (25 mg mL−1) was injected into the suspension in order to reduce the resultant Schiff bases. The as-prepared mAb–MB conjugates were collected by using an external magnet and dispersed into 1 mL of PBS (pH 7.4) containing 1.0 wt% BSA and 0.1 wt% sodium azide.2.3 Synthesis of Red-SiNPs
Red-SiNPs were synthesized according to an inverse microemulsion method described in a previous paper with slight modification.13,14 The details of the procedure are described in the following: 8 mL cyclohexane, 2 g Triton X-100, 2 mL 1-hexanol, 150 μL Cy2 (100 mg mL−1) and 400 μL of water were added into a 25 mL Erlenmeyer flask and stirred for 15 min to ensure the water was completely dispersed into cyclohexane. Afterwards, 100 μL TEOS and 20 μL APTMS were added to this inverse microemulsion system followed by 100 μL ammonia (25–28 wt%) to catalyze the hydrolysis of TEOS. After stirring for 48 h, the microemulsion was broken by adding 10 mL acetone. The Red-SiNPs were separated from the supernatant by centrifugation at 8000 rpm for 10 min and washed with ethanol three times followed by washing twice with ultrapure water.2.4 Covalent immobilization of antibody and GOx onto Red-SiNPs surface
30 mg Red-SiNPs were suspended in 20 mL ultrapure water, and 1.4 mL acetic acid and 200 μL APTMS were added into the Red-SiNPs solution for the post-coating treatment. The APTMS was allowed to hydrolyze under stirring for 1 h at room temperature. After the hydrolysis reaction of APTMS, amino groups were introduced onto the surface of Red-SiNPs. The amino-modified Red-SiNPs (Red-SiNPs-NH2) were isolated by centrifuging at 8000 rpm for 5 min and washing three times with DMF. Carboxyl-modified Red-SiNPs were synthesized by reacting Red-SiNPs-NH2 with glutaric anhydride. 20 mg Red-SiNPs-NH2 were dispersed in 5 mL DMF containing 200 mg glutaric anhydride, and then the solution was stirred under N2 gas for 6 h. These carboxyl-modified Red-SiNPs were centrifuged at 8000 rpm for 10 minutes and washed with PBS (10 mM, pH 7.3). The pAb and GOx were directly immobilized onto the functionalized Red-SiNPs with well-established EDC/NHS coupling chemistry. The immobilization protocol was as follows: 20 mg Red-SiNPs-COOH were resuspended in 5 mL MES (0.1 M, pH 6.7). The Red-SiNPs-COOH suspension was mixed with 5.0 μL of pAb (1.0 mg mL−1) and 50 μL of GOx (1.0 mg mL−1) solution, followed by the addition of 1 mL of 10 mM EDC and 10 mM NHS solution. After 2 h incubation at room temperature, the free pAb and GOx were removed by centrifugation at 8000 rpm at 4 °C for 5 minutes and washed with PBS. The modified Red-SiNPs were resuspended in 3 mL of PBS containing 1% BSA to block non-specific adsorption sites on the nanoparticles. The prepared resultant GOx- and antibody-conjugated Red-SiNPs (GOx–Red-SiNPs-pAb) were dispersed in 1 mL of PBS and stored at 4 °C for subsequent use.2.5 Monitoring of GOx activity using the hemin/4-AAP/phenol system
Fig. 1a displays the assay mechanism of the hemin/4-AAP/phenol system. Initially, 10 μL of GOx with specified concentration (from 0 to 1000 μg mL−1) was added in 50 μL of PBS (0.5 mM, pH 7.0) containing 4 mM glucose. The resulting solution was incubated for 30 min at 37 °C. Then 70 μL of 0.1% 4-AAP, 70 μL of 0.123 M phenol, 35 μL of 0.05 mg mL−1 heme chloride solution and 350 μL buffer solution (K2HPO4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NaOH, pH = 10.6) was added into the above-prepared mixture and reacted for 10 min at room temperature. After interaction, the color of the solution was observed and at the same time the absorbance was recorded at λ = 505 nm with a Lambda 35 UV-visible spectrophotometer.
NaOH, pH = 10.6) was added into the above-prepared mixture and reacted for 10 min at room temperature. After interaction, the color of the solution was observed and at the same time the absorbance was recorded at λ = 505 nm with a Lambda 35 UV-visible spectrophotometer.
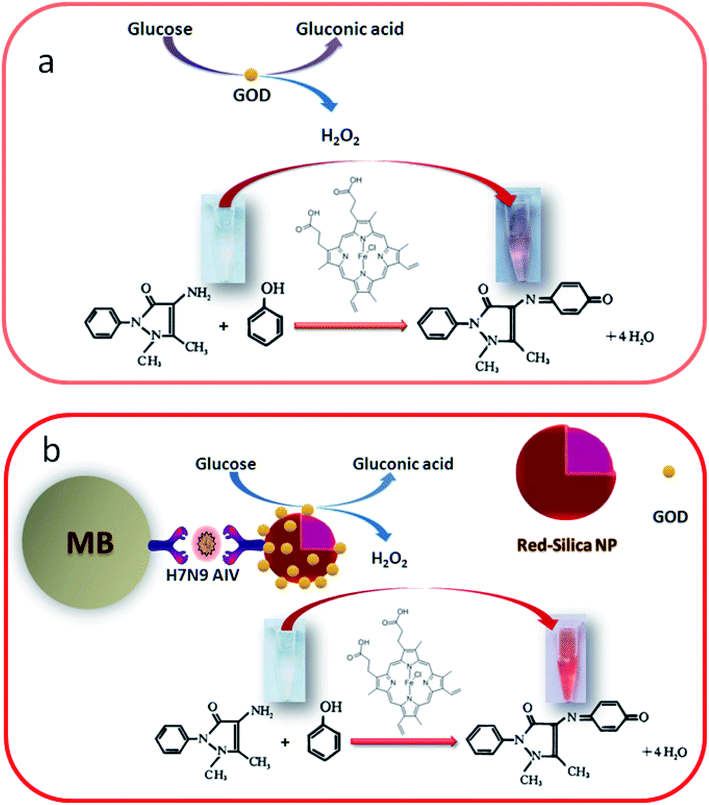 | ||
| Fig. 1 Schematic illustration of (a) H2O2-stimulated colorimetric assay for monitoring of GOx activity and (b) principle of the colorimetric MB–MEMSCI. | ||
2.6 Immunoassay for target H7N9 AIV using MB–MEMSCI
Fig. 1b presents the immunoassay process toward target H7N9 AIV by coupling with the hemin/4-AAP/phenol system. The detailed procedure is summarized as follows: (i) 50 μL of H7N9 AIV standards or samples with different concentrations and 25 μL of mAb–MB suspension (6 mg mL−1) were initially added into a 0.5 mL centrifuge tube, and the mixture was then incubated for 30 min at 37 °C on a shaker to form the antigen–antibody complex; (ii) 50 μL of GOx–Red-SiNPs-pAb as prepared above was injected into the centrifuge tube and incubated for another 30 min at 4 °C to form the sandwiched immunocomplex; (iii) 50 μL of glucose (4 mM) in pH 7.0 PBS was added into the centrifuge tube and incubated for 30 min at 37 °C on the shaker for enzymatic reaction; (iv) 70 μL of 1.0 mg mL−1 4-AAP, 70 μL of 0.12 M phenol, 35 μL of 0.05 mg mL−1 heme chloride solution and 350 μL buffer solution (K2HPO4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NaOH, pH = 10.6) were added to the centrifuge tube in turn; and (v) the color of the solution was observed and at the same time the absorbance was recorded at λ = 505 nm with a Lambda 35 UV-visible spectrophotometer at room temperature. Note that the resulting mixture was separated with an external magnet and washed with pH 7.4 PBS after steps (i) and (ii). Control tests with normal (negative) samples and the evaluations for clinical specimens were performed accordingly. All measurements were done at room temperature (25 ± 1.0 °C).
NaOH, pH = 10.6) were added to the centrifuge tube in turn; and (v) the color of the solution was observed and at the same time the absorbance was recorded at λ = 505 nm with a Lambda 35 UV-visible spectrophotometer at room temperature. Note that the resulting mixture was separated with an external magnet and washed with pH 7.4 PBS after steps (i) and (ii). Control tests with normal (negative) samples and the evaluations for clinical specimens were performed accordingly. All measurements were done at room temperature (25 ± 1.0 °C).
2.7 The procedure of H7N9 AIV optical detection by MB–MEMSCI
In order to get a more clear visual effect and a low detection limit, optical detection and UV detection of H7N9 AIV are carried out separately. The optical detection scheme is given in Fig. 2. For a typical sandwich assay, 20 μL MB–mAb (1 mg mL−1) was mixed with 1 mL H7N9 AIV solution in a centrifuge tube. After incubation for 30 min at 37 °C with gentle shaking, the mAb–MB–H7N9 AIV complex was separated magnetically and the clear supernatant was discarded. The immune complex was then washed with PBS three times to remove any unbound species. The mAb–MB–H7N9 AIV complex was then dispersed in 20 μL PBS (pH 7.3), transferred to a well of a 96-well microtiter plate, and 20 μL GOx–Red-SiNPs-pAb (10 mg mL−1) was added to the well and the mixture was subjected to react for 15 min at room temperature. GOx–Red-SiNPs-pAb formed a sandwich structure with H7N9 AIV and MB–mAb via immune reaction. The immune sandwich complex was separated magnetically and the supernatant was discarded. The immune complex was then washed with PBS to effectively remove unbound GOx–Red-SiNPs-pAb. Then 20 μL of glucose (4 mM) in pH 7.0 PBS was added into the centrifuge tube and incubated for 30 min at 37 °C on the shaker for enzymatic reaction. Simultaneously, 28 μL of 1.0 mg mL−1 4-AAP, 70 μL of 0.12 M phenol, 35 μL of 0.05 mg mL−1 heme chloride solution and 140 μL buffer solution (K2HPO4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NaOH, pH = 10.6) were added to the centrifuge tube in turn. The final color of the suspension solution in the well was recorded with a digital camera.
NaOH, pH = 10.6) were added to the centrifuge tube in turn. The final color of the suspension solution in the well was recorded with a digital camera.
3. Results and discussion
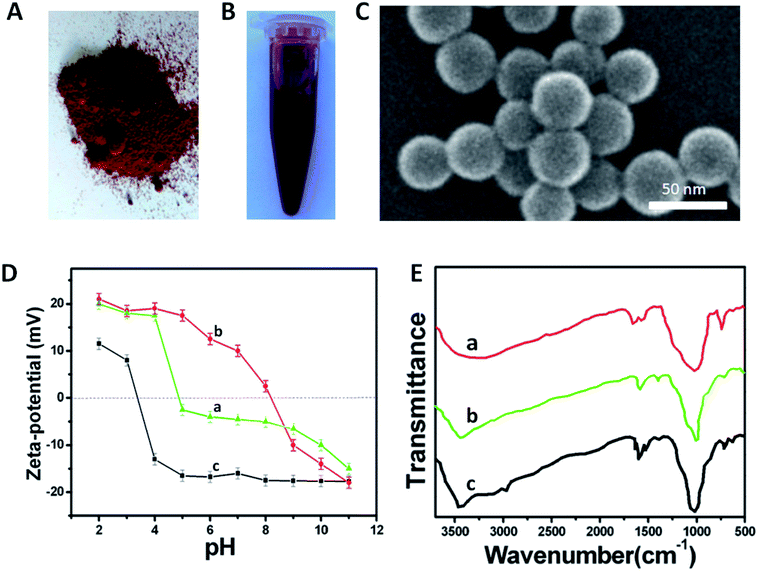
3.1 Characterizations of the multifunctional Red-SiNPs
The inverse microemulsion method was chosen to prepare Red-SiNPs in this study. This method is a simple and diverse preparation method for synthesis of silica nanoparticles in the laboratory, and its use makes it easy to control the morphology of silica nanoparticles. The physical images of Red-SiNPs showed that they were of bright color and formed a good dispersion in aqueous solution (Fig. 3A and B). The size and morphology of Red-SiNPs were characterized by SEM (Fig. 3C). The nanoparticles had uniform particle size and all Red-SiNPs showed a spherical shape and smooth surface. The average diameter of the nanoparticles determined by SEM was approximately 40 ± 5 nm and the size distribution was also quite uniform. The presence of chemical group on the outermost layer of Red-SiNPs was confirmed by zeta potential determination. Zeta potential measurement was carried out using a Zetasizer. For the determination of zeta potential, the pH of the sample was adjusted by the addition of 0.01 M HCl or 0.01 M NaOH. All values shown in this work were the average of three measurements. Fig. 3D displays the zeta potential of Red-SiNPs, Red-SiNPs-NH2 and Red-SiNPs-COOH as a function of pH. The isoelectric point (IEP) of Red-SiNPs was at pH 5.0 (curve a). When Red-SiNPs were in the environment of a neutral solution, the surface potential of Red-SiNPs was about −8 mV (curve a). The particle surface had a negative charge, because of the presence of hydroxyl groups; the IEP of Red-SiNPs-NH2 was shifted to pH 8.2 (curve b). The increase of zeta potential was attributed to the increasing number of protonated amine group on the surface of Red-SiNPs-NH2. Compared to Red-SiNPs-NH2, the IEP of Red-SiNPs-COOH was shifted to pH 3.4 (curve c), which was ascribed to negative charge of the carboxyl group, which made the modification and the bioconjugation of the nanoparticles easier. Chemical composition on the coating layer of Red-SiNPs was also examined by FTIR (as shown in Fig. 3E). Dried sample was measured using the KBr pellet method in the range of 400–4000 cm−1. Strong IR absorption bands in the region 800–1200 cm−1, corresponding to the Si–O–Si of the silica core, were found in spectra of both Red-SiNPs (curve a) and Red-SiNPs-NH2 (curve b). A new band at ∼3090 cm−1 in the Red-SiNPs-NH2 spectrum was assigned to the N–H of the silica surface. Compared to Red-SiNPs, the FTIR spectrum of Red-SiNPs-NH2 had a significant difference in the region 3000–3500 cm−1. These results were consistent with the results of zeta potential. APTMS was thus believed to be successfully introduced onto the surface of the Red-SiNPs. The chemical composition of Red-SiNPs-COOH was also examined using its FTIR spectrum. The stretching band of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O (which is the characteristic band of carboxyl group) appears at 1705 cm−1. Both the symmetric and asymmetric bands of COO arise in this spectrum at 1616 cm−1 and 1510 cm−1, respectively. The spectra indicate the presence of C
O (which is the characteristic band of carboxyl group) appears at 1705 cm−1. Both the symmetric and asymmetric bands of COO arise in this spectrum at 1616 cm−1 and 1510 cm−1, respectively. The spectra indicate the presence of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O (the characteristic band of COOH) which arises from the modification of COOH by glutaric anhydride.
O (the characteristic band of COOH) which arises from the modification of COOH by glutaric anhydride.
3.2 Principle of the MB–MEMSCI
In this work, mAb is immobilized on the MB by using glutaraldehyde as cross-linkage reagent (mAb–MB), which is used as the immunosensing probe for the capture of target H7N9 AIV. MB is not only used as a substrate for the conjugation of mAb antibody but also enables the rapid separation and purification of bionanocomposites after synthesis. Red-SiNPs-COOH heavily functionalized with GOx and pAb antibody (GOx–Red-SiNPs-pAb) is formed possibly owing to the covalent binding between –COOH and free –NH2 of the proteins to form C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N. The as-prepared GOx–Red-SiNPs-pAb is employed as the signal-transduction tag (detection antibody) for the construction of the MB–MEMSCI. In the presence of target H7N9 AIV, the sandwiched immune-conjugates can be formed between MB–mAb and GOx–Red-SiNPs-pAb. Accompanying the GOx–Red-SiNPs-pAb, the carried GOx can trigger the enzymatic catalytic reaction to produce the colored product. Initially, the GOx-biocatalyzed oxidization toward the added glucose leads to the formation of gluconic acid and hydrogen peroxide (H2O2) with the participation of oxygen. The generated hydrogen peroxide is catalyzed by hemin in the 4-AAP and phenol solution to produce the colored product which makes the solution change from colorless to red. The change in the color/absorbance indirectly depends on the concentration of target H7N9 AIV in the sample. By monitoring the shift in the absorbance, we can quantitatively determine the concentration of target H7N9 AIV in the sample. We can also qualitatively judge the H7N9 AIV level by evaluating the change in the visible color. In contrast, GOx–Red-SiNPs-pAb cannot be conjugated onto the functionalized mAb–MB in the absence of target H7N9 AIV; therefore, it cannot trigger the progression of MB–MEMSCI.
N. The as-prepared GOx–Red-SiNPs-pAb is employed as the signal-transduction tag (detection antibody) for the construction of the MB–MEMSCI. In the presence of target H7N9 AIV, the sandwiched immune-conjugates can be formed between MB–mAb and GOx–Red-SiNPs-pAb. Accompanying the GOx–Red-SiNPs-pAb, the carried GOx can trigger the enzymatic catalytic reaction to produce the colored product. Initially, the GOx-biocatalyzed oxidization toward the added glucose leads to the formation of gluconic acid and hydrogen peroxide (H2O2) with the participation of oxygen. The generated hydrogen peroxide is catalyzed by hemin in the 4-AAP and phenol solution to produce the colored product which makes the solution change from colorless to red. The change in the color/absorbance indirectly depends on the concentration of target H7N9 AIV in the sample. By monitoring the shift in the absorbance, we can quantitatively determine the concentration of target H7N9 AIV in the sample. We can also qualitatively judge the H7N9 AIV level by evaluating the change in the visible color. In contrast, GOx–Red-SiNPs-pAb cannot be conjugated onto the functionalized mAb–MB in the absence of target H7N9 AIV; therefore, it cannot trigger the progression of MB–MEMSCI.
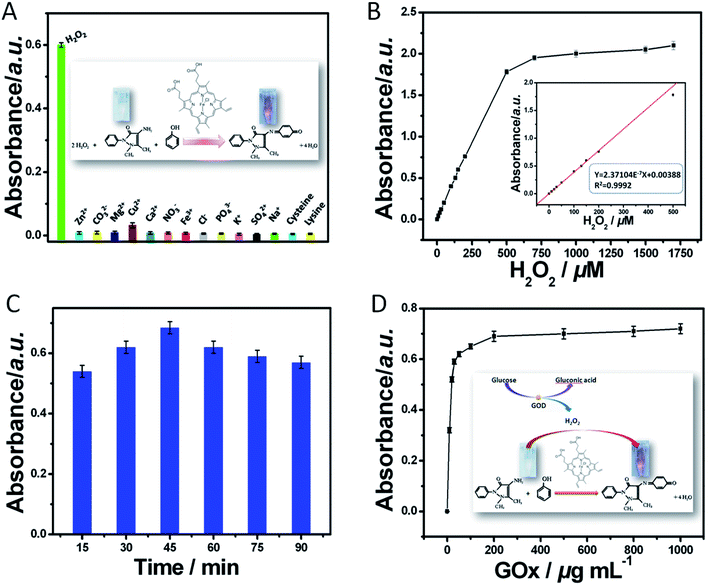
3.3 Control tests for the MB–MEMSCI
For the successful development of MB–MEMSCI, one of the very important preconditions was whether the hemin/4-AAP/phenol system could be smoothly progressed in the presence of GOx, as depicted in Fig. 1a. To demonstrate this concern, several relative control tests were put into effect under the different conditions by using UV-visible absorption spectroscopy (Fig. S1 (ESI†)) and colorimetric measurement (see Fig. S1 (ESI†)) (note: 10 μL of 1 mg mL−1 GOx, 50 μL of 1 mM glucose, 70 μL of 1.0 mg mL−1 4-AAP, 70 μL of 0.12 M phenol, 35 μL of 0.05 mg mL−1 heme chloride solution and 350 μL buffer solution (K2HPO4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NaOH, pH = 10.6) were used in this case). As shown from curve a in Fig S1,† a strong absorption peak at 505 nm was observed after the added GOx reacted with the mixture containing glucose, hemin, 4-AAP and phenol. The reason might be most likely as a consequence of the fact that the H2O2 generated through the catalytic reaction of enzymatic product oxidized 4-AAP to form a dye compound. Moreover, the newly formed dye compound could cause the change in the color of the mixed solution from colorless to pink (photograph a in Fig. S1 (ESI†)). Further, we also found that the absorption increased with increasing GOx concentration under the same conditions (see Fig. 4D). To further investigate the feasibility of the immunoassay system, we also used UV-visible absorption spectroscopy and the visible color of different components in the absence of GOx, glucose, hemin, or 4-AAP. For comparison, we initially monitored the mixture containing GOx, glucose, and hemin (i.e., in the absence of 4-AAP). As seen from curve c in Fig. S1 (ESI†), no absorption peak appeared at 505 nm. Meanwhile, the mixture was colorless (photograph c in the inset of Fig. S1 (ESI†)). Favorably, when glucose (curve b in Fig. S1 (ESI†) and photograph b in the inset of Fig. S1 (ESI†)), hemin (curve d in Fig. S1 (ESI†) and photograph d in the inset of Fig. S1 (ESI†)), or GOx (curve e in Fig. S1 (ESI†) and photograph e in the inset of Fig. S1 (ESI†)) was absent in the detection system, the absorption peak and the color were almost the same as that of curve c and photograph c, respectively. The results revealed that (i) hemin could not cause the appearance of the absorption peak at 505 nm after incubation with the 4-AAP/phenol system without H2O2, (ii) H2O2 could be provided by GOx toward the catalytic reaction of glucose, and (iii) the cascade reaction could be successfully carried out only in the simultaneous presence of glucose/GOx/hemin/4-AAP/phenol system. Hence, we might suspect that when GOx was conjugated onto the detection antibody, the mimic enzyme-chromogenic strategy could be employed for the development of the sandwiched immunoassay by monitoring the change in the absorbance or color.
NaOH, pH = 10.6) were used in this case). As shown from curve a in Fig S1,† a strong absorption peak at 505 nm was observed after the added GOx reacted with the mixture containing glucose, hemin, 4-AAP and phenol. The reason might be most likely as a consequence of the fact that the H2O2 generated through the catalytic reaction of enzymatic product oxidized 4-AAP to form a dye compound. Moreover, the newly formed dye compound could cause the change in the color of the mixed solution from colorless to pink (photograph a in Fig. S1 (ESI†)). Further, we also found that the absorption increased with increasing GOx concentration under the same conditions (see Fig. 4D). To further investigate the feasibility of the immunoassay system, we also used UV-visible absorption spectroscopy and the visible color of different components in the absence of GOx, glucose, hemin, or 4-AAP. For comparison, we initially monitored the mixture containing GOx, glucose, and hemin (i.e., in the absence of 4-AAP). As seen from curve c in Fig. S1 (ESI†), no absorption peak appeared at 505 nm. Meanwhile, the mixture was colorless (photograph c in the inset of Fig. S1 (ESI†)). Favorably, when glucose (curve b in Fig. S1 (ESI†) and photograph b in the inset of Fig. S1 (ESI†)), hemin (curve d in Fig. S1 (ESI†) and photograph d in the inset of Fig. S1 (ESI†)), or GOx (curve e in Fig. S1 (ESI†) and photograph e in the inset of Fig. S1 (ESI†)) was absent in the detection system, the absorption peak and the color were almost the same as that of curve c and photograph c, respectively. The results revealed that (i) hemin could not cause the appearance of the absorption peak at 505 nm after incubation with the 4-AAP/phenol system without H2O2, (ii) H2O2 could be provided by GOx toward the catalytic reaction of glucose, and (iii) the cascade reaction could be successfully carried out only in the simultaneous presence of glucose/GOx/hemin/4-AAP/phenol system. Hence, we might suspect that when GOx was conjugated onto the detection antibody, the mimic enzyme-chromogenic strategy could be employed for the development of the sandwiched immunoassay by monitoring the change in the absorbance or color.
3.4 Evaluation and characteristics of the MB–MEMSCI system
In the MB–MEMSCI system, the cascade reaction mainly consisted of two steps: (i) the catalytic reaction of GOx toward glucose and (ii) the redox reaction between hydrogen peroxide and hemin in the 4-AAP/phenol system. Significantly, the H2O2 in the 4-AAP/phenol system toward hemin was specific and selectable. As shown from Fig. 4A, a significant change in the absorbance was observed at 505 nm with H2O2 against 20-fold higher interfering components (e.g., Zn2+, CO32−, Mg2+, Cu2+, Ca2+, NO3−, Fe3+, Cl−, PO43−, K+, SO42−, Na+, cysteine and lysine). Although Cu2+ ion could cause an increase in the absorbance, the density was less. Hence, the hemin/4-AAP/phenol system could be used for the detection of H2O2. To realize the subsequent MB–MEMSCI, the system must have the ability to exactly differentiate H2O2 with various concentrations. Next, we investigated the analytical performance of the hemin/4-AAP/phenol system toward H2O2 with various concentrations under the same conditions. As shown in Fig. 4B, the absorbance increased with increasing H2O2 concentration. Moreover, a linear tendency could be fitted in the working range of 7 μM to 500 μM, as shown from the inset in Fig. 4B, and the detection limit could be lowered to 2.5 μM. The results revealed that the system was feasible for quantitative monitoring of H2O2 in the sample. For bioactive GOx enzyme, however, the catalytic efficiency toward glucose relied on the catalytic time and temperature to some extent. Usually, normal body temperature (37 °C) is suitable for enzymatic reaction. At this condition, we monitored the effect of different incubation times between GOx and glucose on the absorbance (note: 10 μL of 1 mg mL−1 GOx, 50 μL of 1 mM glucose, 70 μL of 1.0 mg mL−1 4-AAP, 70 μL of 0.12 M phenol, 35 μL of 0.05 mg mL−1 heme chloride solution and 350 μL buffer solution (K2HPO4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NaOH, pH = 10.6) were used in this case). As indicated in Fig. 4C, the absorbance increased within the initial 30 min and then tended to slightly decrease. The reason might be attributed to the fact that H2O2 can be decomposed by light irradiation. Therefore, 30 min was selected for the enzymatic reaction in this work. The enzymatic reactivity of GOx in the cascade reaction system was also studied. Various amounts of GOx were added to the system, and the absorption spectra were recorded. In this case, 50 μL of 1 mM glucose, 70 μL of 1.0 mg mL−1 4-AAP, 70 μL of 0.12 M phenol, 35 μL of 0.05 mg mL−1 heme chloride solution and 350 μL buffer solution (K2HPO4
NaOH, pH = 10.6) were used in this case). As indicated in Fig. 4C, the absorbance increased within the initial 30 min and then tended to slightly decrease. The reason might be attributed to the fact that H2O2 can be decomposed by light irradiation. Therefore, 30 min was selected for the enzymatic reaction in this work. The enzymatic reactivity of GOx in the cascade reaction system was also studied. Various amounts of GOx were added to the system, and the absorption spectra were recorded. In this case, 50 μL of 1 mM glucose, 70 μL of 1.0 mg mL−1 4-AAP, 70 μL of 0.12 M phenol, 35 μL of 0.05 mg mL−1 heme chloride solution and 350 μL buffer solution (K2HPO4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NaOH, pH = 10.6) were used for detection of 10 μL of GOx with various concentrations. As seen from Fig. 4D, the absorbance increased with an increase of GOx concentration in the sample. At a low concentration of GOx, the absorbance exhibited a strong shift, and the detection limit was ∼1 μg mL−1 GOx. On the basis of these results, we might draw a conclusion that the designed cascade reaction strategy based on the hemin/4-AAP/phenol system could be utilized for the determination of GOx by coupling with the change in the color or absorbance, and simultaneously, the results revealed that the system was feasible for quantitative monitoring of H7N9 AIV in the sample, because its concentration changes can indirectly lead to concentration changes of GOx.
NaOH, pH = 10.6) were used for detection of 10 μL of GOx with various concentrations. As seen from Fig. 4D, the absorbance increased with an increase of GOx concentration in the sample. At a low concentration of GOx, the absorbance exhibited a strong shift, and the detection limit was ∼1 μg mL−1 GOx. On the basis of these results, we might draw a conclusion that the designed cascade reaction strategy based on the hemin/4-AAP/phenol system could be utilized for the determination of GOx by coupling with the change in the color or absorbance, and simultaneously, the results revealed that the system was feasible for quantitative monitoring of H7N9 AIV in the sample, because its concentration changes can indirectly lead to concentration changes of GOx.
3.5 Analytical performance of colorimetric immunoassay using MB–MEMSCI
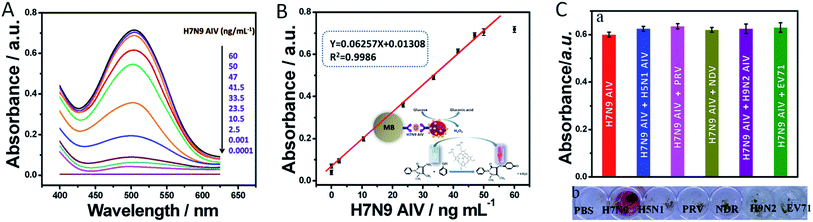
To further investigate the capability of the developed MB–MEMSCI for colorimetric immunoassay, the as-prepared GOx–Red-SiNPs-pAb was employed as secondary antibody for the detection of target H7N9 AIV with a sandwich-type immunoassay format on MB–mAb by using the hemin/4-AAP/phenol system. Under the optimal conditions, H7N9 AIV standards with different concentrations were monitored based on the designed immunoassay protocol. As shown in Fig. 5A and B, the increase of absorbance is proportional to the H7N9 AIV antigen concentration in the range of 0.01–50.0 ng mL−1, and the detection limit was 3.5 pg mL−1 (S/N = 3) which is obviously superior to that of the colorimetric immunoassay without color enhanced multifunctional silica nanoparticles (Fig. S5A and B) (ESI†). Moreover, Table S1 (ESI†) shows the linear range and detection limit of previously reported immunosensors15–22 and Table S2 (ESI†) shows the colorimetric immunoassay without color enhanced multifunctional silica nanoparticles. Compared to other methods, the presented immunosensor has a relatively large linear range and low detection limit. We suspect that improvement of the sensitivity might be attributed to the highly efficient mimic enzyme-chromogenic substrate system as a result of the strong reaction in the hemin/4-AAP/phenol system and the color enhancement of the multifunctional SiNPs.3.6 Reproducibility, selectivity and stability of colorimetric immunoassay using MB–MEMSCI
The reproducibility and precision of the colorimetric immunoassay using MB–MEMSCI were evaluated by calculating the intra- and inter-batch variation coefficients (CVs). Results revealed that the CVs of the intra-assay with this method were between 4.0% and 7.5% (n = 5) in all cases. The batch-to-batch reproducibility of MB–MEMSCI was also monitored by assaying 35 ng mL−1 H7N9 AIV (as an example) for six times on different days, and the obtained CV was about 7.8%. The low CVs might be attributed to the specific antigen–antibody reaction and the strong interaction in the hemin/4-AAP/phenol system. Thus, the precision and reproducibility of the colorimetric immunoassay were acceptable.Further, the specificity of the colorimetric immunoassay was also investigated for other viruses, such as H5N1 AIV, PRV, NDV, H2N9 AIV and EV71. The selectivity of the GOD/NECF biosensor was also studied. Fig. 5C(a) presents the absorbance of the biosensor with 40 ng mL−1 H7N9 AIV, including a 5-fold concentration of interfering substances, such as H5N1 AIV, PRV, NDV, H2N9 AIV and EV71. No obvious interference was observed, and the results clearly indicated the high specificity of the colorimetric immunoassay using the MB–MEMSCI. The stability of the colorimetric immunosensor was also tested, as shown in Fig. S2 (ESI†). When the immunosensor was not in use, it was stored in PBS (pH 7.4) at 4 °C. The absorbance response of the MB–MEMSCI only had a change of 5.0% after 30 days. This indicates the effective retention of the activity of the immobilized H7N9 AIV antibody, and further confirmed the reaction conditions of mimic enzyme are not harsh.
3.7 The development of H7N9 AIV optical detection by MB–MEMSCI
In order to get a more clear visual effect and low detection limit, optical detection and UV detection of H7N9 AIV are carried out separately. In this method, we used the naked eye to detect H7N9 AIV by MB–MEMSCI. mAb–MBs were added to H7N9 AIV solution; the H7N9 AIV was enriched and isolated in a magnetic field and transferred to wells of a 96-well microtiter plate. GOx–Red-SiNPs-pAb, acting as an indicator of the presence of H7N9 AIV, was added and reacted for 15 min. When the sample solution contained enough H7N9 AIV, mAb–MBs and GOx–Red-SiNPs-pAb were effectively combined on the surface of H7N9 AIV, forming red sandwich complexes through double antibody sandwich binding reaction, and producing red compounds in the glucose/hemin/4-AAP/phenol system to amplify the detection signal. The red suspension was clearly observed in the well with the naked eye and GOx–Red-SiNPs-pAb played a role of color indicator.Aiming to prove the superiority of Red-SiNPs in visual detection, pAb-AuNPs and pAb-Red-SiNPs were both used to detect H7N9 AIV, and the preparation of pAb-AuNPs is shown in ESI (Fig. S3 (ESI†)).
The procedure was the same as mentioned in Section 2.7. H7N9 AIV was detected by our sandwich assay method using pAb-AuNPs and pAb-Red-SiNPs separately, and PBS was used as negative control. As shown in Fig. S4 (ESI†), when pAb-AuNPs was used as signal label, the positive and negative results were difficult to distinguish with the naked eye. Red plaque was observed when pAb-Red-SiNPs were used as signal label to detect H7N9 AIV. This result showed that Red-SiNPs were more suitable to be used as signal label in optical immunoassay.
We examined the sensitivity of this optical sandwich assay method and the detection limit for H7N9 AIV. The positive and negative results were distinguished by observing the bottom of the well with the naked eye. Red suspension in the well represents positive result, and brown plaque in the bottom of the well with colorless transparent liquid represents negative result. The original H7N9 AIV solution was diluted to ten concentrations along a gradient from 10 ng mL−1 to 0.1 ng mL−1 using 0.85% stroke-physiological saline solutions. In the optical sandwich assay, pAb–MBs and pAb-Red-SiNPs were sequentially mixed with different concentrations of H7N9 AIV solution. The lowest concentration of H7N9 AIV which can produce a positive result is defined as the detection limit of this method. Fig. S6A (ESI†) shows that a red turbid solution was observed when the concentration of H7N9 AIV was as low as 0.5 ng mL−1. Brown color of pAb–MBs was observed in the well containing 0.4 ng mL−1 H7N9 AIV solution. The low detection limit (0.5 ng mL−1) is due to the enrichment effect of pAb–MBs and the function of vision signal enhancement from the Red-SiNPs in the red reaction product solution. The sensitivity of the optical MB–MEMSCI is related to the volume of the sample and we can improve the detection sensitivity by increasing the sample volume. In order to prove this, we conducted a control experiment, while the other conditions were unchanged; 400 μL H7N9 AIV solutions of different concentrations were detected with this sandwich assay. As shown in Fig. S6B (ESI†), when the concentration of H7N9 AIV was equal to or greater than 2 ng mL−1, the red color of Red-SiNPs and the red enzyme-catalyzed reaction solution started to be observed. In this experiment, the detection limit for H7N9 AIV is 2 ng mL−1. By increasing the volume of H7N9 AIV solution, more H7N9 AIV was enriched by IgG-MBs, and this made the optical sandwich assay more sensitive. Taking into account the impact of blood collection on patients in clinical applications and cost, we chose 1 mL as the sample volume for this test.
3.8 Specificity of H7N9 AIV optical detection by MB–MEMSCI
To demonstrate the potential of our optical assay in biomarker discrimination, we examined the specificity of this optical MB–MEMSCI. The procedure for the specificity test was the same as mentioned in Section 2.7. H7N9 AIV, H5N1 AIV, PRV, NDV, H9N2 AIV and EV71 were all detected by this optical sandwich assay method. The concentration ratio of H7N9 AIV and interferents was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5, and PBS was used as negative control. As shown in Fig. 5C(b), a red signal was only observed for H7N9 AIV but not for H5N1 AIV, PRV, and NDV, confirming this optical assay could distinguish H7N9 AIV from other biomarkers. This result showed that this method had good specificity for H7N9 AIV.
5, and PBS was used as negative control. As shown in Fig. 5C(b), a red signal was only observed for H7N9 AIV but not for H5N1 AIV, PRV, and NDV, confirming this optical assay could distinguish H7N9 AIV from other biomarkers. This result showed that this method had good specificity for H7N9 AIV.
3.9 Stability of H7N9 AIV optical detection by MB–MEMSCI
The stabilities of mAb–MBs and GOx–Red-SiNPs-pAb were studied by the detection of H7N9 AIV solution (8 ng mL−1) with the Red-SiNPs based optical MB–MEMSCI after storing at 4 °C for 1, 10, 20 and 30 days. These two kinds of immune nanoparticles were found to retain similar reaction activity after storage at 4 °C for at least 30 days (as shown in the inset of Fig. S2 (ESI†)).4. Detection of H7N9 AIV in real samples
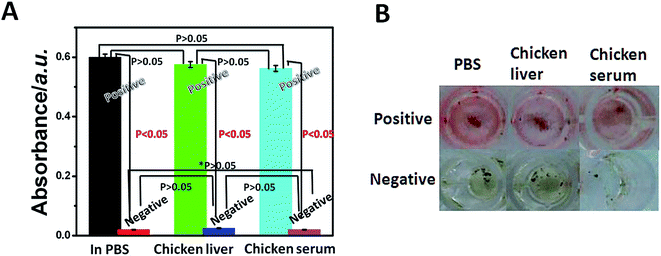
It is reported that avian influenza virus (AIV) could replicate in the organs of infected humans or animals,23,24 so detection methods with good anti-interference ability and high specificity are needed to detect AIV in complex samples such as tissues and feces. Taking advantages of the very good separation and enrichment ability of MBs, we investigated the capability of this method for complex samples. An amount of H7N9 AIV was added to chicken serum and ground chicken liver to prepare H7N9 AIV positive samples. The insoluble solids were removed by centrifuging. And the negative samples were prepared in the same way without adding H7N9 AIV. As shown in Fig. 6, the H7N9 AIV ultraviolet absorption (Fig. 6A) and optical detection (Fig. 6B) also had high signal-to-noise ratio (p < 0.05) and recognition even in these media, which suggested the feasibility of the immunosensor in complex matrices of real samples directly with strong anti-interference ability. And the detection ability in real samples has been verified by the avian influenza virus (H7N9) Real Time RT-PCR Kit (Table S3 (ESI†)). By comparing with the current standard method, we confirmed the accuracy (Table S3 (ESI†)) and practicability (Table S4 (ESI†)) of this MB–MEMSCI.Compared with gold nanoparticles, the most used nanoprobes in sandwich assay method, Red-SiNPs are cheaper and more stable than gold nanoparticles. Moreover, for bright color of Red-SiNPs, our sandwich assay method can be read out easily by the naked eye.
5. Conclusions
In this work, we for the first time demonstrate the ability of unconventional ELISA product for exceptional application in colorimetric immunoassay based on colored nanoparticles and mimic enzymatic formation of the chromogenic compound. The signal was amplified through an enzyme-catalyzed cascade reaction and colored nanoparticles. Experimental results indicated that the visible color of the hemin/4-AAP/phenol system could be successfully triggered by the catalytic product of GOx toward glucose. Compared with traditional enzyme-based colorimetric immunoassay, MB–MEMSCI was low cost, sensitive, rapid, stable and feasible. Moreover, the MB–MEMSCI system is not susceptible to interference and changes in the assay conditions during the color generation stage. Importantly, the MB–MEMSCI system can be further extended for the qualitative and quantitative detection of other low-abundance proteins or biomarkers by controlling the target antibody. Moreover, MB–MEMSCI can be suitable for use in the mass production of miniaturized lab-on-a-chip devices and open a new opportunity for protein diagnostics and food security.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (81403106 and 81460594), Natural Science Foundation of Jiangxi Province (20142BAB215064) and Scientific Research Foundation of Jiangxi University of Traditional Chinese Medicine (2016ZR002).References
- R. Gao, B. Cao, Y. Hu, Z. Feng, D. Wang, W. Hu, J. Chen, Z. Jie, H. Qiu and K. Xu, N. Engl. J. Med., 2013, 368, 1888–1897 CrossRef CAS PubMed.
- W.H.O, Website, Human Infections With Avian Influenza A (H7N9) Virus, Update 2 October 2014, http://www.who.int/influenza/human_animal_interface/influenza_h7n9/riskassessment_h7n9_2Oct14.pdf.
- W.H.O, Website, Human Infections With Avian Influenza A (H7N9) Virus, Update 22 July 2016, http://www.who.int/csr/don/22.july.2016.ah7n9.china/en/.
- K. E. Templeton, S. A. Scheltinga, M. F. Beersma, A. C. Kroes and E. C. Claas, J. Clin. Microbiol., 2004, 42, 1564–1569 CrossRef CAS PubMed.
- J. S. Park, M. K. Cho, E. J. Lee, K. Y. Ahn, K. E. Lee, J. H. Jung, Y. Cho, S. S. Han, Y. K. Kim and J. Lee, Nat. Nanotechnol., 2009, 4, 259–264 CrossRef CAS PubMed.
- D. Chen, N. A. Sarikaya, H. Gunn, S. W. Martin and J. D. Young, Clin. Chem., 2001, 47, 747–749 CAS.
- R. M. Wilson and S. J. Danishefsky, J. Am. Chem. Soc., 2013, 135, 14462–14472 CrossRef CAS PubMed.
- H. Lin, Y. Liu, J. Huo, A. Zhang, Y. Pan, H. Bai, Z. Jiao, T. Fang, X. Wang and Y. Cai, Anal. Chem., 2013, 85, 6228–6232 CrossRef CAS PubMed.
- Y. Song, W. Wei and X. Qu, Adv. Mater., 2011, 23, 4215–4236 CrossRef CAS PubMed.
- B. Zhang, D. Tang, I. Y. Goryacheva, R. Niessner and D. Knopp, Chemistry, 2013, 19, 2496–2503 CrossRef CAS PubMed.
- X. M. Huang, M. Zhu and H. X. Shen, Microchim. Acta, 1998, 128, 87–91 CrossRef CAS.
- Y. Song, Y. Shen, J. Chen, Y. Song, C. Gong and L. Wang, Electrochim. Acta, 2016, 211, 297–304 CrossRef CAS.
- R. P. Bagwe, C. Yang, L. R. Hilliard and W. Tan, Langmuir, 2004, 20, 8336–8342 CrossRef CAS PubMed.
- Q. Sun, G. Zhao and W. Dou, Sens. Actuators, B, 2016, 226, 69–75 CrossRef CAS.
- Z. Wu, C. H. Zhou, J. J. Chen, C. Xiong, Z. Chen, D. W. Pang and Z. L. Zhang, Biosens. Bioelectron., 2015, 68, 586–592 CrossRef CAS PubMed.
- C. H. Zhou, Y. M. Long, B. P. Qi, D. W. Pang and Z. L. Zhang, Electrochem. Commun., 2013, 31, 129–132 CrossRef CAS.
- W. M. Hassen, V. Duplan, E. Frost and J. J. Dubowski, Electrochim. Acta, 2011, 56, 8325–8328 CrossRef CAS.
- T. M. P. Hewa, G. A. Tannock, D. E. Mainwaring, S. Harrison and J. V. Fecondo, J. Virol. Methods, 2009, 162, 14–21 CrossRef PubMed.
- K. K. Deng, X. J. Wang, M. Y. Zheng and K. Wu, Mater. Sci. Eng., A, 2013, 560, 824–830 CrossRef CAS.
- U. Jarocka, R. Sawicka, A. G. Sochacka, A. Sirko, W. Z. Ostoja, J. Radecki and H. Radecka, Sensors, 2013, 14, 15714–15728 CrossRef PubMed.
- U. Jarocka, R. Sawicka, A. G. Sochacka, A. Sirko, W. Dehaen, J. Radecki and H. Radecka, Sens. Actuators, B, 2016, 228, 25–30 CrossRef CAS.
- M. F. Diouani, S. Helali, I. Hafaid, W. M. Hassen, M. A. Snoussi, A. Ghram and N. J. Renault, Mater. Sci. Eng., C, 2008, 28, 580–583 CrossRef CAS.
- G. F. Rimmelzwaan, T. Kuiken, G. V. Amerongen, T. M. Bestebroer and R. A. M. Fouchier, J. Virol., 2001, 75, 6687–6691 CrossRef CAS PubMed.
- Y. Yu, Z. Xi, B. Zhao, Y. Sun, X. G. Zhang, T. Bai, J. Lu, Z. Li, L. Liu, D. Y. Wang, Y. L. Shu, J. F. Zhou and K. Qin, J. Virol. Methods, 2017, 247, 58–60 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra06273g |
| This journal is © The Royal Society of Chemistry 2017 |